MCAT Biochemistry - Enzymes
1/84
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
85 Terms
hypertension
high blood pressure; increases risk of stroke, heart failure, kidney failure
angiotensin-converting enzyme (ACE) inhibitors
anti-hypertensive medication; inhibits ACE
angiotensin-converting enzyme (ACE)
catalyses a reaction that converts angiotensin I to angio tensin II
angiotensin II
causes constriction of blood vessels; stimulate sthe release of aldosterone
aldosterone
hormone that activates the kidneys to reabsorb more water into the blood stream; increase in blood volume increases blood pressure
enzymes
biological catalysts; used to regulate homeostatic mechanisms in every organ system and are highly regulated themselves by environmental conditions, activators, and inhibitors; may be naturally occurring or may be given as a drug
all catalytic properties +
Are pH- and temperature-sensitive, with optimal activity at specific pH ranges and temperatures
Are specific for a particular reaction or class of reactions
catalysts
Lower the activation energy
Increase the rate of the reaction
Do not alter the equilibrium constant
Are not changed or consumed in the reaction
Do not affect the overall ΔG of the reaction
enzyme specificity
a given enzyme will only catalyze a single reaction or class of
reactions with these substrates
substrate
the molecules upon which an enzyme acts
urease
catalyses the breakdown of urea
chymotrypsin
cleaves peptide bonds around the amino acids that contain aromatic rings—phenylalanine, tryptophan, and tyrosine.
-ase
suffix for most enzymes
Oxidoreductases
catalyzes the transfer of electrons between molecules
electron carrier
often a cofactor for oxidoreductases
e.g. NAD+, NADP+
reductant
electron donor
oxidant
electron acceptor
dehydrogenase
name suggesting oxidoreductase (1)
reductase
name suggesting oxidoreductase (2)
oxidase
name suggesting oxidoreductase with oxygen as the final electron acceptor
transferases
catalyze the movement of a functional group from one molecule to another
aminotransferase
convert aspartate and α-ketoglutarate toglutamate and oxaloacetate by moving amino group →
kinase
catalyze the transfer of a phosphate group (esp. from ATP) to another molecule
hydrolases
catalyze the breaking of a compound into two using hydrolysis; using naed only for substrate
hydrolysis
breaking a compound/polymer using the addition of water molecules
phosphatase
hydrolase that cleaves a phosphate group
peptidases
hydrolase that cleaves a polypeptide
nucleases
hydrolase that cleaves a nucleic acid
lipases
hydrolase that cleaves a lipid
lyases
catalyze the cleavage of a single molecule into two products without water
may also be a synthase
synthase
catalyzes the synthesis of two (esp. small) molecules into a single one
may also be a lyase
isomerase
catalyze the rearrangement of bonds within a molecule; can be classified as another type of enzyme depending on mechanism
ligase
catalyze addition or synthesis reactions, generally between large similar molecules, often require ATP
e.g. nucleic acid synthesis
endergonic reaction
requires energy input (ΔG > 0)
exergonic reaction
energy is given off (ΔG < 0)
activation energy
the minimum amount of energy that must be available to reactants for a chemical reaction to occur; make it easier for substrates to achieve transition state
enzyme-substarte complex
the physical interaction between the enzyme and the molecule it acts upon; transition state
active site
teh location within the enzyme where the substrate is held during the chemical reaction
lock and key theory
active site is always in the appropriate conformation for the substrate to bind
induced fit theory
the substrate induces a change in the shape of the enzyme; conformation is endergonic, release is exergonic; enzymes return to original shape when not bound
Thiamine (Vitamin B1)
essential cofactor for enzymes in cellular metabolism and nerve conduction
defienciency from excess alcohol consumption, poor diet
Wernicke-Korsakoff syndrome
disease stemming from thiamine deficiency, esp. from alcoholism
symptoms: neurologic deficits, including delirium, balance problems, the inability to form new memories
cofactors
inorganic molecules or metal ions, often ingested as dietary minerals, that aid enzymes; small, bound to the active site of enzyme and participate in catalysis by carrying charge (ionization, (de)protonation); low concentrations in cells, recruited when needed
coenzymes
small organic groups, often vitamins or derivatives, that aid enzymes; small, bound to the active site of enzyme and participate in catalysis by carrying charge (ionization, (de)protonation); low concentrations in cells, recruited when needed
e.g. NAD+, FAD, coenzyme A
water-soluble vitamins
must be replenished regularly, easily excreted
e.g. B complex, ascorbic acid/C
fat-soluble vitamins
regulated by partition coefficients
e.g. A, D, E, K
partition coefficients
quantify the ability of a molecule to dissolve in a polar vs. nonpolar environment
apoenzymes
enzymes without necessary cofactors/coenzymes
holoenzymes
enzymes containing necessary cofactors/coenzymes
riboflavin (vitamin B2)
involved in energy metabolism, cellular respiration, and antibody production, as well as normal growth and development
niacin (vitamin B3)
converted within the body to NAD+
Pantothenic acid (vitamin B5)
synthesize coenzyme A (CoA), which is essential for cellular energy production and for the synthesis and degradation of proteins, carbohydrates, and fats
Pyridoxal phosphate (vitamin B6)
all transamination reactions, and in certain decarboxylation, deamination, and racemization reactions of amino acids
Biotin (vitamin B7)
involved in the catabolism of amino acids and fatty acids, synthesis of fatty acids, and gluconeogenesis
Folic acid/folate (vitamin B9)
required for the body to make DNA and RNA and metabolise amino acids necessary for cell division and maturation of blood cells
(Cyano)cobalamin (vitamin B12)
required for DNA synthesis, in both fatty acid and amino acid metabolism, the synthesis of myelin, and in the circulatory system in the maturation of red blood cells in the bone marrow
saturation
every enzyme is busy/occupied; increasing substrate concentration cannot increase velocity further
maximum velocity (vmax)
the highest rate of catalysis at a given concentration of enzymes can perform
synonymous with max activity/rate
Michaelis-Menten equation
As the amount of substrate increases, the enzyme is able to increase its rate of reaction until it reaches vmax; once vmax is reached, adding more substrate will not increase the rate of reaction.
v = vmax [S] / Km + [S]
When rate = ½ vmax, Km = [S]
v = kcat[E][S] / Km + [S] ≈ kcat / Km ([E][S])
![<p>As the amount of substrate increases, the enzyme is able to increase its rate of reaction until it reaches <em>v<sub>max</sub></em>; once <em>v<sub>max</sub></em> is reached, adding more substrate will not increase the rate of reaction.</p><p><em>v = v<sub>max</sub> [S] / K<sub>m</sub> + [S]</em></p><p>When rate = ½ <em>v<sub>max</sub>, K<sub>m</sub> = [S]</em></p><p><em>v = k<sub>cat</sub>[E][S] / K<sub>m</sub> + [S] ≈ k<sub>cat </sub>/ K<sub>m</sub> ([E][S])</em></p>](https://knowt-user-attachments.s3.amazonaws.com/6991522e-4725-4167-9e7f-921fc40e2f0a.png)
Michaelis constant (Km)
a measure of the affinity of the enzyme for its substrate; higher Km = lower affinity for substrate, requires a higher substrate concentration to be half-saturated
substrate turnover (kcat)
measures the number of substrate molecules converted to product per enzyme molecule per second
vmax=[E]kcat
catalytic efficiency
the ratio of the kcat / Km of a particular enzyme
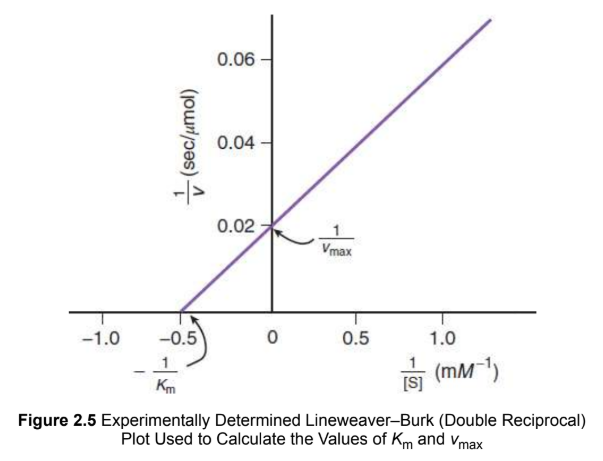
Lineweaver-Burk plot
double reciprocal graph of the Michaelis–Menten equation
x-intercept = −1/Km
y-intercept = 1/vmax
Cooperativity
Binding of the substrate to one subunit encourages the transition of other subunits from the low affinity, tense (T) state to the high-affinity, relaxed (R) state, which increases the likelihood of substrate binding by these other subunits
sigmoidal (S-shaped) Michaelis-Menten plot
e.g. haemoglobin, phosphofructokinase-1 (glycolysis)
Hill’s coefficient
quantifies cooperativity, indicates the nature of binding by the molecule
>1 - positively cooperative binding
<1 - negatively cooperative binding
=1 - no cooperative binding
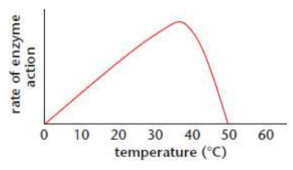
Enzymes and temperature
double in velocity per 10°C inc. until optimum temp. then activity falls sharply due to denaturation
optimal temp. is typically body temp. (37°C/98.6°F/310 K in humans)
tyrosinase
enzyme responsible for pigmentation
mutated in Siamese cats: ineffective @ body temperature, but active at cooler temperatures
Enzymes and pH
pH affects the ionization of the active site, also changes in pH can lead to denaturation of the enzyme
e.g. ideal blood pH 7.4, pepsin pH 2 (stomach), pancreatic enzymes pH 8.5 (small intestine)
Enzymes and salinity
Increasing levels of salt can disrupt hydrogen and ionic bonds, causing a partial change in the conformation of the enzyme and causing denaturation
feedback regulation
regulation by products further down a given metabolic pathway
feed-forward regulation
regulated by intermediates that precede the enzyme in the pathway
Feedback inhibition/negative feedback
helps maintain homeostasis; once we have enough of a given product, we want to turn off the pathway that creates that product rather than creating more; competitive inhibition
methanol (wood alcohol)
enzymatically converted to toxic metabolites in the body; may cause blindness or death
treatment: intravanous ethanol, with competes for active sites
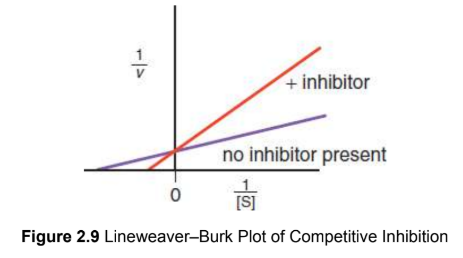
competitive inhibition
occupancy of the active site; overcome by adding more substrate
binding site: active
Km: increases
vmax: unchanged
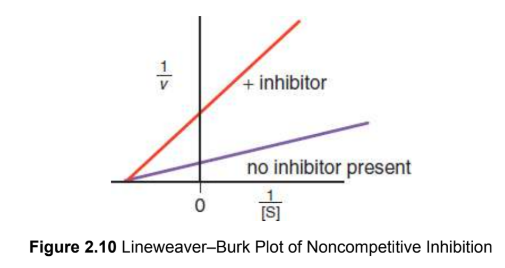
noncompetitive inhibition
bind to allosteric site that induces a conformational change; no extra substarte with form a complex; less enzyme available to react
binding site: allosteric
Km: unchanged
vmax: decreases
mixed inhibition
inhibitor can bind to either the enzyme or the enzyme–substrate complex, but has different affinity for each
binding site: allosteric
Km: increases (favours enzyme) or decreases (favours complex)
vmax: decreases
On a Lineweaver–Burk plot, the curves for the activity with and without the inhibitor intersect at a point that is not on either axis
uncompetitive inhibition
inhibitors bind only to the enzyme–substrate complex and essentially lock the substrate in the enzyme, preventing its release
binding site: allosteric
Km: decreases
vmax: decreases
On a Lineweaver–Burk plot, the curves for activity with and without an uncompetitive inhibitor are parallel
Allosteric site
non-catalytic regions of the enzyme that bind regulators
irrevirsible inhibition
the active site is made unavailable for a prolonged period of time, or the enzyme is permanently altered
e.g. aspirin
Acetylsalicylic acid (aspirin)
irreversibly modifies cyclooxygenase-1, which now can no longer bind its substrate (arachidonic acid) to make its products (prostaglandins), which are involved in modulating pain and inflammatory responses
Allosteric enzymes
alternate between an active and an inactive form
allosteric activators
result in a shift that makes the active site more available for binding to the substrate
allosteric inhibitors
result in a shift that makes the active site less available for binding to the substrate
phosphorylation/dephosphorylation
addition/removal of a phsophate group; depending on the enzyme, either could activate or deactivate it
glycosylation
the covalent attachment of sugar moieties can tag an enzyme for transport within the cell or can modify protein activity and selectivity.
zymogens
inactive form of an enzyme; often when enzymes are digestive; regulatory domain must be either removed or altered to expose the active site; often ends in -ogen
e.g. tyrpsinogen (→ trypsin), pepsinogen (→ pepsin)