MCB 2050 2nd Half Lectures
1/178
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
179 Terms
Nuclear compartments
Nucleoplasm (subdomains), matrix, envelope, lamina
Functions of each nuclear subdomain
Envelope: boundary between cytoplasm and nucleus
Pores: ‘doorways’ in envelope; regulate transport
Nucleolus: site of ribosome synthesis
Nucleoplasm: ordered architecture; chromatin found here, RNA processing
Prokaryotes have nucleoid only - less DNA, less DNA packaging, little to no RNA processing
Spatial/temporal regulation in prok. vs euk.
Prok: Simultaneous translation and transcription
Euk: post-transcriptional processing/splicing of RNA, then transport to cytoplasm or ER for translation; envelope limits TF access to genome
Nucleoplasm and nucleolus
Nucleoplasm: fluid-filled interior of nucleus; organized into >30 subdomains (not membrane bound)
Eg. Nucleolus: irregular shape, dense, granular. 1-5 per cell. Ribosome production (rDNA txn., rRNA processing, initial ribosomal assembly). Final ribosome assembly in cytoplasm.
Internal nuclear organization: chromosomes in interphase
Most active genes (euchromatin) at periphery of subdomains
Interchromosomal channels prevent unwanted DNA-DNA and DNA-protein interactions
Internal nuclear organization: chromosomal subdomains
Euchromatin from different subdomains or chromosome regions extend into these channels, forming txn. factories (TFs concentrated).
Interchromosomal interactions/kissing chromosomes: regulatory region from one chromosome activate another.
Internal nuclear organization: nuclear speckles
Subdomains where mRNA splicing factors are concentrated; often located in channels (next to txn. factories), numerous and highly dynamic (can move around, grow/shrink, increase/decrease)
Internal nuclear organization: nuclear matrix
Insoluble fibrillar protein network/mesh throughout nucleoplasm. Analogous to cytoskeleton network, which contains microfilaments, microtubules, intermediate filaments.
Internal nuclear organization: nuclear matrix functions
Structural role to maintain nucleus shape
Scaffold that organizes and anchors protein factors (TFs, RNA processing, DNA repl., etc)
Internal nuclear organization: nuclear envelope
Serves as barrier to regulate passage between nucleus and cytoplasm
Establishes unique nucleus composition, spatial regulation of gene expression
Structural framework
3 main parts: nuclear membranes, nuclear lamina, NPCs.
Nuclear membranes: definition
Inner and outer membranes (phospholipid bilayers) parallel to each other
These are separated by nuclear envelope lumen (10-50nm)
Membranes serve as barriers
Nuclear membranes: details/outer vs inner
Outer continuous with RER; ribosomes attached to cytoplasmic surface of outer membrane (similar function to RER)
Inner has unique protein composition compared to outer
Both highly curved/joined at NPCs
Nuclear lamina: definition + function
Located on inner surface of inner membrane (ie. nucleoplasmic side)
Mesh of long, filamentous proteins, ex. ABC nuclear lamins (evolutionarily related to cytoskeleton intermediate filament proteins)
Provides mechanical support to envelope (binds to inner integral proteins)
Scaffold for chromatin and nuclear matrix attachment to envelope
Nuclear lamina: mutations
Mutations in LAMIN genes responsible for diseases such as Hutchinson-Gilford Progeria: point mutation in LAMIN A, leading to truncation and subsequent breakdown of nuclear lamina. Causes premature aging and early death.
Nuclear pore complexes (NPCs): definition and functions
Channels/doorways in envelope
Regulate all trafficking between nucleus and cytoplasm: small polar molecules (ex. nucleotides), RNAs, proteins
3000-4000/nucleus (related to nuclear activity)
NPCs: structural elements
Large, highly complex structure (~30x more than ribosome)
~40 different proteins, known as nucleoporins/Nups
—>highly conserved, includes both integral and peripheral inner/outer proteins
—>several related to COPII proteins (ER vesicle formation); same function of curving membranes
Overall structure: 8-fold symmetrical structure with a large, central aqueous channel
NPCs: parts
Central scaffold: composed of integral/transmembrane nucleoporins
—>anchors NPC to membranes, at junction of outer and inner
Forms central aqueous channel (20-40nm wide pore)
Inner surface of channel lined by FG nucleoporins: contain hydrophilic polypeptides with short repeats of hydrophobic domains that include FG AAs
—>unique, highly disordered structure
FG nucleoporins
FG domains extend into central channel, forming a gel-like mesh that limits diffusion of molecules >~40kDa
Small molecules freely diffuse through NPC
Bigger molecules must be selectively transported
Size-exclusion limit of NPC determined with microinjected gold particles
NPCs: other elements
Y-complexes: contain cytoplasmic and nuclear rings, both composed of structural Nups
—>Linked to central scaffold, cytoplasmic filaments or nuclear basket
Cytoplamic filaments: long, filament-shaped structural Nups that extend into cytoplasm
—>involved in nuclear receptor-cargo protein recognition and import (from cytoplam)
Nuclear basket: basket-like structure of structural Nups, linked to Y-complex nuclear ring; involved in nuclear receptor-cargo protein import and export
Nucleocytoplasmic transport via NPC: definition
One of most congested bi-directional trafficking pathways
Variety of import and export pathways, with wide range of cargo
All proteins needed for DNA repl., txn., splicing, ribosome assembly, histones/chromatin packing, nuclear matrix, lamins, etc. imported from cytoplasm
all RNA, partially assembled ribosomes, some proteins exported out of nucleus into cytoplasm
Typical animal cell: 100 histones, 180 ribosomal proteins, 6 ribosomal subunits/NPC/min
Cytoplasm-to-nuclear transport/NLSs
Most contain NLS: specific stretch of AAs recognized by nuclear receptor proteins, that target protein to nucleus
2 types: classic (short stretch of +ve/basic AAs), bipartite (two short stretches of basic AAs, with 7-10 AA long spacer between)
Proteins can have multiple NLSs and NESs; identified based on mutational analyses
NLS defined as AA sequence that is both necessary (LOF; mutation = mislocalization to cytoplasm) and sufficient (GOF; linking sequence to non-nuclear protein causes redirection of fusion protein to nucleus)
NLS identification experiment in ARC1
ARC1 is required for plant pollination; shuttled between nucleus and cytoplasm via NLS and NES.
Experiment 1: mutation of residues 261-266 results in myc-tagged ARC1 mislocalizing to cytoplasm
Experiment 2: Fusing 261-266 to cytoplasmic CAT protein results in redirection to nucleus
Cytoplasm-to-nucleus transport: other factors needed
Transport receptors: mobile proteins that move cargo across envelope
Karyoferins: large family of receptor proteins that move large molecules into (importins) or out of nucleus (exportins)
Step 1-4 of cytoplasm-to-nucleus transport
Cargo with NLS recognized in cytoplasm by importin — alpha + beta dimer; alpha binds to residues in NLS
Cargo-importin receptor complex moves toward nucleus (via importin binding to cytoskeleton); at nucleus surface, beta binds to cytoplasmic filaments in the NPC
Complex translocated through NPC central channel, likely through interactions with FG Nups (=untangling)
Complex associates with nuclear basket, binds to Ran-GTP via beta, resulting in NPC release and disassembly (of importin alpha and cargo) in nucleoplasm; NLS not cleaved
Ran protein
GTP-binding; Ran-GTP higher in nucleus and Ran-GDP higher in cytoplasm. GEF maintains high Ran-GTP and GAP maintains high Ran-GDP. Hydrolysis provides energy needed for transport
Step 5 of cytoplasm-to-nucleus transport/GTP gradients
Importin beta, bound to Ran GTP, moves back to cytoplasm due to gradient (high Ran-GTP in nucleus)
In cytoplasm, GTP hydrolyzed and beta released from Ran-GDP; now available for another round of transport
Ran-GDP moves back to nucleus, due to high Ran-GDP in cytoplasm, then converted to Ran-GTP by GEF
Steps of nucleocytoplasmic transport
Exportin binds importin alpha (as cargo disassembly exposes NES in it); exportin can also bind to other cargo proteins with an NES (several types; most common is leucine-rich motif LxxLxxL…)
Exportin-importin alpha/cargo complex binds Ran-GTP (high in nucleus), which promotes stability of the complex
Importin alpha-exportin-Ran GTP complex transported via NPC into cytoplasm (down Ran-GTP gradient)
In cytoplasm, GAP hydrolyzes Ran-GTP to Ran-GDP, which leads to disassembly of the complex
—>importin alpha used for another round of import, Ran-GDP moves back into nucleus (down gradient) and converted to Ran-GTP, exportin moves back into nucleus (via exposed NLS and importin) for another round of export
Nucleocytoplasmic transport: piggybacking, ARC1
Piggyback import: protein without NLS binds to protein with NLS, then imported as normal via importin
Many protein shuttle back and forth, contain both NLS and NES (distribution controlled by relative strength of each); phosphorylation can modify this
Ex. ARC1: before pollination, NLS>NES so mostly in nucleus; during self-pollination, NLS phosphorylated and disrupted, so localized mostly in cytoplasm instead
Co-IP assay info
Bait = epitope-tagged protein missing its NLS
Prey = importin (alpha/beta)
Mix bait and prey, add IgGs that bind to epitope, centrifuge, SDS-Page + coomassie blue staining to visualize. Can compare protein with and without NLS to see what importin binds to (… no NLS = no binding)
Checkpoints in cell cycle
Mid G1/START: cell commits to DNA replication and organelle duplication in S
end of G2: commits to entering mitosis
end of metaphase: commits to chromosome segregation
Mitotic cyclin-CDK levels in cell cycle; phos. of target proteins at end of G2
Early G1: cyclin level low, CDK activity low
End of G2/start of M: cyclin level high, CDK activity high, leading to phos. of target proteins
—>histones + condensins: leads to chromatin packing, chromosome condensation
—>lamins: leads to disassembly of nuclear lamina
—>Nups: leads to NPC disassembly
Nuclear structure during mitosis
Dynamic process, but nucleus completely disassembled by metaphase. In prophase: both nuclear membranes break down, lamins + NPCs disassemble, membrane-bound and soluble nuclear proteins released into ER and cytoplasm respectively
Opposite effects at end of mitosis (telophase): low cyclin/CDK; dephos. of Nups and lamins = reformation of lamina, nuclear envelope, NPCs; soluble NLS-proteins reimported
Decrease in mitotic cyclins after start of M phase: why?
Decreased synthesis of new cyclins and degradation of existing cyclins
Existing cyclins also prevented from targeting to nucleus
Mitotic cyclin B1 localization throughout cell cycle
* cyclins generally contain both NLS and NES
Cyclin B1:
up to and during G2: NES>NLS, primarily in cytoplasm
end of G2/start of M: NES phos. = primarily in nucleus, activates CDKs
after start of M: NES dephos. = primarily in cytoplasm
Brightfield microscope: details
Light diffracted by specimen, undiffracted light focused by objective lens
Image usually captured by camera - more sensitive to low light intensities
Software can manipulate images, ex. deconvolution: removes background and unfocused light (=better contrast and clarity)
Resolution
Minimum distance that two separate points can be identified as such (and not as one object); most important aspect
Dependent on wavelength of light and numerical aperture=NA (light-gathering quality of objective lens and specimen medium)
How to maximize? Use shorter wavelength or increase NA by changing medium; limit for most standard brightfield/CLSM is ~200nm (ie. can only be used for larger organelles) — electron microscope is higher
Limitations of brightfield microscopy
Poor contrast
Specimens usually fixed (causes cell death), embedded + sectioned (can lead to ‘artifacts’), stained (limited molecular-specific stains available)
Fluorescence microscopy: details, pros and cons
Visualize fluorescent molecules in living or fixed specimens
Based on autofluorescence, applied fluorescent dyes or conjugated antibodies (immunofluorescence), or autofluorescent proteins (ex. GFP)
Pro: increased contrast, 3D image, can study fine structures or dynamic processes (non-fixed only)
Con: thick specimens can result in blurry image due to out-of-focus fluorescence
Confocal Laser-Scanning Microscopy (CLSM): definition and basic details
Similar to standard brightfield light microscope, but one or more lasers at specific wavelengths excite fluorescent molecules in specimen. Emitted light specifically focused to get a detailed image.
Usually living specimen - fixing not necessary
Can view dynamic processes in real time
Lasers can penetrate into thicker specimens
CLSM: mechanism and image produced
Specimen rapidly scanned with laser at specific excitation wavelength
Emitted fluorescence from one focal plane only is focused through pinhole for viewing
Out-of-focus fluorescence excluded (doesn’t pass through pinhole)
Yields a clear 2D z-section/optical slice; z-sections can be collected at different depths and combined into a 3D z-stack
CLSM: cons
Rapid, but can’t capture extremely dynamic processes
Laser light can photobleach fluorescent molecules or damage live cells through phototoxicity
Not efficient for deep imaging of thick specimens/tissues
Limited spatial resolution (~200nm)
Super-resolution CLSM
10x better resolution (~20nm) than standard CLSM
Different techniques (change wavelength, angles, beam widths) + image combined and processed for increased resolution
Pro: useful for smaller intra-cellular structures
Cons: longer scanning time, not efficient for deep imaging or very dynamic processes
Endomembrane system + components
Dynamic, interconnected network of organelles (other than mitochondria and chloroplasts): ER, ER-derived organelles (nucleus + peroxisomes + lipid bodies), Golgi, endosomes, lysosomes/vacuoles, secretory vesicles/granules, plasma membranes. Material trafficked between via transport vesicles (small, membrane-bound).
General steps of endomembrane trafficking
Cargo-containing vesicles buds off donor compartment; vesicle coat proteins select membrane + cargo proteins that enter vesicle, regulate formation and budding
Vesicle transported through cytoplasm to acceptor membrane; coat proteins regulate trafficking to proper acceptor along with molecular motors and cytoskeleton highways
Vesicle fuses with acceptor and cargo proteins incorporated into compartment; receptor proteins regulate vesicle-acceptor fusion.
Budding and fusion process repeated and can occur in reverse. Other proteins regulate recycling of ‘escaped’ proteins back to donor.
Biosynthetic pathway
Transport from ER to Golgi, endosomes, then lysosomes (and vacuoles in plants) OR via exosomes from endosomes to plasma membrane and extracellular space (ie. secretory pathway… overlap)
Two types of secretion
Constitutive: continual transport from Golgi to pm and/or released via exocytosis in secretory vesicles (to outside of cell; membrane incorporated into pm and cargo released)
—> exocytosis: trafficking to pm, fusion, release of contents
Regulated: only in specialized cells. ER-derived materials from Golgi stored in secretory granules, which fuse with pm and release cargo outside cell in response to a signal
Endocytic pathway
Opposite direction of secretory pathways: uptake of materials into cell. Materials from pm/extracellular space enter cell via endocytosis, and are transported to endosomes and lysosomes/vacuoles.
Secretory pathway: pancreatic and other cells
Amount of secretion varies, some higher than others: yeast + plant cells producing cell wall materials, pancreatic acinar cells (digestive enzymes), small intestine epithelial cells (produce mucus)
Pancreatic + intestine epithelial cells highly polarized, organelles specifically organized: basal end = nucleus + RER, central region = Golgi + lysosomes, apical end (at duct) = secretory granules with enzymes/mucus
Autoradiography/pulse-chase experiments
Used to view movement through secretory pathway
Pulse with labelled/radioactive AAs that are incorporated into new proteins (‘pulse’)
Tissue washed and incubated with non-radioactive AAs for varying amount of time
Synthesis continued, labelled proteins traffic through cell (‘chase’)
Tissue fixed/killed and X-rayed (autoradiography)
Live-cell imaging with autoflorescence
Gene encoding autofluorescent protein (GFP, RFP, etc) linked to gene-of-interest, ex. for a protein of a certain organelle. Recombinant gene fusion introduced via cloning, and protein visualized with CLSM as it traffics through cell
Modern pulse-chase labelling (temperature sensitive)
Live-cell imaging (CLSM + autofluorescence)
temperature-sensitive viral glycoprotein VSVG fused to GFP, transfected into cell
VSVG mutation is reversible: at 40C (restrictive temp.), protein misfolded and stays in ER due to quality control processes; at 32C (permissive temp.), it folds properly and is transported from ER to…
Results: 40C = in rough ER (where protein synthesized), 32C = in Golgi (near nucleus - site of protein modification/folding); longer times = found in pm (cell surface)
Subcellular fractionation through centrifugation
Separating and purifying specific organelles based on their size and/or density — allows for study of organelle structure and function
Cell/tissue disrupted by homogenization, while organelles remain intact
Homogenate filtered (removes unbroken cells or big fragments) and differential centrifugation is done
—>600 g * 10min isolates nuclei as a pellet
—>remaining supernatant (liquid at top) subjected to 15K g * 5min, produces mitochondria + lysosomes
—>remaining supernatant subjected to 100K g* 60min: gives pm and ER microsomes
Microsomes: fragments of ER membrane or pm that fuse and form small, spherical vesicles
Subcellular fractionation: equilibrium density-gradient centrifugation
15K g * 5min pellet from previous step added to top of sucrose solution w/gradient (increasing density from top to bottom), then centrifuged
Individual organelles migrate to known sucrose densities
Layers of gradient can be purified and organelles identified by EM and/or marker proteins/enzymes
Cell-free systems
Characterizing activity of specific endomembrane protein components in vitro
Isolated proteins incubated with liposomes and mixed with purified proteins, effectively recreating the endomembrane trafficking pathways
Mutant phenotype analysis
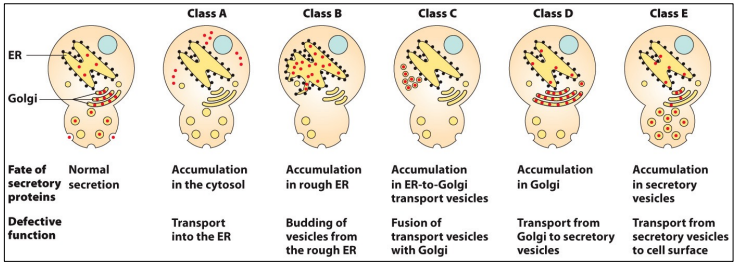
Identifying genes/proteins and trafficking steps by screening for mutant phenotypes, ex. with secretory mutants of yeast. Temperature sensitive - only secrete proteins at permissive temperature.
Types of sec yeast mutants
5 classes
A = accumulation in cytosol (defect in co-translation/translocation)
B = accumulation in ER (defect in ER vesicle formation), ex. sec12 (has large, expanded ER)…
Double mutants indicate order of steps in pathway - B + D = B mutant, since ER budding occurs before Golgi budding
Other mutants can indicate biogenesis events
Endomembrane system: transport pathways
Biosynthetic, secretory, endocytic pathways + transport vesicles; ER is starting point for secretory and biosynthetic (site of protein + lipid synthesis, protein folding, processing/quality control)
need to add image
Endoplasmic reticulum
Complex network of membrane-enclosed, rod-like tubules and sheet-like cisternae (flattened sacs); organelle with largest SA
Endoplasmic reticulum: lumen + more details
Lumen = aqueous space inside ER tubules and cisternae
Tubules and cisternae shapes mediated by reticulons
ER integral membrane proteins possess hairpin structure that regulate membrane curvature and overall shape
Endoplasmic reticulum: 2 main subdomains
Distinct regions with unique morphology and/or functions
RER: mostly cisternae with bound ribosomes, involved in protein + membrane phospholipid synthesis
SER: mostly curved tubules with no ribosomes, involved in Ca2+ storage and hormone synthesis
Endoplasmic reticulum: other subdomains
>20 other subdomains, with unique proteins and lipids
Nuclear envelope: continuous with RER, contains Nups + attached ribosomes
*Mitochondria + plasma membrane-associated membranes (MAM & PAM respectively): regions that make contact with mitochondria/pm, involved in membrane protein and lipid exchange
ER Exit Sites (ERES): regions where transport vesicles bud off and move to Golgi
*add/review diagram?
RER and main sites for translation (protein synthesis)
1. Free ribosomes in cytoplasm — proteins either remain in cytoplasm, or target to proper organelle (nucleus, mitochondria, etc). Can be soluble or membrane-bound.
2. ER membrane-bound ribosomes in RER — proteins can:
Remain in RER or localize to another subdomain (ex. nuclear envelope)
Localize to other ER-derived organelles (ex. peroxisomes - organelles bud off from ER)
Target (via transport vesicles) to another post-ER compartment of endomembrane system (ex. Golgi, lysosomes, pm, etc)
Co-translational translocation of soluble protein into RER lumen: steps 1/2
mRNA translation on free ribosome in cytoplasm.
N-T of growing polypeptide emerges from ribosome, contains signal sequence: stretch of 8-15 hydrophobic AAs that serves as RER targeting signal
Signal recognized by SRP particle (consists of 6 proteins + 1 small RNA), which binds to ribosome and stops translation.
Co-translational translocation: step 3
SRP targets complex (ribosome, stalled polypeptide, mRNA) to surface of RER and binds to SRP receptor
SRP receptor is a heterodimer, with cytoplasmic domains that serve as ‘docking site’ for SRP
Interaction strengthened by GTP binding to both.
Co-translational translocation: step 4
GTP hydrolysis results in release of SRP and SRP receptor (later reused)
Simultaneously, polypeptide and ribosome move to cytoplasmic side of Sec61 translocon
Sec61: multi-protein complex with ER integral membrane subunits (Sec 61a/b/y), forming an hourglass-shaped aqueous channel
Co-translational translocation: step 4 continued
During transfer to translocon, polypeptide N-T inserted into opening of channel
Translation continues, elongating polypeptide passes through translocon channel (towards ER lumen).
add diagram?
Sec61 translocon: more details
Contains pore ring: 6 hydrophobic AAs at narrowest part of channel, serving as a gate/seal
Channel also blocked by short alpha-helix plug (additional ‘gate’) that is pushed out of the way during translocation
Co-translational translocation: steps 5 and 6
N-T signal sequence enters ER lumen, cleaved by signal peptidase and degraded
—> an integral membrane protease associated with translocon that recognizes cleavage sequence motif at C-T end of signal sequence
Co-translation then continues
Co-translational translocation: steps 7 and 8
Translation completed: ribosome released from translocon, remainder of protein enters ER lumen
Translocon closes via pore plug moving back into channel
Nascent protein glycosylated and folded by reticuloplasmins
—>ER molecular chaperones that mediate folding and oligomeric assembly, ex. BiP, calnexin, calreticulin
Co-translational insertion of integral protein into RER
Most membrane proteins (resident ER proteins, all other post-ER compartments) synthesized on RER membrane-bound ribosomes, except for mitochondria and chloroplast proteins
Similar insertion process as soluble protein, but instead protein anchors in ER membrane with a specific orientation/topology
Integral membrane topology
Number of transmembrane domains and orientation
TMD: usually alpha-helix of 16-25 hydrophobic AA (favorable in hydrophobic interior of phospholipid bilayers)
Several different classes
Overview of classes of ER integral membrane proteins
review diagram + orientations + flipping
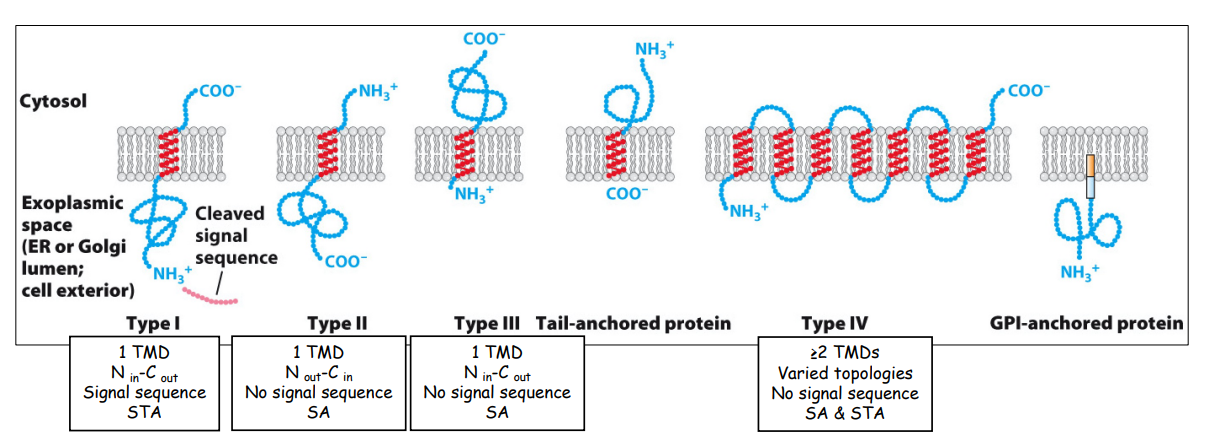
Type I membrane protein (NER lumen, Ccytosol)
Similar targeting process, but:
First and only hydrophobic TMD enters translocon, serving as a stop-transfer anchor (STA) sequence that stops further translocation
STA moves laterally out of translocon and anchors in adjacent phospholipid bilayer.
As translation continues, polypeptide extends into cytosol and diffuses laterally into ER membrane bilayer once translation complete
Type II membrane protein (Ncytosol, CER lumen)
Opposite orientation as Type I, with no N-T signal sequence
Has internal signal-anchor (SA) sequence — only TMD, both targets SRP + complex to translocon AND anchors protein in membrane
SA enters translocon and gets flipped so that polypeptide faces cytosol
—>mediated by several +ve AAs just upstream of SA, which determine orientation of most RER membrane proteins: positive-outside rule
Remaining steps are the same as type I
Type III membrane protein (NER lumen, Ccytosol)
Same orientation as Type I, but have internal SA sequence and no N-T signal sequence
Same steps as Type II, but polypeptide not flipped towards cytosol
Multi-spanning/Type IV membrane protein
Multiple TMDs + no N-T signal sequences
Internal SA (targets protein to ER, orient protein based on positive-outside rule)
AND internal STA (stop transfer of protein and anchor it into ER membrane)
*review SA + STA function differences
Membrane biosynthesis at ER
Membranes must arise from pre-existing membranes, cannot form de novo
Most membrane proteins + lipids synthesized at ER, except for glycolipids in Golgi and chloroplast + mitochondrial proteins + lipids
These can move to other ER subdomains (lateral diffusion through bilayer) OR downstream organelles (via transport vesicles).
Membrane biosynthesis: protein + lipid distribution
Asymmetrical distribution in lipid bilayer
Integral = different regions on cytoplasmic and exoplasmic faces
Peripheral = cytoplasmic or lumenal side
Membrane phospholipids = unequal distribution between cytoplasmic and exoplasmic faces
Asymmetry maintained throughout endomembrane system — orientations stay the same throughout
Processing of newly-synthesized proteins at ER: steps
Signal sequence cleavage (removing N-T signal sequence)
Initial glycosylation: addition of carbohydrate side chains to specific AAs on amino acids (allows for proper folding, protein-protein interactions)
Protein folding + assembly: protein folded into 3D conformation and oligomerically assembled by chaperones/reticuloplasmins
Quality control: misfolded or improperly assembled proteins recognized and degraded
ER is optimal for these functions as it is the first endomembrane system compartment (for biosynthetic + secretory pathways)
Protein processing: glycosylation
Most proteins are glycoproteins: one or more sugar molecules added to specific AAs on polypeptide
—> needed for proper folding and provides binding sites for other molecules
Most common type is N-linked glycosylation: specific short sugar chains added to terminal amino group of asparagine
—>two stages: core glycosylation and core modification
—>some glycoproteins transported to post-ER compartments, so core modification continues in Golgi instead
Core glycosylation
Various ER membrane-bound glycosyltransferases synthesize core oligosaccharide: 14 sugar residues, including mannoses and terminal 3-glucose-long branch (needed for quality control)
First sugar added to dolichol phosphate - acts as membrane anchor and sugar carrier
Glycosyltransferases continue to add sugars — tunicamycin blocks this
Final step: transfer from dolichol to lumenal portions of nascent protein (containing -N-x-S/T- sequence) via Sec61 pathway
Empty dolichol recycled for another round of synthesis
Core modification: steps
Second stage of N-linked glycosylation
14-sugar core oligosaccharides are trimmed and modified
2 of 3 terminal glucose units removed, last unit removed and re-added (important for proper folding/assembly)
All done by lumenal glucosidases
Core modification: enzymes and folding
Protein rapidly folded into proper 3D conformation during glycosylation and modification.
Reticuloplasmins: chaperones including BiP, calreticulin and calnexin that reversibly bind to proteins to prevent misfolding or aggregation
Protein disulfide isomerase (PDI): catalyzes formation of intra- and intermolecular disulfide bonds between cysteine residues, which stabilize proper 3D confirmation
Core oligosaccharides also contribute to stability and protein quality control.
ER protein quality control: release from glycosylation
Reticuloplasmins and PDI bind to nascent glycoprotein (with 1 remaining glucose)
Lumenal glucosidase removes last glucose, releasing protein from reticuloplasmins
ER protein quality control: proper vs improper folding responses
If properly folded: 1 mannose unit removed by lumenal mannosidase, then protein functions as ER resident protein OR transport from ER to Golgi for further modification (and potentially transported further)
If misfolded: UGGT glycosyltransferase recognizes hydrophobic residues (should be buried in protein) and adds terminal glucose back to oligosaccharide core
—>chaperones bind again to mediate proper folding (repeats the process)
ERAD pathway + ubiquitination
If proteins continue to fold incorrectly, they are degraded by ER-associated degradation (ERAD) pathway
Involves AAA ATPase p97, an ER membrane protein that uses ATP hydrolysis to pull proteins across membrane into cytosol
Oligosaccharide chains then removed and protein poly-ubiquitinated — serves as a degradation signal for proteasome
Mono-UB signal also targets membrane proteins into intralumenal vesicles in late endosomes/multivesicular bodies.
Proteasome and protein degradation
Barrel-shaped, multi-subunit machine located in cytoplasm (… and nucleus)
Poly-UB binds to cap/lid of proteasome and is removed/recycled
Protein threaded into proteasome and degraded by proteolysis
AA products reused for new protein synthesis.
ER protein quality control: ER stress and UPR response
In certain conditions (CF, Alzheimer’s, etc), misfolded proteins can accumulate and overwhelm ERAD pathway, leading to ‘ER stress’ (can lead to toxicity and cell death).
Stress activates 1 of 3 UPR pathways, with unique protein sensors:
—>Ire1, PERK, ATF6
PERK and ATF6 UPR pathways
PERK and ATF6 both membrane-bound with lumenal-facing stress-sensing domains, which bind to chaperones (ex. BiP) in ER lumen
Normal (no stress): PERK and ATF6 bound to BiP, inactive
Stress conditions: pathways activated
PERK-mediated UPR pathway
BiP is released from PERK to aid in proper folding of misfolded ER proteins, PERK dimerizes/activates
Cytoplasmic kinase domains of PERK dimers phos. eIF2alpha, an initiation factor needed to start translation
Causes decrease in protein synthesis (including at RER), so that chaperones can focus on misfolded proteins
ER stress alleviated OR cell death occurs
ATF6-mediated UPR pathway
Like PERK, BiP released from ATF6
Active ATF6 moves from ER to Golgi (via transport vesicles from ERES)
at Golgi, cytoplasmic-facing TF domain of ATF6 cleaved by Golgi-associated protease, exposing NLS which targets ATF6 to the nucleus
In nucleus, ATF6 TF upregulates genes for reticuloplasmins, ER export components (assist in export of properly-folded proteins out of ER), ERAD components (assist in degrading remaining misfolded proteins in ER)
Stress alleviated OR cell death occurs
Fate of newly-synthesized proteins in ER
Properly folded/assembled/glycosylated proteins at RER either:
Stay in ER: localized to RER OR move laterally through ER lumen or bilayer to another subdomain (ex. SER)
Exit from ER: quickly move to ERES for transport to Golgi
ER exit sites (ERES)
Usually located next to cis-face of Golgi
Contains machinery for formation/budding of transport vesicles bound for Golgi
Also responsible for packing vesicles with correct lumenal and membrane-bound proteins — very selective
Resident ER proteins (ex. BiP) usually prevented from entering these transport vesicles
Transport vesicle assembly at ERES
Vesicles have distinct morphology: small (20-50nm) diameter and fuzzy appearance on EM
Due to layer of soluble coat proteins (COPs) attached to cytoplasmic surface of vesicle membrane (assemble at cytoplasmic ERES surface)
Two main functions of COPs: mediate membrane curvature + vesicle formation, and concentrate specific components into vesicles (proteins, lipids, Rabs, SNAREs)
Coat proteins
Three major classes:
COPII: forward/anterograde transport from ERES to Golgi
COPI: backward/retrograde transport from Golgi to ER, and backward within Golgi
Clathrin: transport from Golgi or plasma membrane to endosomes
COPs assemble sequentially to form coat/curved scaffold on surface of transport vesicle
COPII-coated vesicle assembly at ERES: step 1
Sar1 GTPase recruited from cytoplasm to ERES membrane via Sec12 binding (functions as GEF, catalyzes GDP—>GTP exchange on Sar1)
GTP binding causes conformational change to expose amphipathic hydrophobic N-T (anchors into ERES membrane)
Sar1-GTP integrated in cytoplasmic leaflet of ERES bilayer
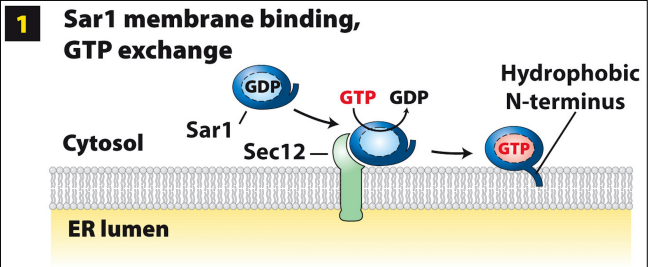
COPII-coated vesicle assembly at ERES: step 2
Sar1-GTP recruits several other COPII proteins from cytosol to ERES surface, starting with Sec23 + 24, which act as scaffolding and promote outward bending (towards cytosol) of ERES membrane
Sec24 also involved in vesicle protein selection by binding to cytoplasmic domains of membrane proteins:
cargo (exit ERES for Golgi)
cargo-receptor (bind to soluble lumenal cargo destined to exit ERES for Golgi)
trafficking (required for trafficking and vesicle docking with acceptor, ex. v-SNAREs)
COPII-coated vesicle assembly at ERES: step 2 continued
Sec24 protein selection mediated by ER export sorting signal
—>di-acidic — most common is Asp-X-Glu located in cytoplasmic domains of Sec24 selected proteins
Not found on ER resident proteins
All Sec24-bound proteins (and those bound by cargo-receptor proteins) are concentrated within growing COPII vesicle.
COPII-coated vesicle assembly at ERES: step 2 continued
Sec23 + 24 recruit additional soluble COPII components from cytoplasm to outer surface
Sec13 + 31 self-assemble into outer, cage-like lattice and act as structural scaffolding for bud
Promote outward bending and eventual release/scission of vesicle into cytosol
COPII-coated vesicle assembly at ERES: steps 3 and 4
After release of vesicle, Sec23 promotes hydrolysis of Sar1-GTP to Sar1-GDP
This results in disassembly of COPII coat — Sar1-GDP and all soluble COPII proteins released into cytoplasm. Results in nascent, uncoated vesicle.