AP bio 5
1/44
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
45 Terms
DNA (deoxyribonucleic Acid)
a polymer made of nucleotides and it has three parts: a phosphate group (PO4), deoxyribose sugar, and a nitrogenous base
Pyrimidines
single ring and is cytosine, thymine, and uracil
Purines
double ring and is guanine and adenine
Antiparallel structure
one strand runs 3’ to 5’ and the other 5’ to 3’. The 5’ end has a phosphate group and the 3’ group has a sugar.
Hydrogen bonding in DNA
Adenine and thymine have two hydrogen bonds connecting them while guanine and cytosine makes three hydrogen bonds
Phosphodiester bonds
formed between sugars and phosphate groups
What does the order of nucleotides do?
stores information
What happens when a gene is expressed and when it isn’t?
the order of nucleotides are “read” and then used to build proteins so if a gene is “turned on” it is expressed and the protein is produced and if the gene is “turned off” the gene is not expressed and no protein is produced
Gene regulation
control of gene expression (turning genes on or off). Not all genes are transcribed all the time so most are regulated and only transcribed at certain times and under certain conditions when the product is needed. Some genes are always needed so are always being transcribed.
Enzymes in DNA replication
Helicase - unwinds DNA at the replication fork (so it breaks hydrogen bonds holding base pairs together)
Single-strand binding proteins - holds the stands apart
topoisomerase - it works before helicase unwinds the DNA to prevent tangling and relieve pressure
DNA polymerase - builds the new, complementary DNA
primase - builds RNA primer as “starting blocks” (the RNA primer will eventually be removed and replaced by DNA)
DNA ligase - fuses short DNA fragments during discontinuous replication
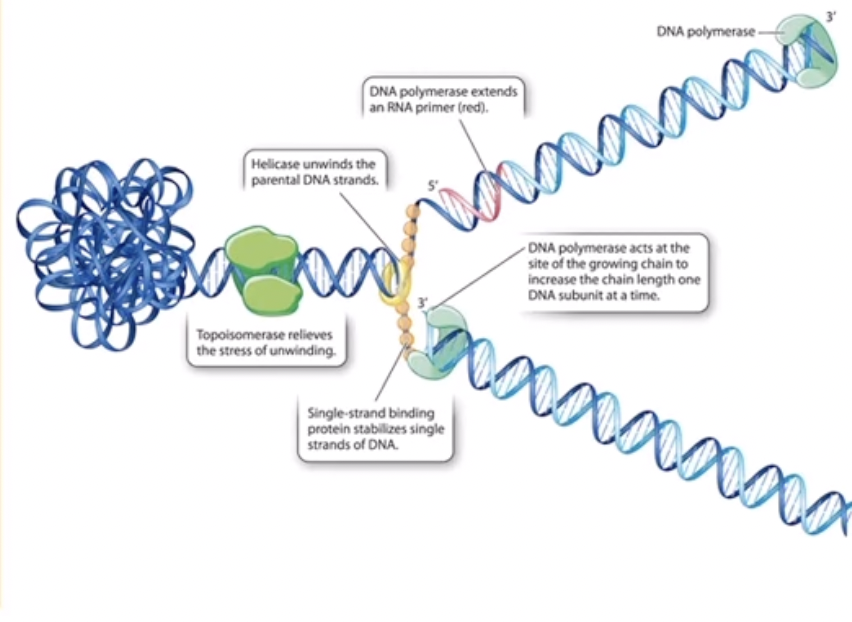
2 rules of DNA polymerase
DNA polymerase can only add nucleotides onto other nucleotides so primase builds a section of RNA primer to that polymerase has something to build onto
DNA polymerase can only add nucleotides onto the 3’ end of a nucleotide so only one strand of DNA can be replicated continuously, building from 5’ to 3’. One strand faces the right way (so the strand is built continuously) but the other faces “backwards” (so the strand is built discontinuously).
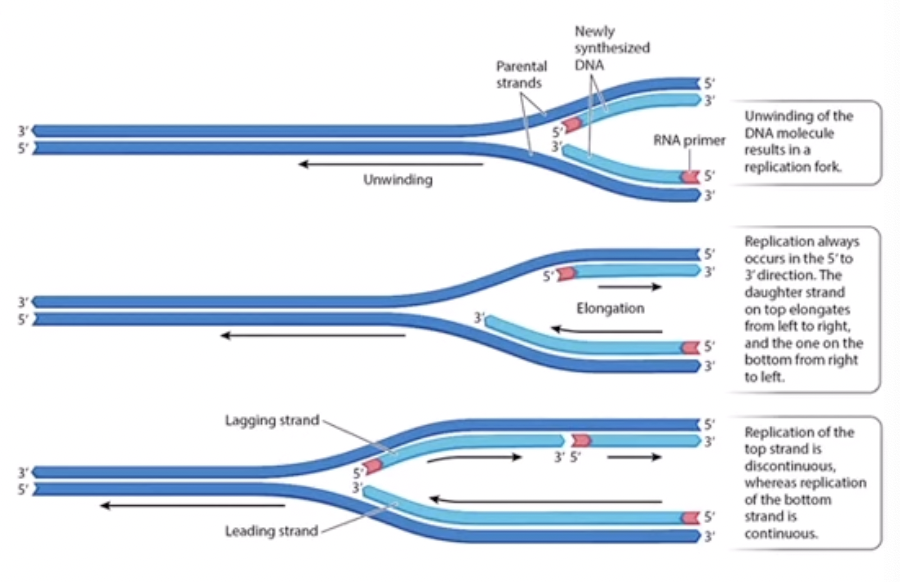
Continuous Replication
The parental 3’ to 5’ is replicated continuously so DNA polymerase builds from 5’ to 3’ (towards the replication fork) always adding nucleotides to the 3’ end. The continuous strand is sometimes called the leading strand because it is faster so will be ahead of the other. The continuous strand will have the 3’ template strand.
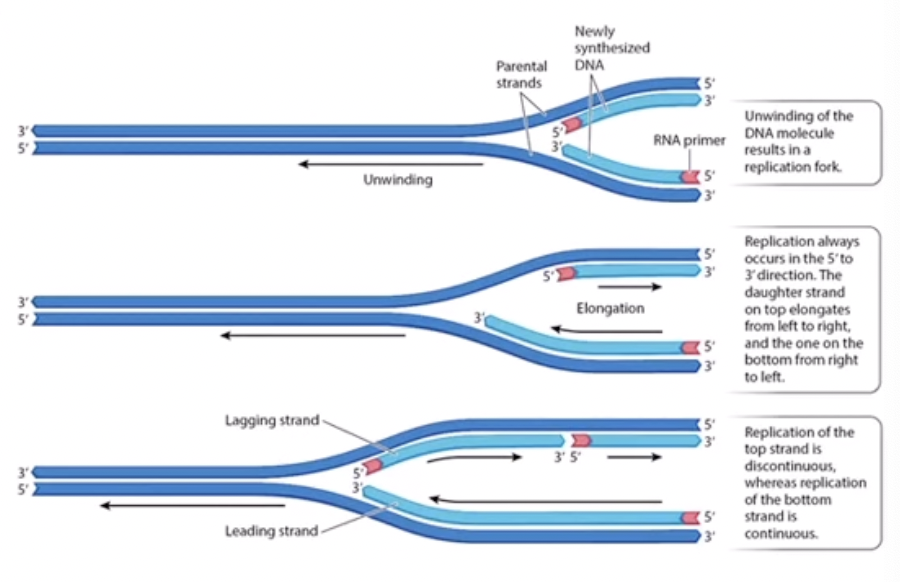
Discontinuous replication
because DNA polymerase cannot build on the 5’ end, it will jump ahead about 1000 nucleotides closer to the replication fork, where a RNA primer will be built, and then add nucleotides going away from the replication fork. Once it has completed building that, it will jump again so it can build on the 3’ end. Even though it builds away from the replication fork, the overall movement is towards it because it keeps jumping forward. This strand is called the lagging strand because it is more complicated and thus slower. The discontinuous strand has the template strand of 5’.
How are mutations in DNA caused?
If DNA polymerase makes a mistake and doesn't fix it while replication DNA
Semi-conservative DNA
this means that DNA is half new and half old (old is called the parental strand)
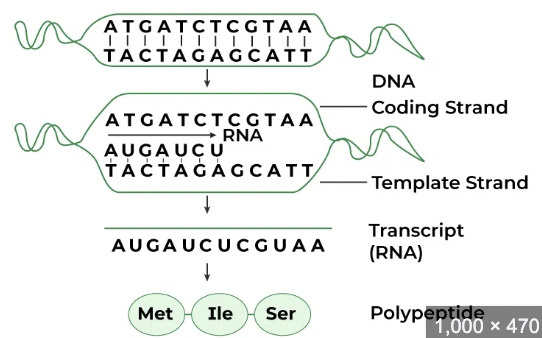
RNA Transcription
DNA is read and copied into a section of messenger RNA, in eukaryotic cells this occurs in the nucleus and in prokaryotic cells in the cytoplasm. 3 steps:
Initiation
RNA polymerase binds to DNA & separates the strands, transcription factors (a protein) also bind with the RNA polymerase
RNA polymerase begins pairing RNA nucleotides to the template DNA strand (only one strand will be copied)
Promoter sequence signals start of gene (this is what the RNA polymerase recognizes and binds to) The promoter sequence is a sequence of nucleotides that includes 5’ - TATAAA - 3’ and it is known as the TATA box, transcription begins about 25 base pairs away from the TATA box.
Elongation
RNA polymerase adds new RNA nucleotides to the 3’ end of a growing transcript as it moves along the template strand
RNA transcript grows in 5’ to 3’ end but is read 3’ to 5’ direction
Termination
Occurs when RNA polymerase reaches a terminator nucleotide sequence in the DNA that signals the end of transcription
RNA polymerase will release DNA
DNA strand goes back together like normal
Transcription in prokaryotic vs eukaryotic cells
Prokaryotic - the product of transcription is mRNA and can immediately translate into protein
Eukaryotic - the product of transcription is a primary transcript that then undergoes processing to become mRNA
RNA Processing
in the nucleus of eukaryotes, this process converts primary RNA transcript into mRNA which brings the message from DNA to ribosomes. It makes 3 modifications:
GTP cap (a modified guanine nucleotide) is added to 5’ end and allows the ribosome to recognize the strand and begin translation
Poly-A tail (repeating adenine nucleotides) added to 3’ end and it helps prevent digestive enzymes in cytoplasm from breaking it down before it reaches the ribosome. It does this by providing a buffer, the digestive enzymes will break down the buffer nucleotides so it gives the strand time to reach a ribosome.
Introns (sections of the RNA) removed and exons (what's left after introns removed) are spliced together
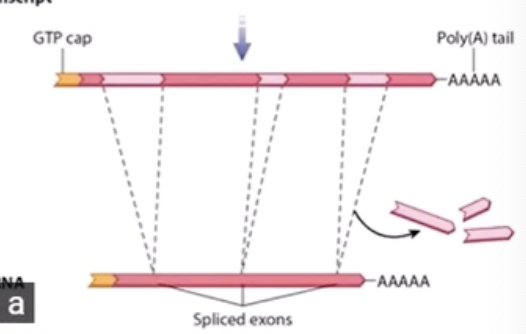
RNA splicing
removes introns (regions that are not expressed as proteins) and stitches together exons (regions that are expressed as proteins). The amount of introns varies per gene.
Alternative RNA splicing
Which exons are stitched together allows for different mRNA strands and thus different proteins
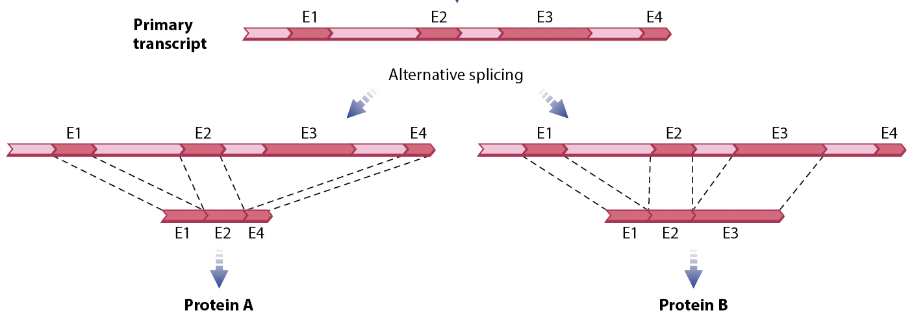
3 types of noncoding RNA
these do not code for proteins but are all made through the process of transcription
Ribosomal RNA (rRNA) - along with protein it makes up the ribosome
Transfer RNA (tRNA) - transports amino acids to the ribosome and positions them at the right place on the polypeptide chain
Small RNAs - regulates gene expression
Translation
the instructions of the messenger RNA are used to build the primary protein, this occurs in the cytoplasm on the surface of a ribosome. Has three steps:
Initiation - begins at the start codon and the first amino acid is added
Initiation factors (proteins) locate the start codon
tRNA attaches with the amino acid MET (methionine). Methionine is always going to be the first amino acid in a protein
Establishes reading frame (establishes what the first three nucleotides are so the ribosome will always move over three nucleotides from there)
Ribosome assembles around mRNA (like a burger bun!🍔 )
Elongation - amino acids are added one by one and it is the longest part of the process
The ribosome moves over three nucleotides
A new tRNA attaches on the open codon and a peptide bond forms by dehydration synthesis (adding a new amino acid to the chain with the addition of energy) and the tRNa is released
This process repeats
Termination - top codon ends the addition of amino acids and the protein is released
Stop codon is reached and has no corresponding amino acid
The release factor protein binds to ribosome and causes the pieces of the ribosome to fall apart to release the mRNA
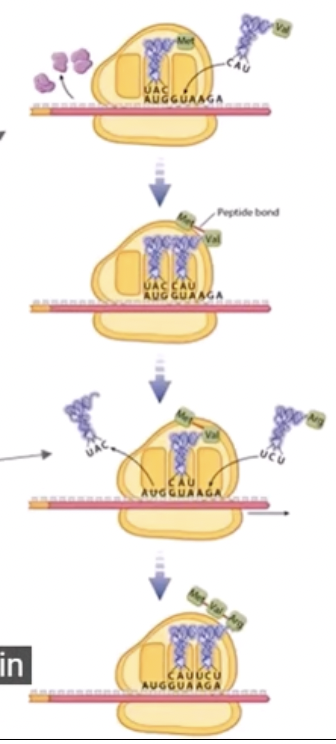
Picture of an overview of both transcription and translation
What happens after mRNa strand has been read?
another ribosome can bind to it and create another protein or it can be degraded and the nucleotides are reused
Codon
a set of three nucleotides on a mRNA strand that corresponds to one amino acid in the chain
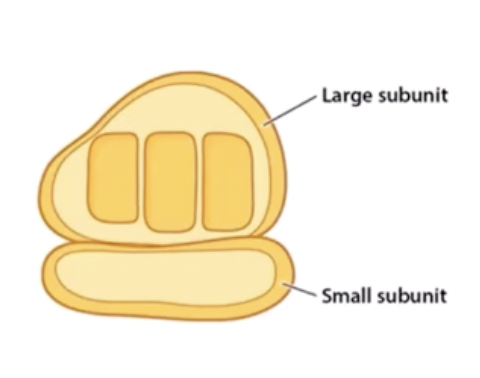
Ribosome
attaches to an mRNA strand and reads it to catalyze the joining of amino acids into a protein. It is made of protein and rRNA and has two parts, which are only attached when translating mRNA. It is not membrane bound so all cells (eukaryotes and prokaryotes) have them.
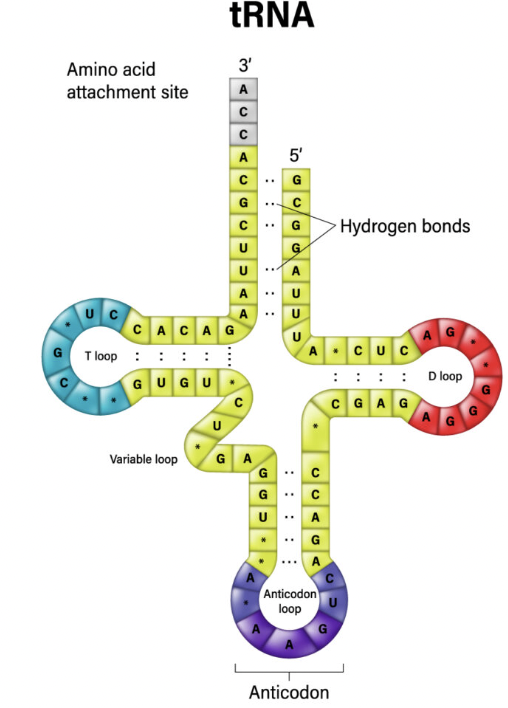
Transfer RNA
tRNA binds to specific amino acids on the 3’ end and on the other end it has the anticodon (made of three nucleotides) that is complementary to the codon. tRNA acts as a bridge between the mRNA and the protein
Genetic code
the relationship between the codon found on the mRNA strand and the amino acid it specifies. It is redundant so more than one codon specifies the same amino acid (this allows for some room to mutate) and it is used by all living things (another example of common ancestry).
What is the start codon, anticodon, and amino acid?
codon - AUG
anticodon - UAC
amino acid - MET
What are the stop codons?
UAA, UAG, UGA
Is DNA transcription and translation the same for all cells?
Yes, and this is more evidence of common ancestry
DNA in prokaryotes
mainly consists of one circular chromosome and small extra pieces called plasmids that contain only a few genes. Because there is no nuclear membrane transcription and translation can happen at the same time in the same place.
DNA in Eukaryotes
more DNA so has multiple chromosomes and are condensed more tightly to fit. To condense, DNA is coiled around histone proteins.
RNA polymerase
an enzyme similar to DNA polymerase that makes a single strand of RNA that is complementary to DNA (but uses uracil instead of thymine)
What process is used to add nucleotides to the 3’ end?
dehydration synthesis
How do you read a template strand and in what direction does the RNA strand grow?
Grow 5’ to 3’ but read the template strand 3’ to 5’
Mutation
change in the DNA that can be positive, negative, or neutral.
Gene mutation
change in nucleotide sequence of a particular gene (one gene involved)
Chromosomal mutation
change in the structure of chromosome or the number of chromosome (many genes involved)
Nucleotide substitution
may or may not change the amino acid, this is a point mutation because change in a single nucleotide
Silent mutation - DNA changed but no effect on amino acid or protein
Nonsense mutation - changes amino acid to a stop codon which results in an incomplete codon
Missense mutation - DNA change and leads to different amino acid and this different protein
Conservative - changes to a similar amino acid (Ex. if the right amino acid is polar and the new one is also polar then it is conservative)
Nonconservative - changes that are not similar (Ex. if the right amino acid is nonpolar and the new one is polar then it is nonconservative)
Nucleotide insertion
frameshift mutation, if inserted in multiples of 3, 1 amino acid added. This usually leads to a nonfunctional protein
Nucleotide deletion
frameshift mutation, if deleted in multiples of 3, 1 amino acid(s) deleted. This usually leads to nonfunctional protein
Nondisjunction
change in the number of chromosomes because they didn’t separate properly
Causes of mutation
errors in DNA replication or repair
external factors like radiation or reactive chemicals
and errors in meiosis (nondisjunction resulting in trisomy (3 copies of a particular chromosome) or polyploidy (multiple sets of chromosomes)). Animals can handle having maybe 3 chromosomes but plants can handle having many.