Lecture 20 Repair and Recombination
1/111
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
112 Terms
mutations
permanent change sin the nucleotide sequence
-linked to cancer and aging
substitution
replacement of one base pair with another
insertion
the addition of 1+ base pairs
Deletion
the deletion of 1+ base pairs
silent mutations
only affect nonessential DNA or has a negligible effect on gene function
genes dedicated to DNA repair
nearly 200 genes in the human genome
DNA repair processes tend to be
energetically inefficient
-but it is irrelevant because the integrity of the genetic information is more important
how does the body fix DNA muations?
-Methyl-directed Mismatch repair (MMR)
-Base Excision Repair (BER)
-Nucleotide Excision Repair (NER)
-Direct Repair
Mismatches
break pairing rules
-e.g., A=G, T=C, T=G, C=A
Can be correct to reflect the proper template strand information
mismatch correction in prokaryotes
template strand are distinguished from the newly synthesized strand by the presence of methyl tags from Dam methylase on the template DNA
model for MMR
-Error Recognition
-Methylation Status Determination
-Mismatch Detection
-Strand Discrimination
-Excision
-DNA Resynthesis
-Ligation
-Methylation
error recognition (MMR)
mismatches often result from DNA polymerase replication errors
Methylation Status Determination (MMR)
methylation distinguishes the newly synthesized DNA strand from the parental strand
Mismatch Detection (MMR)
MutS mismatch repair proteins bind the mismatched base pair or indel mutation (nucleotide insertion or deletions)in the unmethylated DNA strand
-MutS forms a complex with MutL
Strand Discrimination (MMR)
the MutH binds to methyl groups on the parental DNA strand
-very important as it reads the methylated strand as the correct code and not the one with the mutation
Excision (MMR)
the MutS-MutL complex combines with MutH and recruits exonucleases and other proteins to remove a section on the 3' end of the unmethylated DNA strand
RNA Resynthesis (MMR)
DNA polymerase III and other repair-associated proteins are recruited and synthesize a new DNA strand using the intact parental strand as a template
-this step correct the mismatch by replacing the erroneous DNA segment with the correct sequence
Ligation (MMR)
DNA ligase seals the DNA backbone, making the repaired DNA a continuous strand
Methylation (MMR)
DNA methyltransferases add methyl groups to the newly synthesized strand
eukaryotic mismatch repair systems
-several proteins are structurally and functionally analogous to the bacterial MutS and MutL proteins
-many details of eukaryotic mismatch repair are unknown
DNA glycosylases
recognize common DNA lesions and remove the affected base by cleaning the N-glucosyl bond in the process of base-excision repair (BER)
-each class of glycosylase is usually specific for only one base
-The backbone remains intact
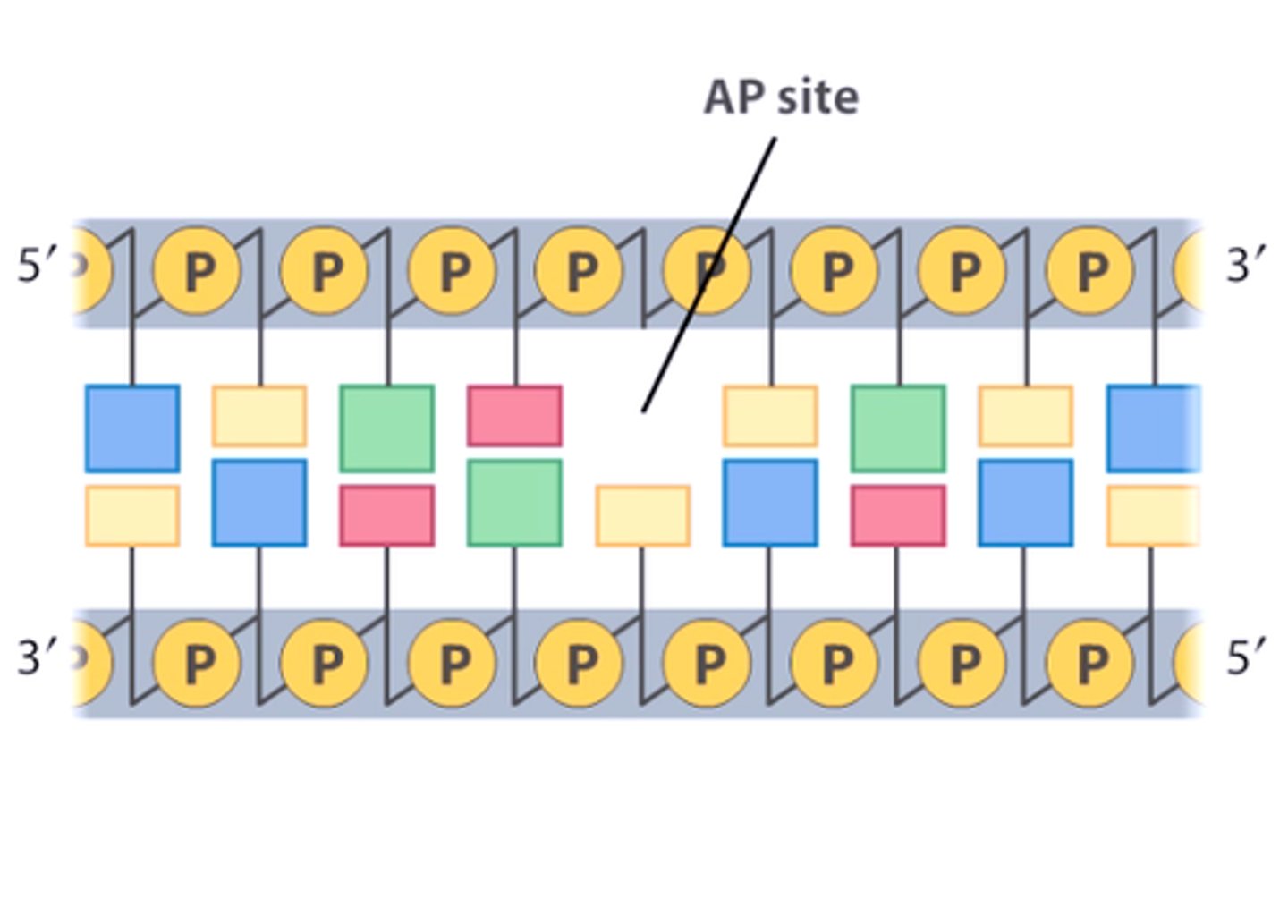
AP site (abasic site)
an apurinic or apyrimidinic site in the DNA resulting from removal of a base by DNA glycosylase
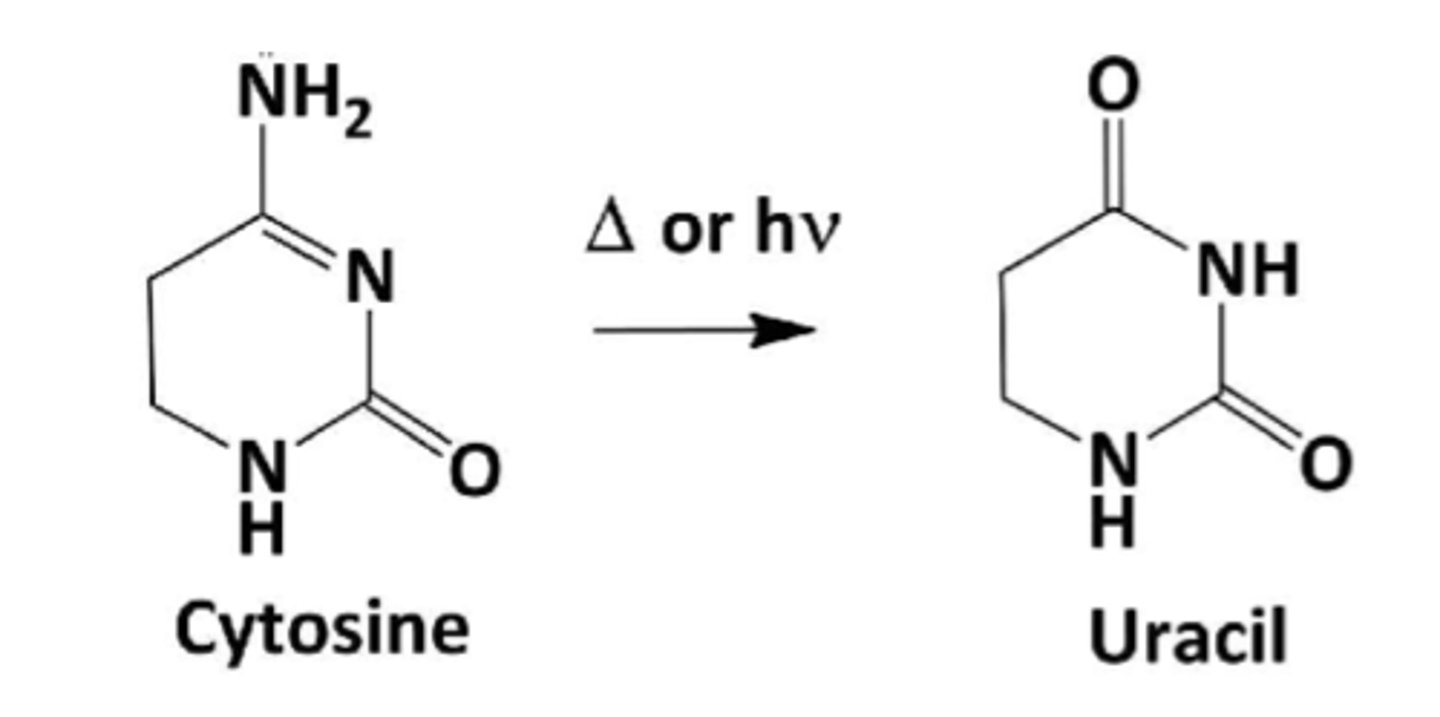
spontaneous deamination of cytosine
results in uracil in DNA
uracil DNA glycosylases
remove the uracil that results from spontaneous deamination of cytosine
-DNA specific (does not happen in RNA)
repairing AP sites in bacteria
the deoxyribose 5'-phosphate left behind is removed and replaced with a new nucleotide
-AP endonucleases cut the DNA strand containing the AP site
-DNA polymerase I removed and replaces the DNA
-DNA ligase seals the nicks
Nucleotide-excision repair (NER)
system that fixes large distortions in the DNA structure
Excinuclease
a multi subunit enzyme that hydrolyzes two phosphodiester bonds, one on either side of the distortion (on the same strand)
-vary by cell type:
--Prokaryotes = ABC exinuclease
--Eukaryotes = eukaryotic exinuclease
the ABC excinuclease
a bacterial enzyme with three components
-UvrA
-UVrB
-UvrC
a fragment is removed as a result
UvrA
a dimeric ATPase that scans DNA and binds to the lesion site
UvrB
binds UvrA and makes and incision on the 3'-side of the lesion after UvrA dissociates from the lesion
UvrC
binds UvrB and makes an incision on the 5'-side of the lesion after UvrB
Eukaryoitc exinuclease
similar mechanism to its bacterial counterpart
-Hydrolyzes the 3'-side and the 5'-side
Produces a larges fragment
Direct Repair
mechanisms that directly repair specific DNA problems
-photodimers (using NDA photolyases)
-Alkylated bases
-Deaminated bases
DNA breaks
in ssDNA and dsDNA arise from
-ionizing radiation
-Oxidative reactions
-A replication fork encountering and unrepaired DNA lesion
repaired by
-Homologous genetic recombination
-Error-prone translesion DNA synthesis (TLS)
SOS response
a cellular stress response to extensive DNA damage
Error-Prone TLS
part of the SOS response
-last ditch effort to save the cell
-use error-prone DNA polymerase
results of error-prone TLS
it is a desperation strategy!
-resulting mutation kill some cells and create deleterious mutations in others
-At least a few mutant daughter cells survive
-Resulting mutations contribute to evolution
DNA polymerase IV and V
activated during the SOS response
DNA polymerase V
SOS proteins include UvrA, UvrB + UmuC, and UmuD
Umu= unmutable
Eliminating umu genes
eliminates error-prone repair
genetic recombination
describe the exchange of genetic material between host cell nucleic acids that leads to progeny with traits that differ form their parent
-critical for genetic diversity, DNA repair, and evolution
3 classes of genetic recombination events
-Homologous genetic recombination
-Site-specific recombination
-DNA transposition
Homologous recombination (HR)
exchange of genetic material between two similar DNA sequences
-important for repairing DNA damage, mitosis, and antibody creation
-used to fix dsDNA breaks or stalled/collapsed replication forks
Recognition of DNA damage (HR)
cell machinery identified a double-strand break
-the replication fork collapses when it encounters DNA damage in a template strand
End Resection (HR)
-Enzymes remove some damaged DNA at the break site
-Creates single-stranded DNA (ssDNA) overhangs
Recruitment of Recombinase proteins (HR)
recombinase proteins RecA (bacteria) or Rad51 (eukaryotes) are recruited to the resected ssDNA regions
Formation of nucleoprotein filament (HR)
-forms after recombinase proteins bind to the ssDNA
-acts as a scaffold for subsequent recombination steps
Search for Homology (HR)
the nucleoprotein filament actively searches for a homologous region in an intact, double stranded DNA molecule
-The intact strand will serve as a repair template
Strand Invasion (HR)
the nucleoprotein filament invades the resected ssDNA strand into the homologous dsDNA molecule
-Results in the formation of a displacement loop of D-loop structure
DNA synthesis and repair (HR)
the invading strand is extended by DNA polymerase, synthesizing a complementary DNA strand using the intact, homologous DNA strand serves as a template
Heteroduplex Formation (HR)
the newly synthesized DNA strand and the complementary strand form the intact DNA molecule creates a heteroduplex region (Holliday structure) where the two DNA molecules are joined together
-This region contains mismatched base pairs
Resolution of Heteroduplex (HR)
the Holliday structure must be resolved to restore the original DNA molecule
Branch Migration (HR)
In some cases, the branch point formed during strand invasion and repair can migrate along the DNA molecules
-Helps ensure complete repair of damaged DNA
Ligation (HR)
sealing nicks or gaps in the phosphodiester backbone of DNA strands
Site-specific recombination
genetic exchanges only at a particular DNA sequence
-used in molecular biology for controlled genetic manipulation
DNA transposition
involves a short segment of DNA with the capacity to move from one location in a chromosome to another (transposons aka "jumping genes")
Meiosis
process by which diploid germ-line cells with two sets of chromosomes divide ad produce haploid gametes
-eukaryotic homologous recombination is critical for proper chromosome segregation during meiosis
meiosis I
homologous chromosomes are segregated into separate daughter cells at the end of
meiosis II
sister chromatids are separated during
prophase I
recombination or "crossing over" occurs during this phase
-occurs at "hot spots" on chromosomes
Chiasmata
points where two pairs of sister chromatids are linked due to crossing over
-this process aligns sister chromosomes for proper segregation and increases genetic diversity
functions of Homologous recombination
-it repairs several types of DNA damage
-It provides transient physical link between chromatids that promotes the orderly segregation of chromosomes at the first meiotic cell division
-It enhances genetic diversity in a population
Nonhomologous end joining (NHEJ)
alternative process for double-strand break repair
-No recombination
Broken chromosome ends are processed and ligated back together
-A mutagenic process (random nucleotide are introduces to try and repair the break)
NHEJ occurs when
recombinational DNA repair is not feasible because replication is not occurring and sister chromatids are not present
-AKA, there is no homolog to use as a template
NHEJ steps
-Recognition of DNA damage
-Binding of Ku Protein
-Recruitment of DNA-PKcs
-End Processing
-Ligation by DNA ligase IV
Recognition of DNA damage (NHEJ)
DSBs are detected by sensor proteins
Binding of Ku Protein (NHEJ)
the Ku70/Ku80 heterodimer, also know as the Ku protein, quickly binds to the broken DNA ends
-Ku protects DNA ends from degradation and recruits other repair factors
Recruitment of DNA-PKcs (NHEJ)
-Ku protein recruits the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to the DSB site
-Ku and DNA-PKcs form a DNA stabilizing complex
End Processing (NHEJ)
the broken DNA ends are not necessarily required to be complementary
-involves removal of damaged or mismatched nucleotides by nucleases and resynthesis by DNA polymerases
-not necessary if the ends are already compatible and have 3' hydroxyl and 5' phosphate termini
Ligation by DNA ligase IV (NHEJ)
DNA ligase IV and cofactor XRCC4 ligate/seal the DNA strands
primary repair mechanism for DSBs
nonhomologous end joining
NHEJ mutations
-occasionally leads to chromosomal rearrangements, especially when multiple DSBs are close together
-can lead to indel mutations at the repair site
--inducing a frameshift mutation
frameshift mutation
mutation that shifts the "reading" frame of the genetic message by inserting or deleting a nucleotide
-destroys gene/protein function!
Cre-lox recombination
is a site-specific DNA recombination system
-uses the Cre recombinase enzyme and specific DNA sequences called loxP sites
-Allows fore precise excision, inversion, or integration of DNA segments flanked by loxP sites in a controlled and reversible manner
LoxP Sites
short DNA sequences recognized by the Cre recombinase
-two inverted repeats separated by an asymmetric core
Cre recombinase
an enzyme that catalyzes the recombination between two loxP sites
Inversions (Cre-Lox Controls)
If the loxP sites are arranged in opposite orientations on the DNA molecules, Cre recombinase flips the DNA segment between the loxP sites, effectively reversing the sequence
Deletion (Cre-Lox Controls)
when two loxP sites are arranged in the same orientation on a DNA molecule, Cre recombinase excises the DNA segment flanked by the loxP sites
Integration/Translocation (Cre-Lox Controls)
Researchers can use two different DNA molecules, each containing one loxP sites, to introduce a specific DNA segment into a target genome
Inducible Expression (Cre-Lox Controls)
Cre recombinase activity is typically controlled by an inducible promoter or system, allowing researchers high levels of genetic control
Gene Knockout (SSR)
SSR is commonly used to eliminate genes and create gene knockout mice or other organisms
Conditional Gene regulation (SSR)
Researchers can use SSR to control the expression of genes in specific tissues
Cell Lineage tracing (SSR)
SSR can be used to trace the lineage of cells in developmental biology and caner research
Gene tagging and reporter systems (SSR)
SSR can be used to insert reporter genes or other sequences of interest at specific genomic loci
genome engineering (SSR)
Researchers can use SSR to engineer genomes by modifying specific DNA segments
Flp-FRT recombination
a site-specific DNA recombination system for controlled genetic manipulation
-precise excision, invert, or integrate DNA segments flanked by FRT sites in a controlled and reversible manner
FRT sites
short DNA sequences recognized by Flp recombinase enzyme
-contain two inverted repeats separated by a central region
-orientation and arrangement of the FRT sites determine the outcome of the recombination event
Flp recombinase
an enzyme the catalyzes recombination between two FRT sites
Transposons
transposable elements the "jump" from one place on a chromosome (the donor site) to another on the same or a different chromosome (the target site)
-Found in virtually all cells
Transposition
describes the movement of transposons
-Essentially random movement
-Tightly regulated and infrequent
-Can have deleterious consequences
Insertion sequences (simple transposons)
only contain the genes for proteins required for transposons
Complex transposons
contain 1+ genes in addition to those needed for transposition
-Example: antibiotic-resistance genes
insertion of a transposon
short repeated sequences at each end of the transposon serve as binding sites for the transposase enzyme
-they become duplicated following transposon insertion
direct transposition
the transposon is excised via cuts on each side and moves to a new location
-a staggered cute is made at the target site and the transposon is inserted into the break
-DNA replication fills in the gaps to duplicate the target-site sequence
-"cut and paste"
Replicative transposition
the entire transposon is replicated, leaving a copy behind at the donor location
-"copy and paste"
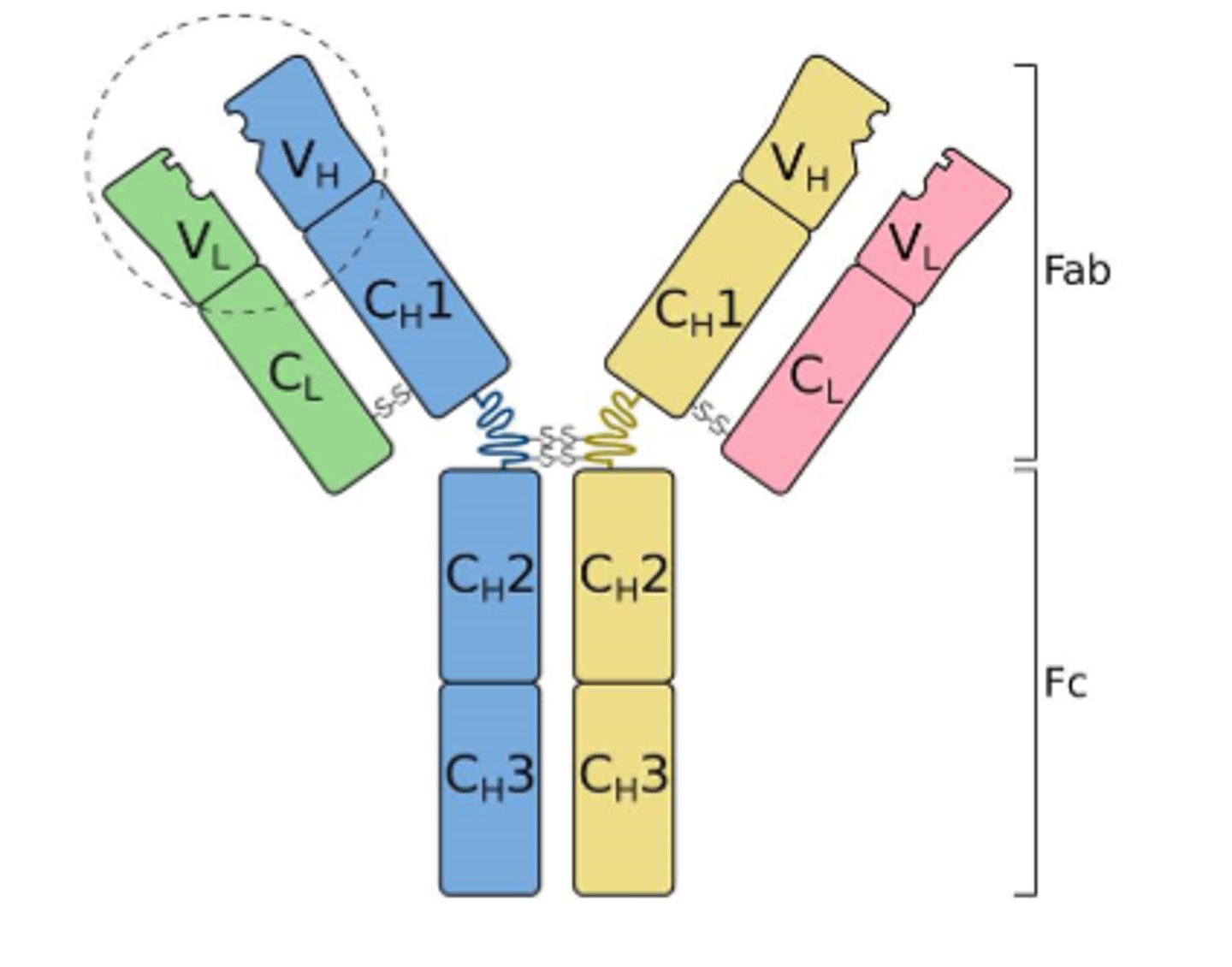
Immunoglobulins (Igs) consist of
-Two heavy (H) polypeptide chains
-Two light (L) polypeptide chains
--can be from kappa or lambda families
-Each chain has variable (V) and constant (C) regions
V(D)J recombination
crucial genetic process in vertebrates that occurs primarily in developing lymphocytes of the immune system
-Responsible for generating the diverse repertoire of antibodies and T-cell receptors (TCRs) that enable the immune system to recognize a wide range of pathogens and foreign substances
Stem cells mature into
B lymphocytes in the bone marrow
-each B lymphocyte produces one kind of antibody
V(D)J provides
specificity and diversity for the adaptive immune response
-allows the immune system to adapt to constant pathogen evolution
why is (V(D)J highly regulated?
because it is a fundamental process for a properly functioning immune system