Lecture 18 and 19 The Mechanism of Translation
1/29
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
30 Terms
14.1 Overview of translation
mRNA+charged-tRNA+ribosome
Ribosome subunits are assembled in the nucleolus.
tRNAs are “charged” with their appropriate amino acids.
All the players in protein synthesis join together in the cytoplasm.
Structure of ribosomes
Ribosomes consist of two subunits, large and small, composed of rRNAs and many ribosomal proteins
Functional motifs on the ribosome
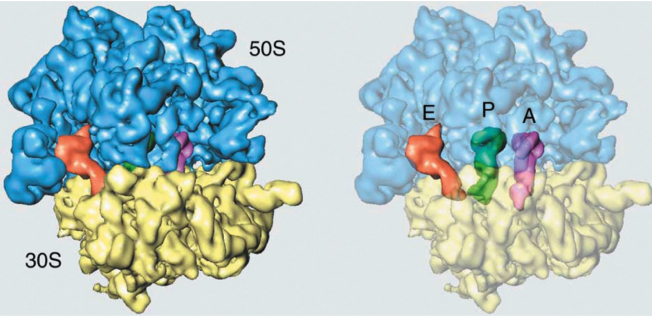
Three tRNA binding sites on the ribosome that bridge the larger and small subunits
(A) Aminoacyl
(P) peptidyl
(E) exit
peptidyl transferase center is in the large subunit
Decoding the mRNA occurs on the small subunit
Svedberg unit
S increases with particle mass and density (related to shape).
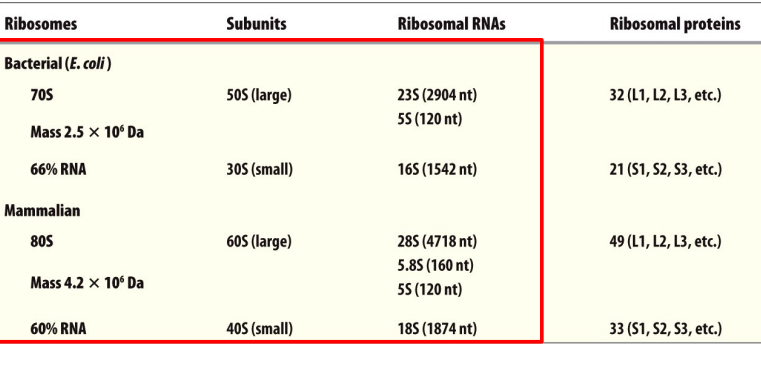
rRNAs and ribosomal proteins
So 23, 5 and 16 is prokaryotic
28, 5.8 and 5, 18 are eukaryotic
5 is both
The nucleolus
non-membrane-bound subcompartment of the nucleus.
Eukaryotic large and small ribosomal subunits are assembled within the nucleolus.
Site of 45S pre-rRNA transcription by RNA polymerase I.
overall fidelity of translation is dependent on the accuracy of two processes
Codon-anticodon recognition during translation
Aminoacyl-tRNA synthesis
Aminoacyl-tRNA charging
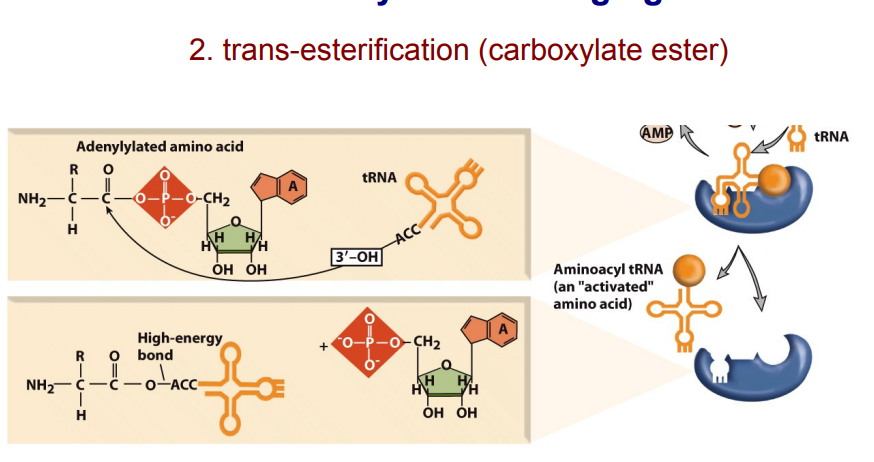
Aminoacyl-tRNA synthetases attach an amino acid to a tRNA by two enzymatic steps:
The amino acid reacts with ATP to become adenylylated and pyrophosphate is released.
AMP is released and the amino acid is transferred to the 3′ end of the tRNA
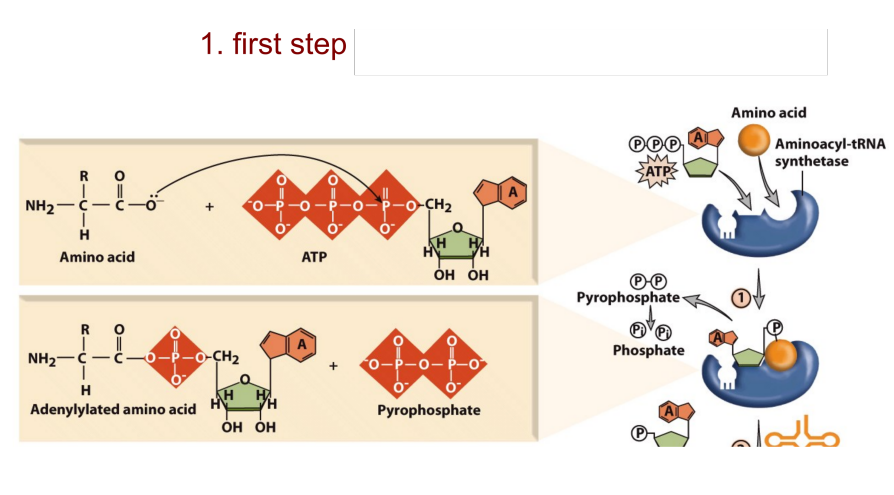
Aminoacyl-tRNA charging 1st adn 2nd steps
tRNA is the translator
High fidelity of aminoacyl-tRNA synthetases
•Initial high fidelity selection
Can easily match AA with corresponding anticodon
Proofreading
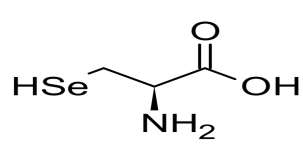
The 21st amino acid
This charged Sec-tRNASec recognizes and translate UGA codon
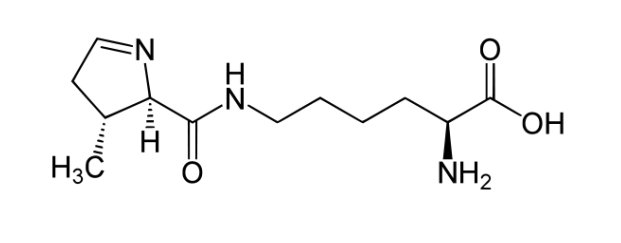
The 22st amino acid
The charged tRNA can recognize and translate UAG stop codon
Overview of translation
• Initiation is the most complex and tightly controlled.
• Focus on eukaryotic protein synthesis.
Initiation is subdivided into four steps
Ternary complex formation and loading onto the 40S subunit.
mRNA Loading.
Scanning and binding start codon.
Joining of the 40S and 60S subunits to form 80S ribosomes.
Ternary complex formation and loading onto the 40S ribosomal subunit
The ternary complex is composed of:
Eukaryotic initiation factor 2 (eIF2)
GTP
The amino acid-charged initiator tRNA (Met-tRNA)
The ternary complex binds the 40S subunit, plus other initiation factors, including eIF4G/E, to form a 43S complex.
mRNA loading onto the 40S subunit
eIF4G and eIF4E
initiation with RNA helicases unwind with secondary and tertiary structures
poly(A)- binding protein (PABP)
bound to 3′-poly(A) tail.
The closed-loop model of translation initiation
The 5′-cap and 3′-poly(A) tail of the mRNA join to form a closed loop with eIF4G serving as the bridge between them.
Some cellular RNAs are translated by a 5′-cap-independent mechanism in which ribosomes are directly recruited by an internal ribosome entry site (IRES)
Scanning and binding start codon
Once the mRNA is loaded, the 43S complex scans along the message from 5′→3′ looking for the AUG start codon.
ATP-dependent mechanism.
AUG is embedded in a Kozak consensus sequence (Shine-Dalgarno sequence in E. coli).
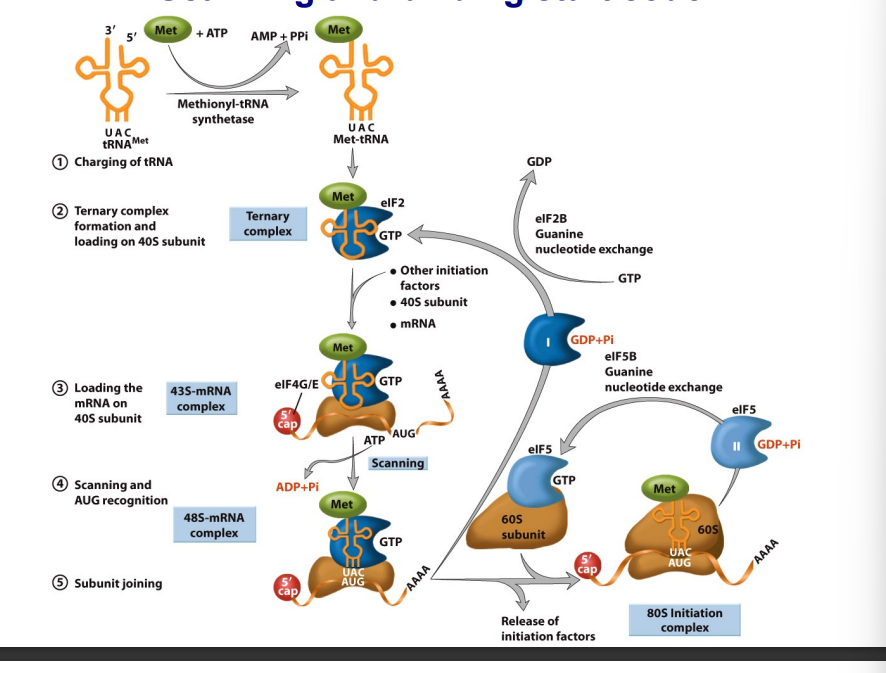
Joining of the 40S and 60S ribosomal subunits to form 80S ribosomes
eIF2-GTP is hydrolyzed into eIF2-GDP and released.
eIF2-GDP is converted to eIF2-GTP through a nucleotide exchange reaction mediated by eIF2B.
The 60S subunit joins with the 40S subunit to form the 80S ribosome initiation complex, in a process that requires a second GTP hydrolysis step.
eIF5-GDP is converted to eIF5-GTP through a nucleotide exchange reaction mediated by eIF5B

14.5 Elongation
needs 2 GTP(total) for initial binding and translacation
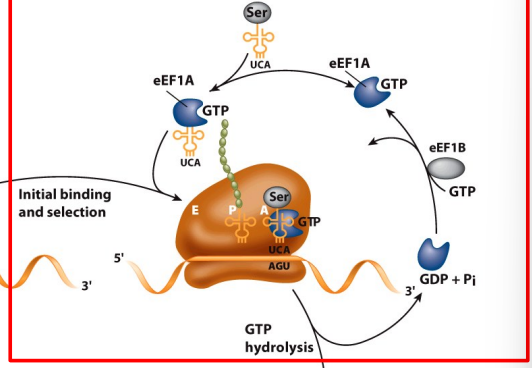
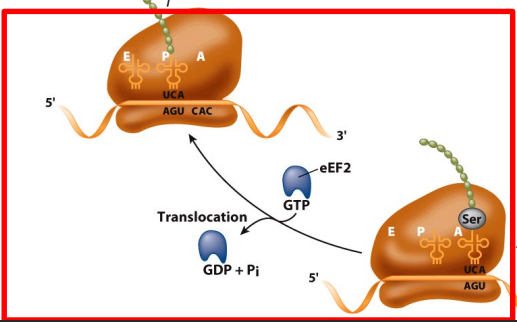
Peptide bond formation and translocation
Peptidyl transferase activity transfers a growing polypeptide chain from peptidyl-tRNA in the P site to an amino acid esterified with another tRNA in the A site.
After the tRNAs and mRNA are translocated and the next codon is moved to the A site, the process is repeated.
Mediated by eEF2; requires GTP hydrolysis.
translocation need 1 GTP faci
A = where oi
Biochemical evidence that 23S rRNA is a ribozyme
“Fragment reaction” used by Harry Noller and colleagues to shown that purified bacterial 23S rRNA has “peptidyl transferase activity” in vitro.
Structural evidence that ribosome is a ribozyme
rRNA forms the catalytic center, decoding site, A, P and E sites, and the intersubunit interface.
Ribosomal proteins are abundant on the exterior of the ribosome.
The peptidyl transferase center is located in domain V of the 23S rRNA.
Cotranslation translocation
Cotranslation translocation pathway from the ribosome to the endoplasmic reticulum (ER) lumen.
Signal recognition particle (SRP) binds to a ribosome translating a polypeptide that bears a signal sequence for targeting to the ER.
The SRP and SRP receptor use a cycle of recruitment and hydrolysis of GTP to control delivery of the ribosome-mRNA complex to ER
14.6 Termination of translation
The stop codons are recognized by release factor eRF1 in association with eRF3.
The completed polypeptide is cleaved from the peptidyl-tRNA
GTP hydrolysis may trigger the release of eRF1 and eRF3.
Phosphorylation of eIF2a blocks ternary complex formation (EXAM)
Hypoxia, viral infection, amino acid starvation, heat shock, etc. trigger the phosphorylation of the a- subunit of eIF2.
Phosphorylation of eIF2a inhibits GDP-GTP exchange. stop hydrolysis so no energy is evailable to continue translation
eIF2a phosphorylation leads to inhibition of translation by blocking ternary complex formation.
Selective translation of a subset of mRNAs continues, which allows cells to adapt to stress conditions.
eIF2 phosphorylation is mediated by four distinct protein kinases
Protein Kinase RNA (PKR)
senser to innitiate phosphorylation of elf2a
dsRNA recognise PKR(senser and receptor) and bind to GDP of elf2-GDP to creating an isolated state
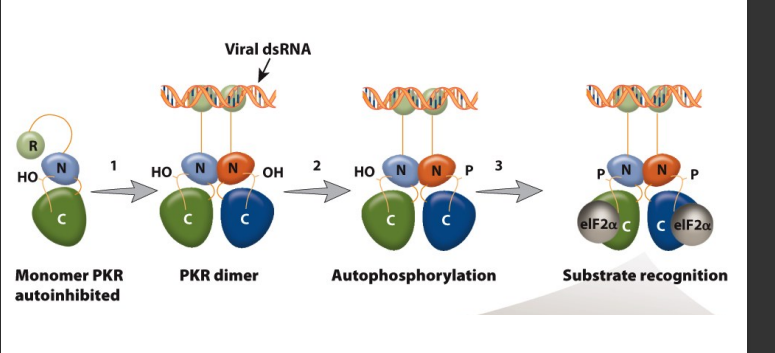
Model of the protein kinase RNA (PKR) activation pathway
Viral double-stranded RNA binds to the RNA binding domains of PKR.
PKR catalytic-domain dimerization.
Autophosphorylation of PKR.
Specific recognition of eIF2a.
Phosphorylation of eIF2a. to stop translation from continueing when a viral infectant is present
pathway on exam