Biom1051 - Module 2 (Principles of Biochemistry)
1/98
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
99 Terms
Chemical insights - info cells body functions
WATER + simple solutes (metal ions: sodium, nerve impulses, heart activity, metabolic process)
AMINO ACIDS - proteins
NUCLEOTIDES - DNA/RNA
VITAMINS carry out them reaction otherwise not possible
Diff cells - own chem for function
- Muscle: contain ATP + used to help do mechanical work
- Epithelial: contain keratin (imp protein for skin)
- Red blood cells (erythrocytes) - make + store haemoglobin (oxygen carrier)
- Neurons: contain neurotransmitters (e.g. adrenalin, serotonin)
LARGE biological molecules
all organisms rely on 4 types: Carbohydrates, lipids, proteins & nucleic acids.
- built from small mol joined together
- mol structure/function inseparable
- large bc # of atoms, naked eye/reg microscope can't see
Macromolecules
Polymers built from monomers
- POLYMERS are a long molecule made up of small building-block molecules called MONOMERS (e.g. metabolites)
3/4 classes of organic mol are polymers:
1) carbohydrates 2) proteins 3) nucleic acids
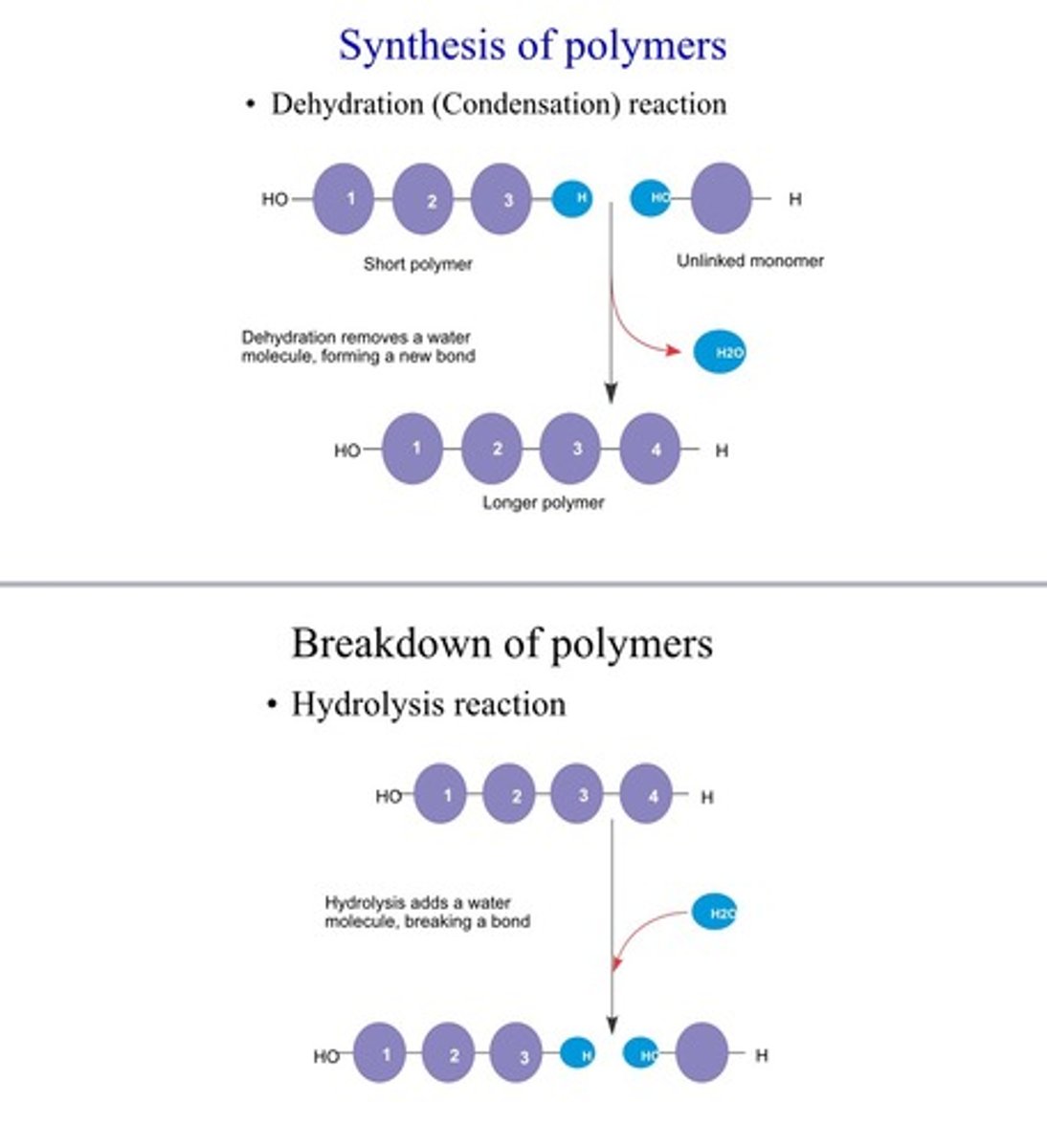
Polymer - Synthesis/breakdown
Synthesis: Condensation/dehydration reaction - two monomers bond together, losing H2O mol.
Breakdown: hydrolysis reaction, adding H2O mol, breaks bond between monomers - disassembles polymers into monomers
- Enyzme's (protein) macromolecules speed up both reactions
Carbohydrates
Serve as fuel/building material; include sugars + polymers of sugars.
Monosaccharides - simplest (single sugar)
e.g glucose (most common)
Carb. macromol. = polysaccharides - sugar polymers
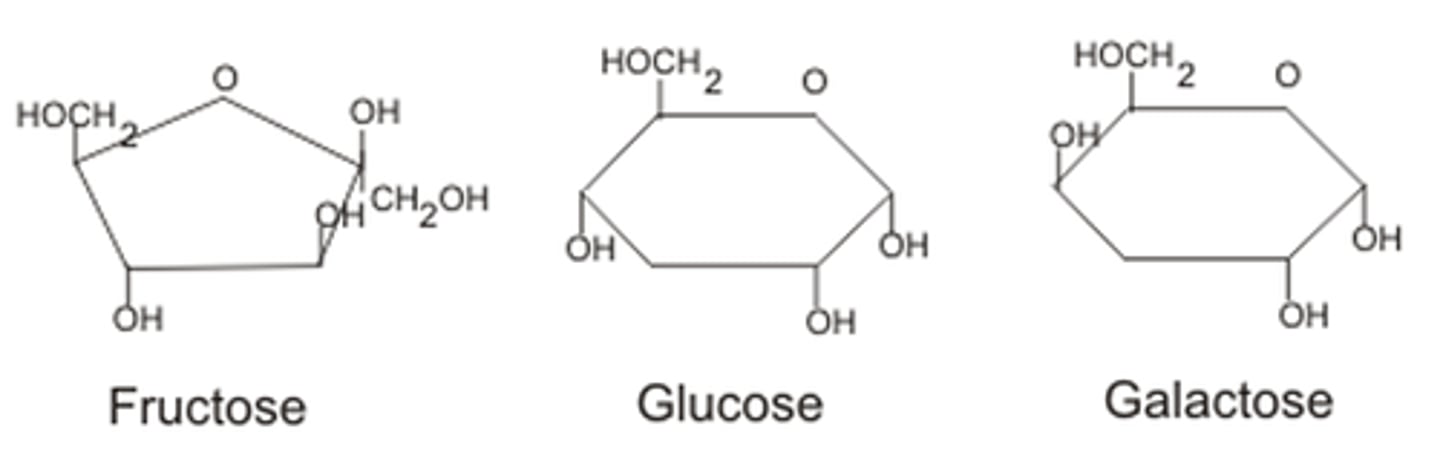
Monosaccharides
E.g. glucose, fructose, galactose
Structure varies - ketones or aldehydes; vary in length (3-6 "C") + all possess carbonyl grp; spatial arrangement (flip OH = glucose vs galactose)
- Can be LINEAR (often drawn as) or CYCLIC (in aq sol many sugar - ring) (Can cyclise)
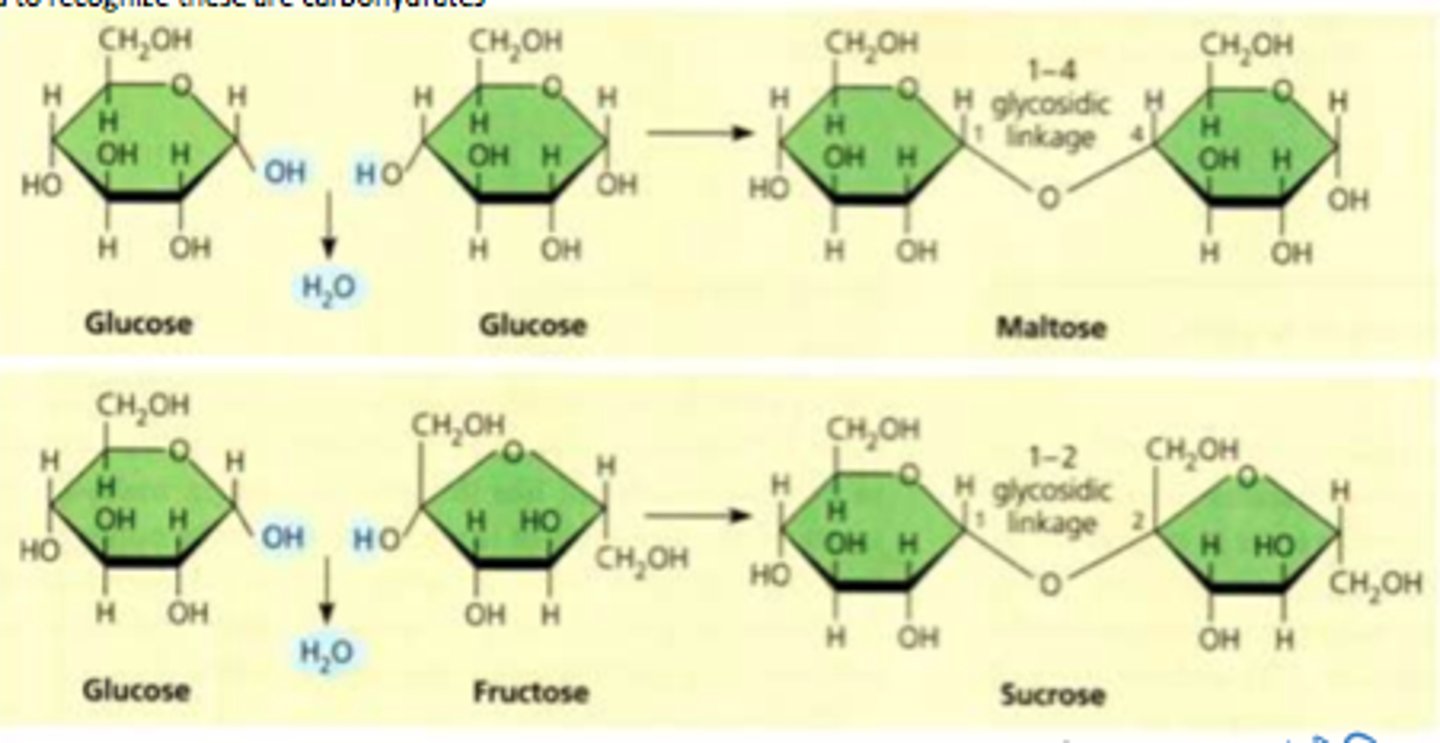
Disaccharides
Formed via dehydration joins 2 mono-.
- COVALENT BOND = GLYCOSIDIC LINKAGE
Polysaccharides
Sugar polymers - storage/structural roles determined by sugar mono + glycosidic linkage positions
Storage - Polysaccharides (plants)
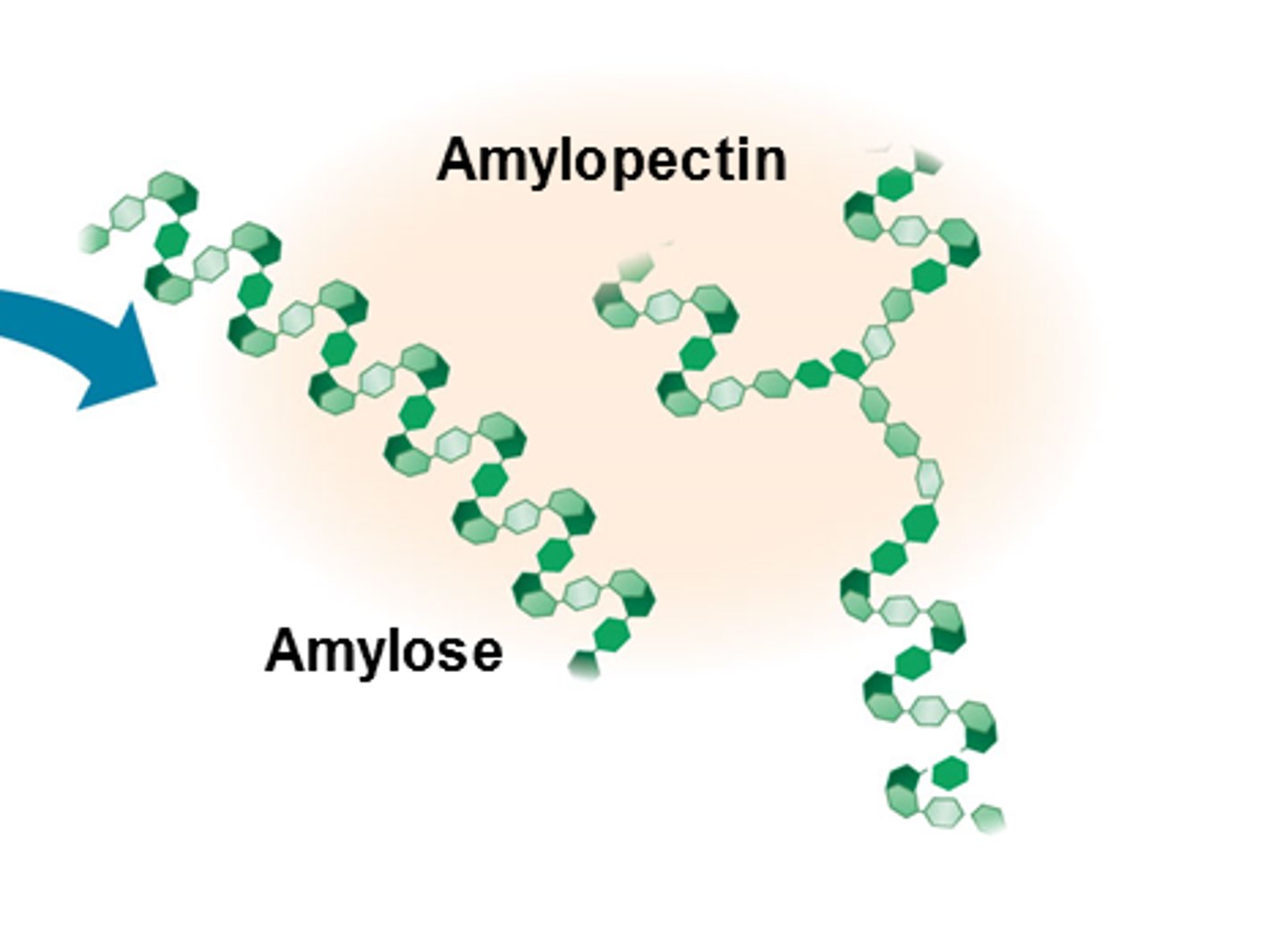
E.g. Starch - storage poly of plants - entirely glucose mono
- surplus starch stored as granules within chloroplasts + other plastids
2 forms: amylose (unbranched), amylopectin (branched)
Cellulose
Important structural component of plant cell walls.
- most abundant organic polymer
- Cellulose conversion -> biofuels e.g. ethanol possible renewable fuel
- derived from glucose units: beta(1->4) glycosidic bonds - makes it indigestible for mammals - no enzyme + beta means alternating.
(starch's is alpha(1->4) glycosidic bonds)
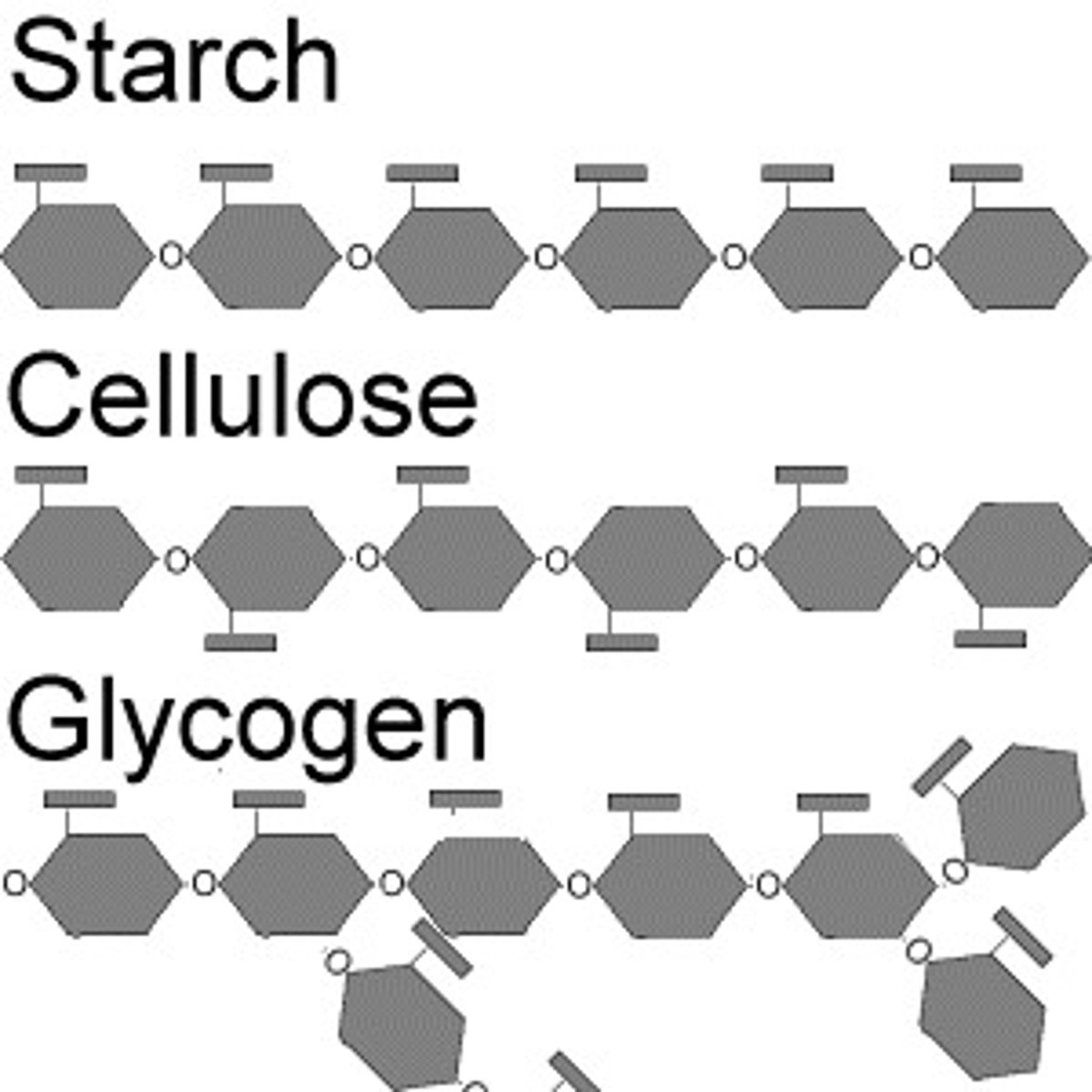
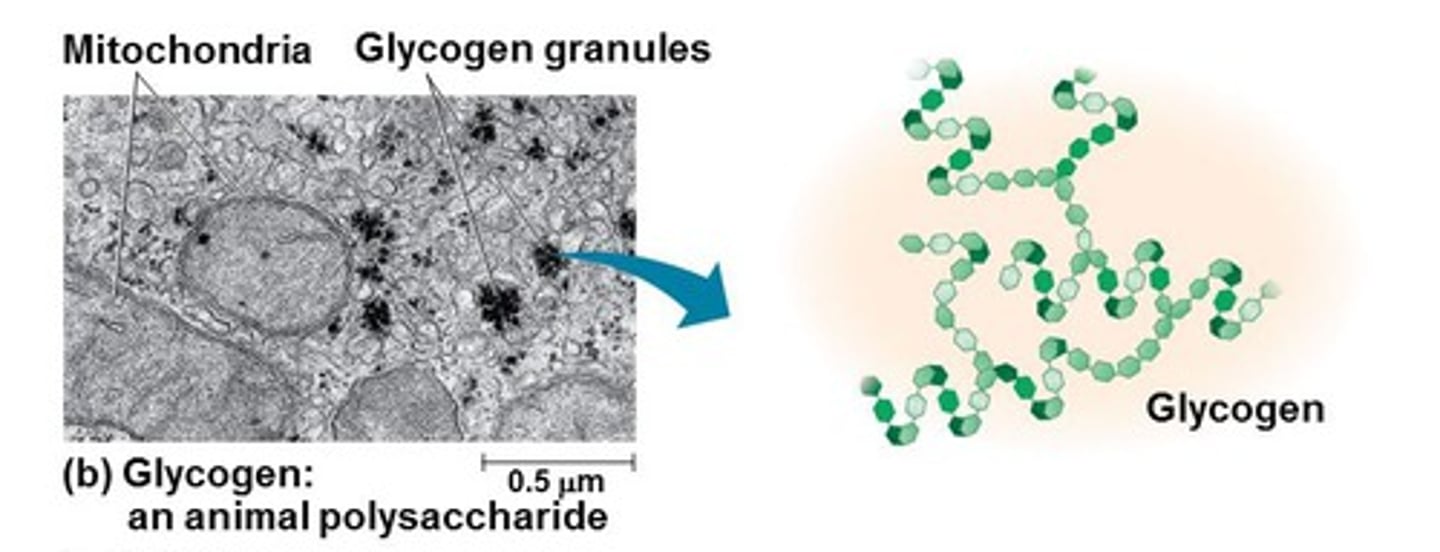
Storage - Polysaccharides (animals)
E.g. Glycogen - storage polysaccharide in animals
- Liver + muscle checks mainly store glycogen (humans +other vertebrates)
Proteins
Complex structures + many diff functions
- 50% of dry mass of most cekks
FUNCTIONS:
- structural (collagen)
- transport (hemoglobin - O2)
- hormonal regulation (insulin - sugar control)
- Enzymes - accelerate chem reactions
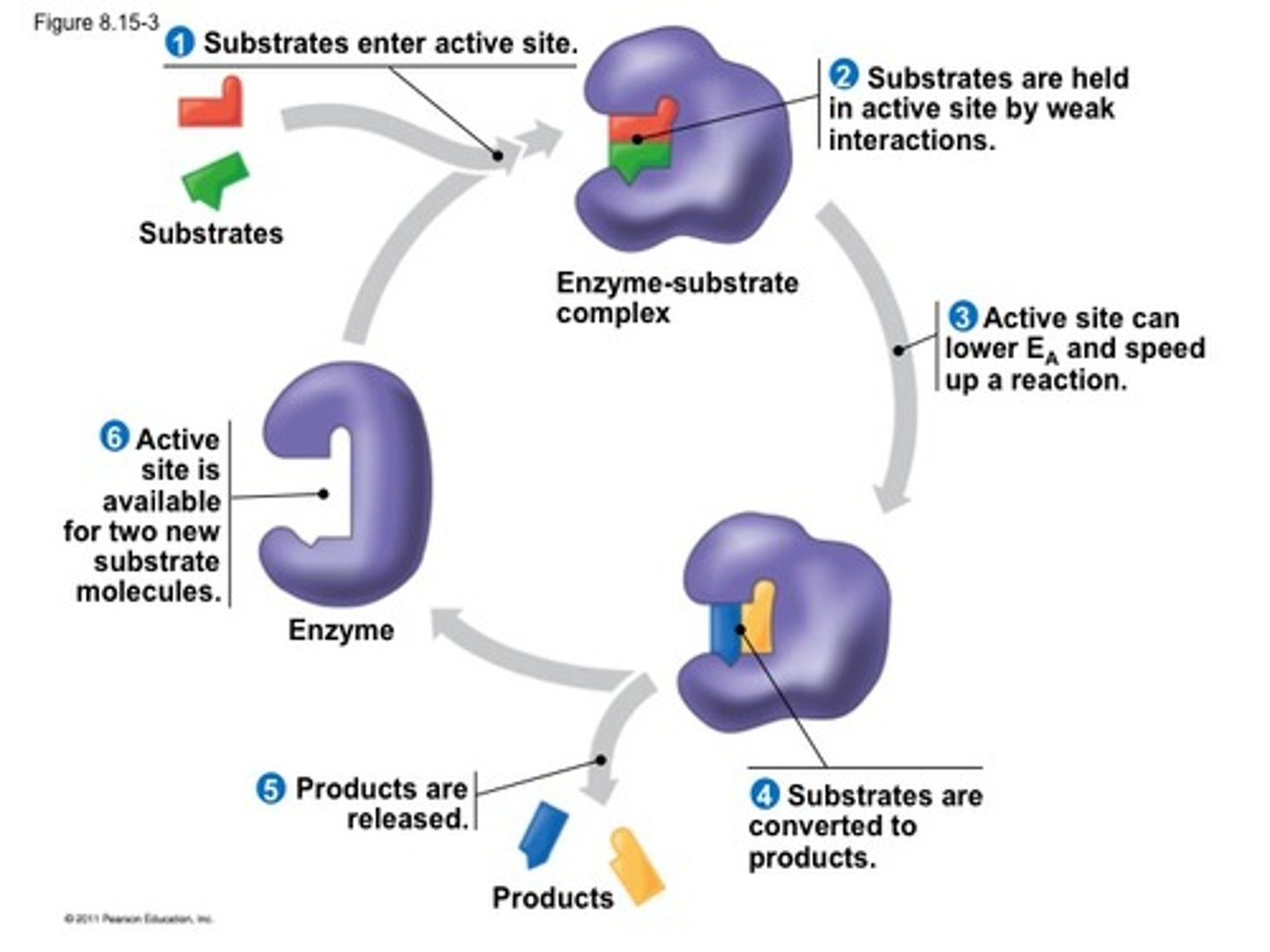
Enzymes
involved in range of biological activities
e.g. food digestion, making DNA + other proteins (carbs), protein processing (proinsulin or pro collagen activation)
are biological catalysts that speed up them reactions
- can repeatedly preform functions - carry out essential life processes
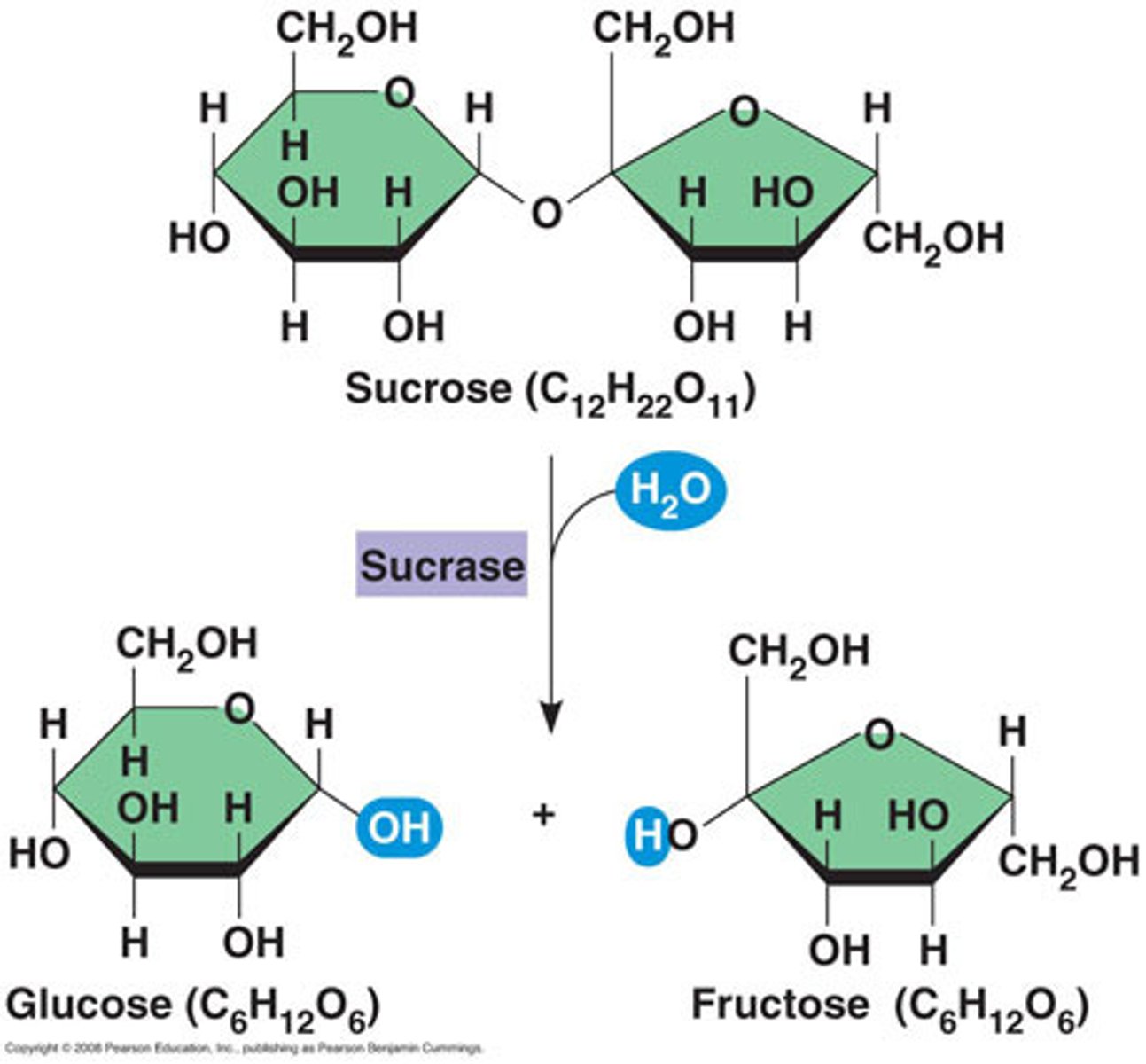
Sucrase
breaks down sucrose (hydrolysis) into glucose and fructose
- Sucrase (example of enzyme-catalysed reaction)
turnover number
Kcat (s^-1) indicates the number of substrate molecules an enzyme can convert into product per second (indicates how fast enzymes)
Larger # better
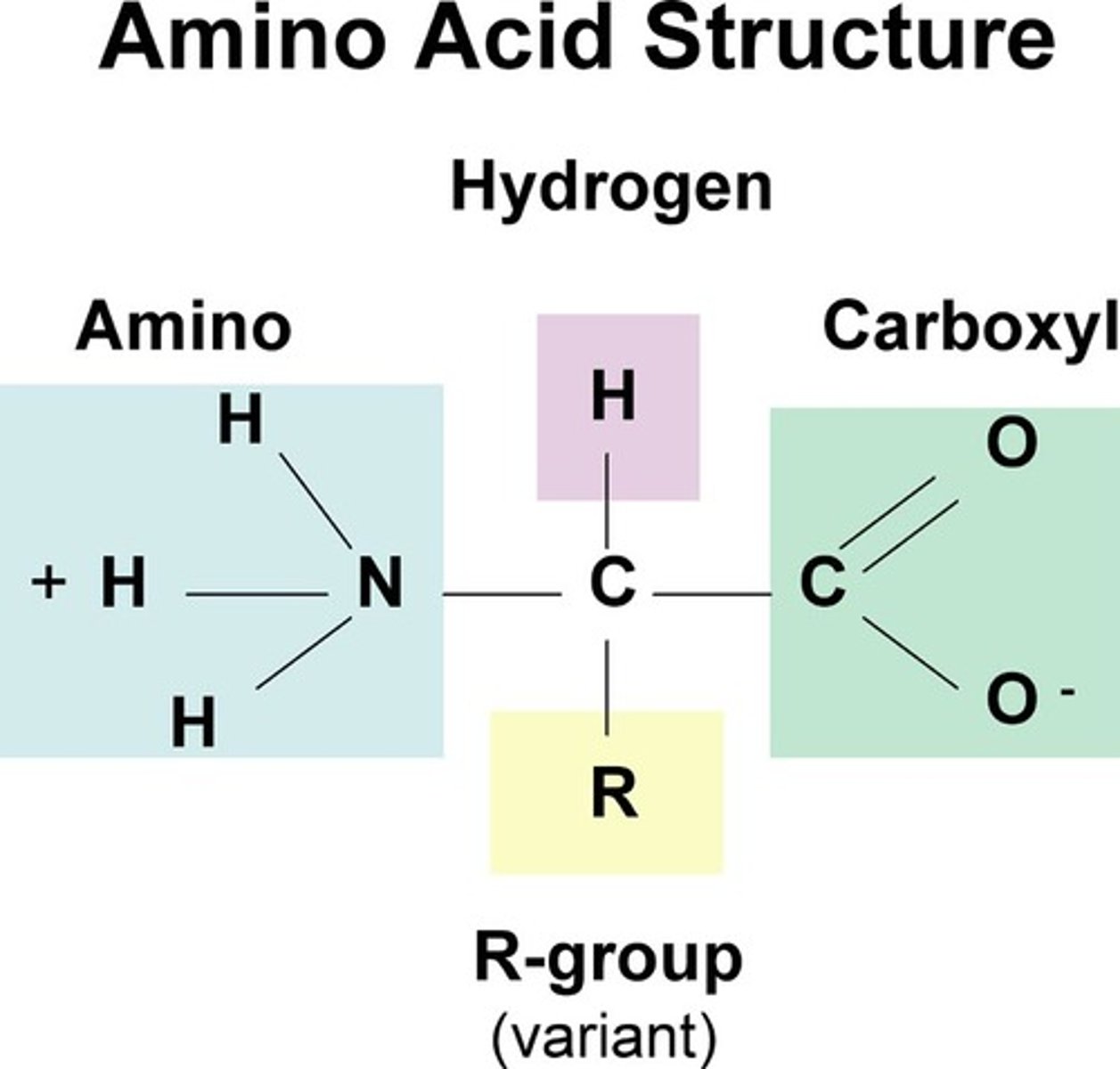
Proteins
made from amino acids
- Amino acids: organic mol w carboxyl + amino grps; differ in properties by diff side chains (R grps)
-Polypeptides: polymers built from set of 20 amino acids
Proteins consist of 1 or more polypeptides
Protein pic for exam
...............COO^-
.................... | (pKa 4)
H3N+ -- C --H
(pKa 9)... |
.................. R
Amino acid - charges
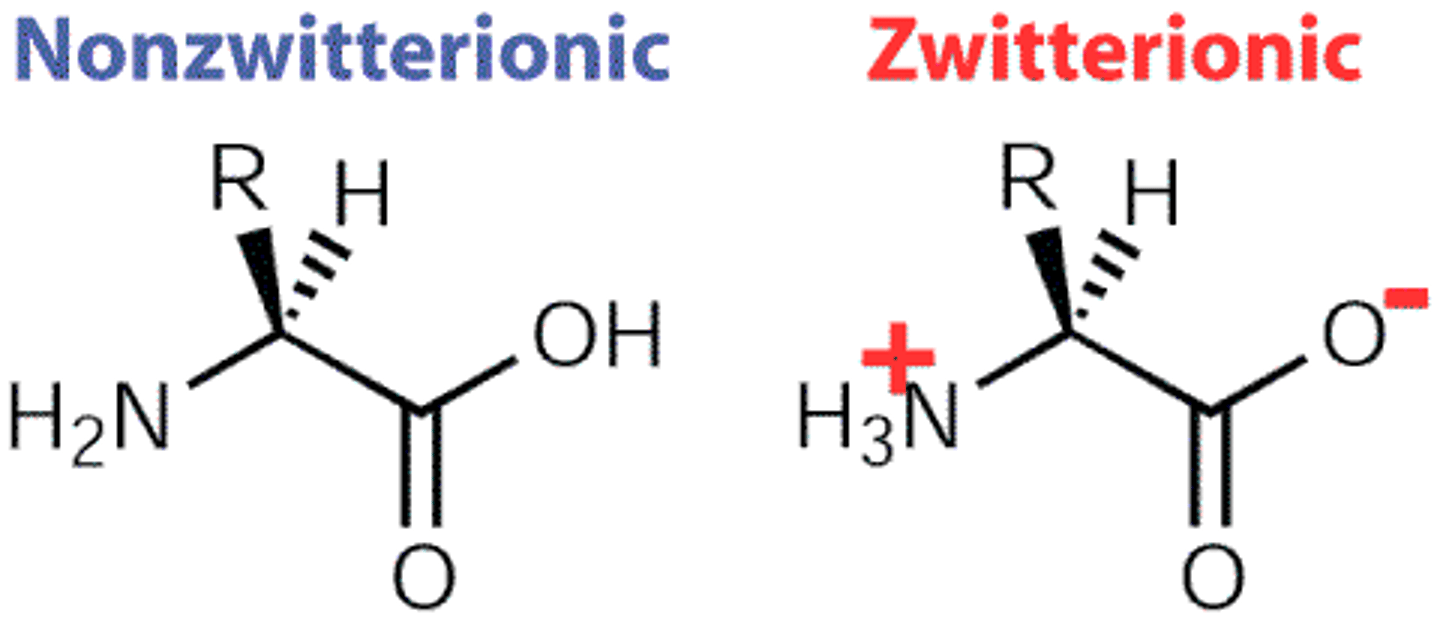
In sol, ionisation state of amino and carboxyl grps change w pH.
- Zwitter ion has both +/-
Low pH--> positive on amino group
medium --> positive on amino group negative on carboxylic acid
High ph --> negative on carboxylic acid
α-amino acid
A molecule containing an amino group and a carboxylic acid group that are separated by one carbon?
- most are chiral ( all 4 substituents are different)
- central α-C has 4 substituents + tetrahedral
- each amino acid - unique fourth substituent (R) e.g. alanine R = CH3
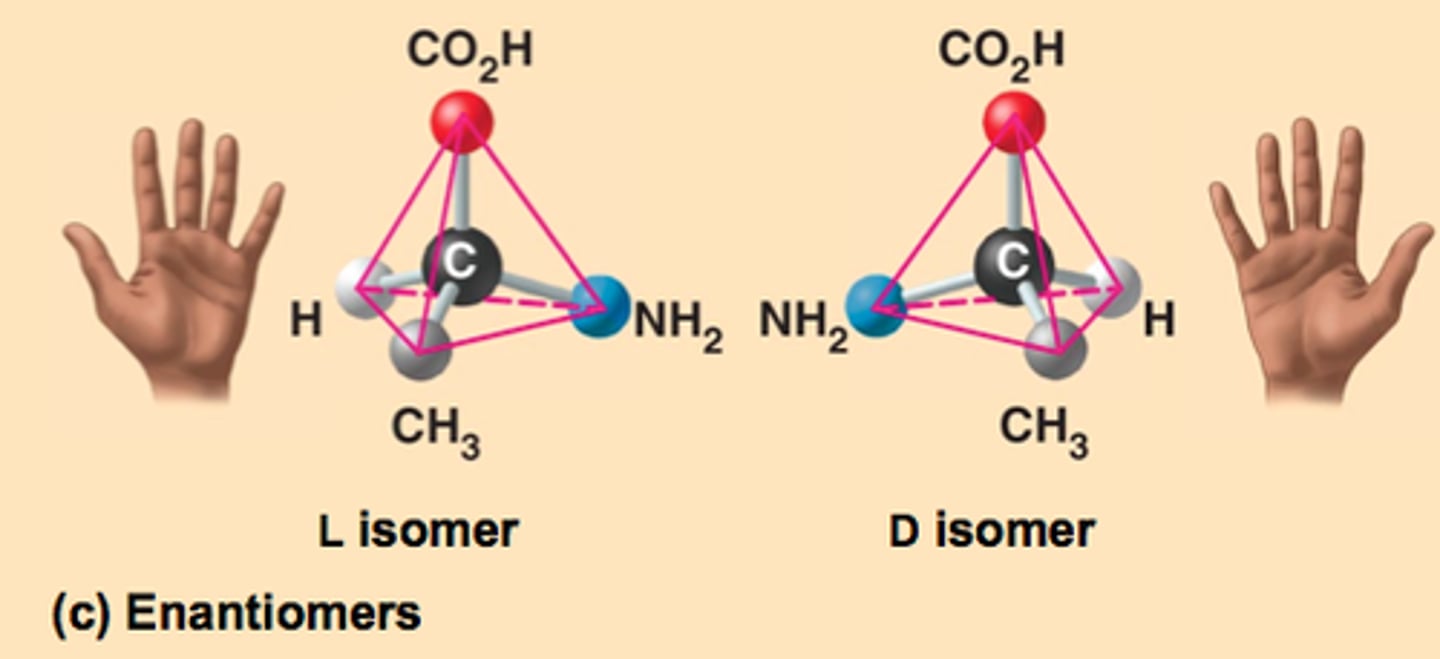
Enantiomers
isomers that are mirror images of each other, bc of dif spatial arrangement.
- cannot be superimposed on eachother
L and D isomers for left and right
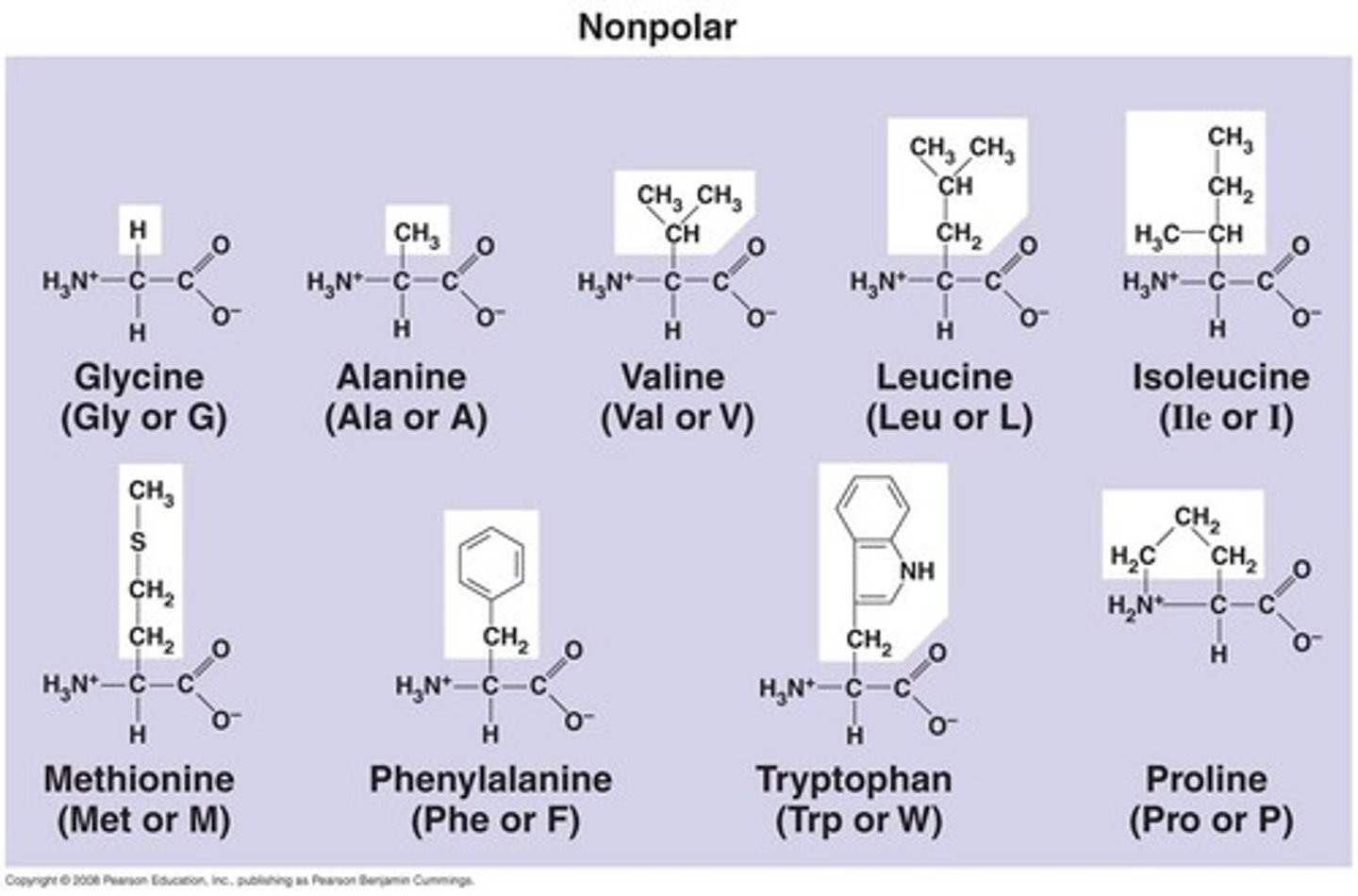
Non-polar amino acid side chains
Non-polar molecules side-chains contain 'C' and 'H'.
- hydrophobic
Uncharged polar side chains
Polar side-chains contain 'C' and 'H' and 'O' and/or 'N'
- hydrophilic
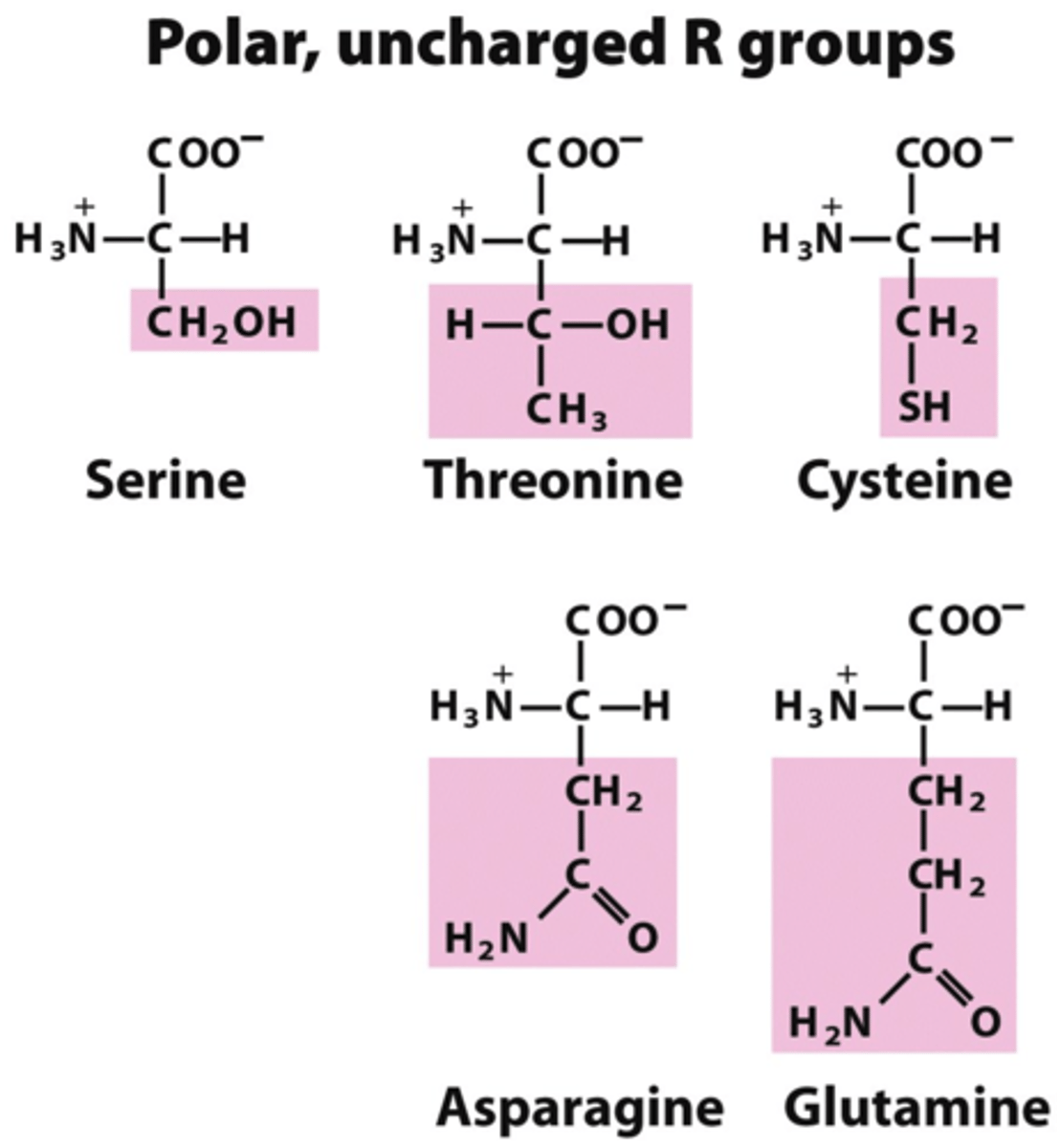
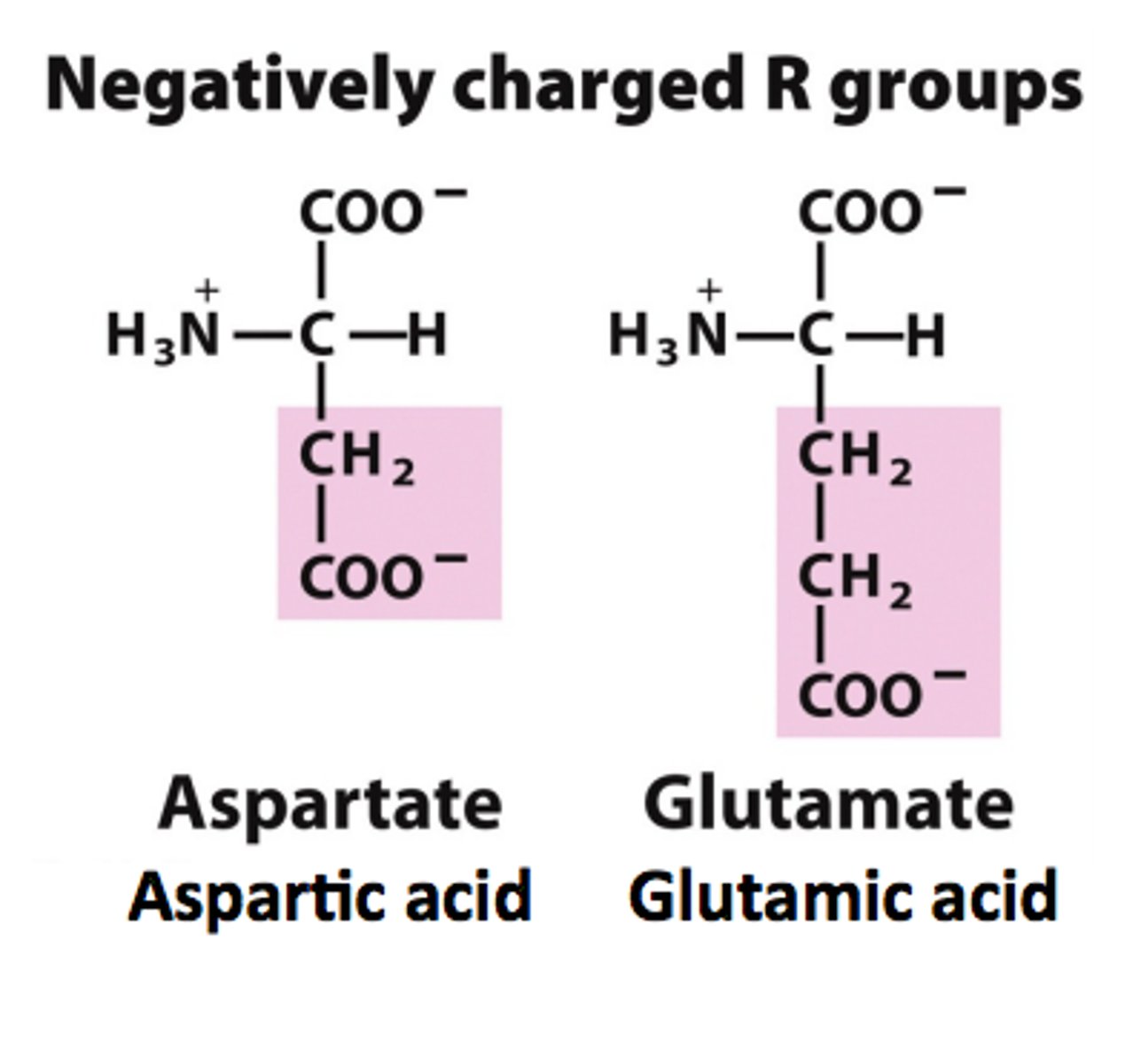
(-)ly charged side chains
Side chains have pKa values around 4.
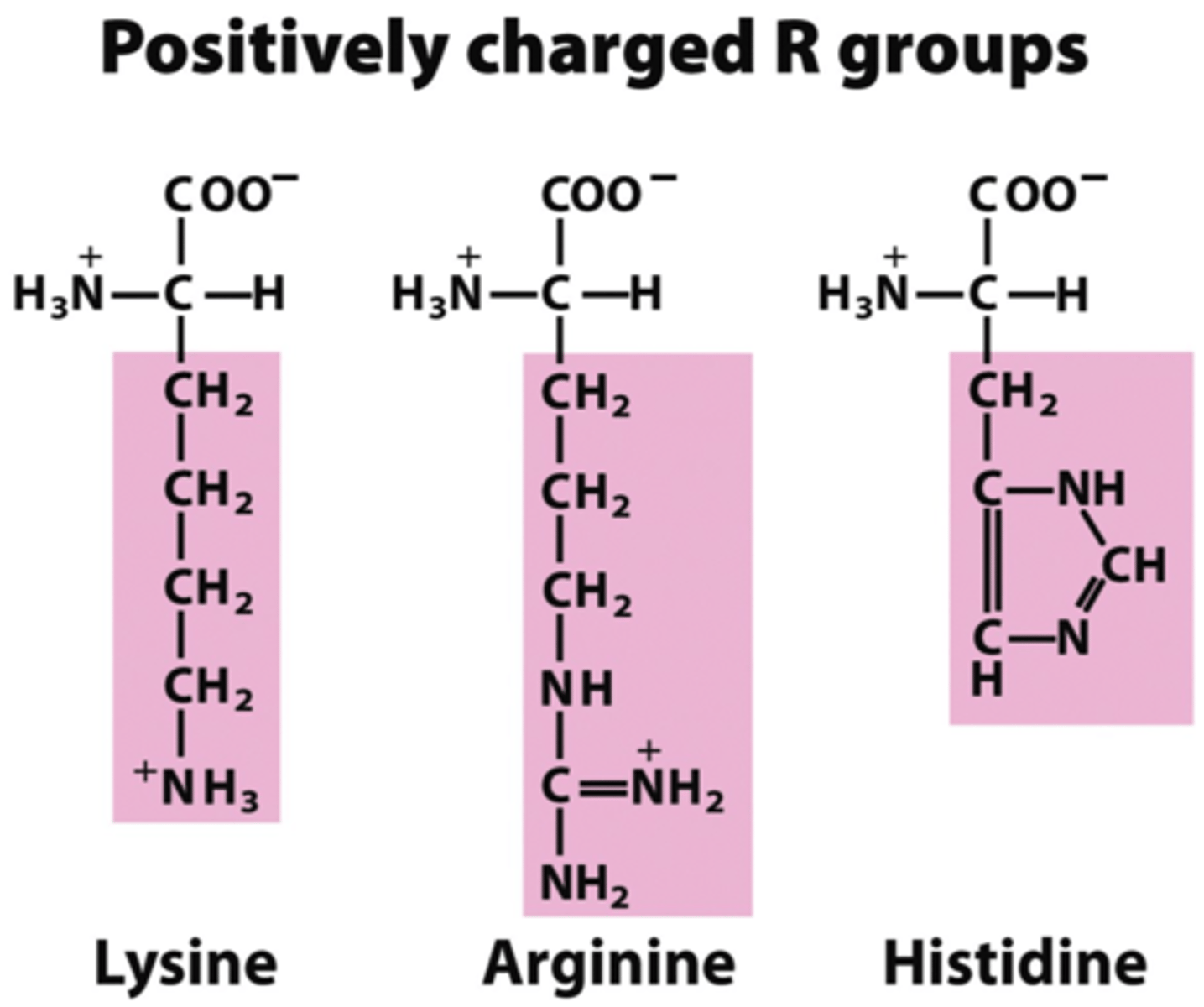
(+)ly charged side-chains
from left to right:
pKa 10, 12, 6.5
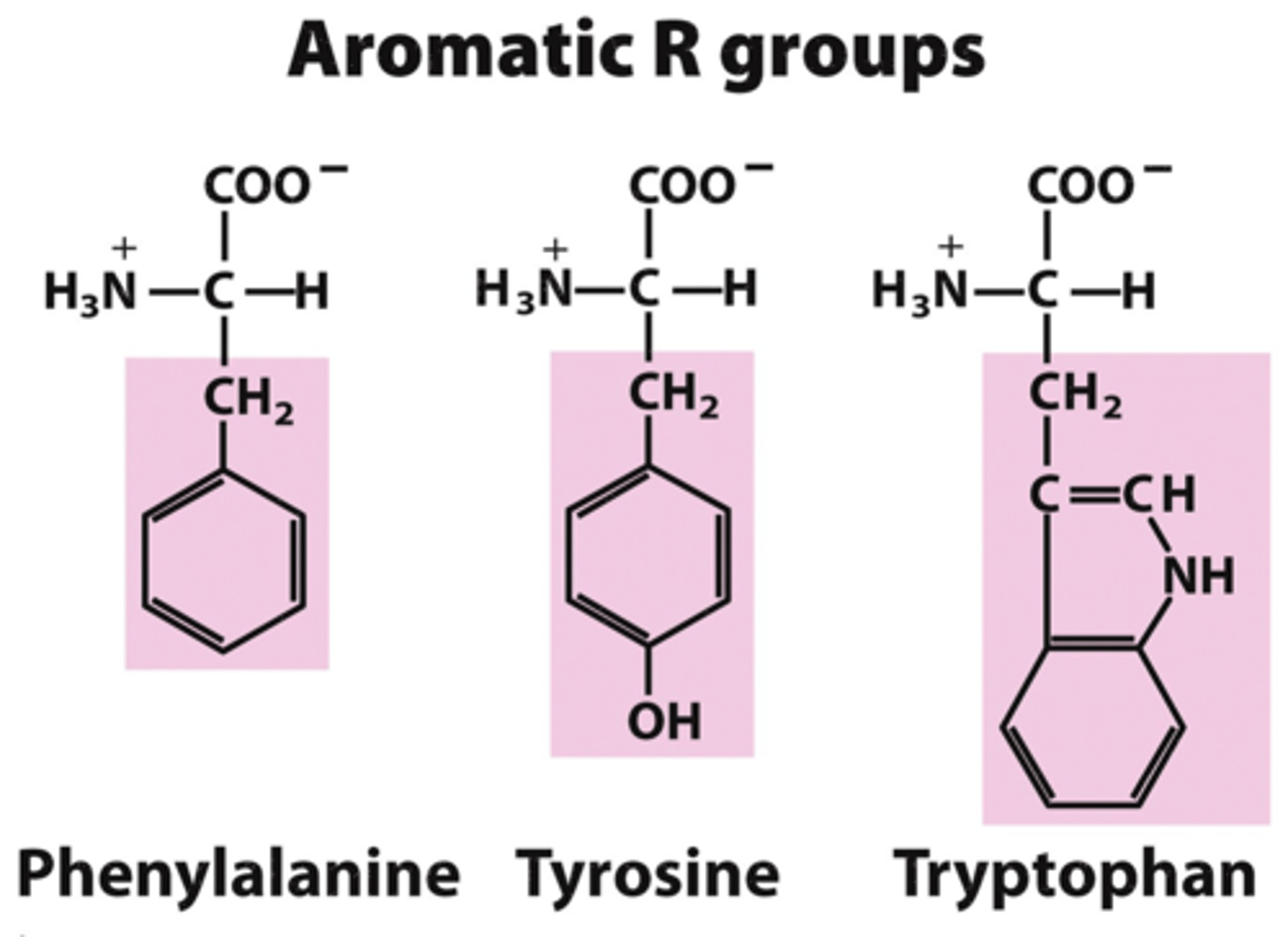
Aromatic side-chains
phenylalanine, tyrosine, tryptophan
->phenylalanine is also hydrophobic
- all have aromatic rings
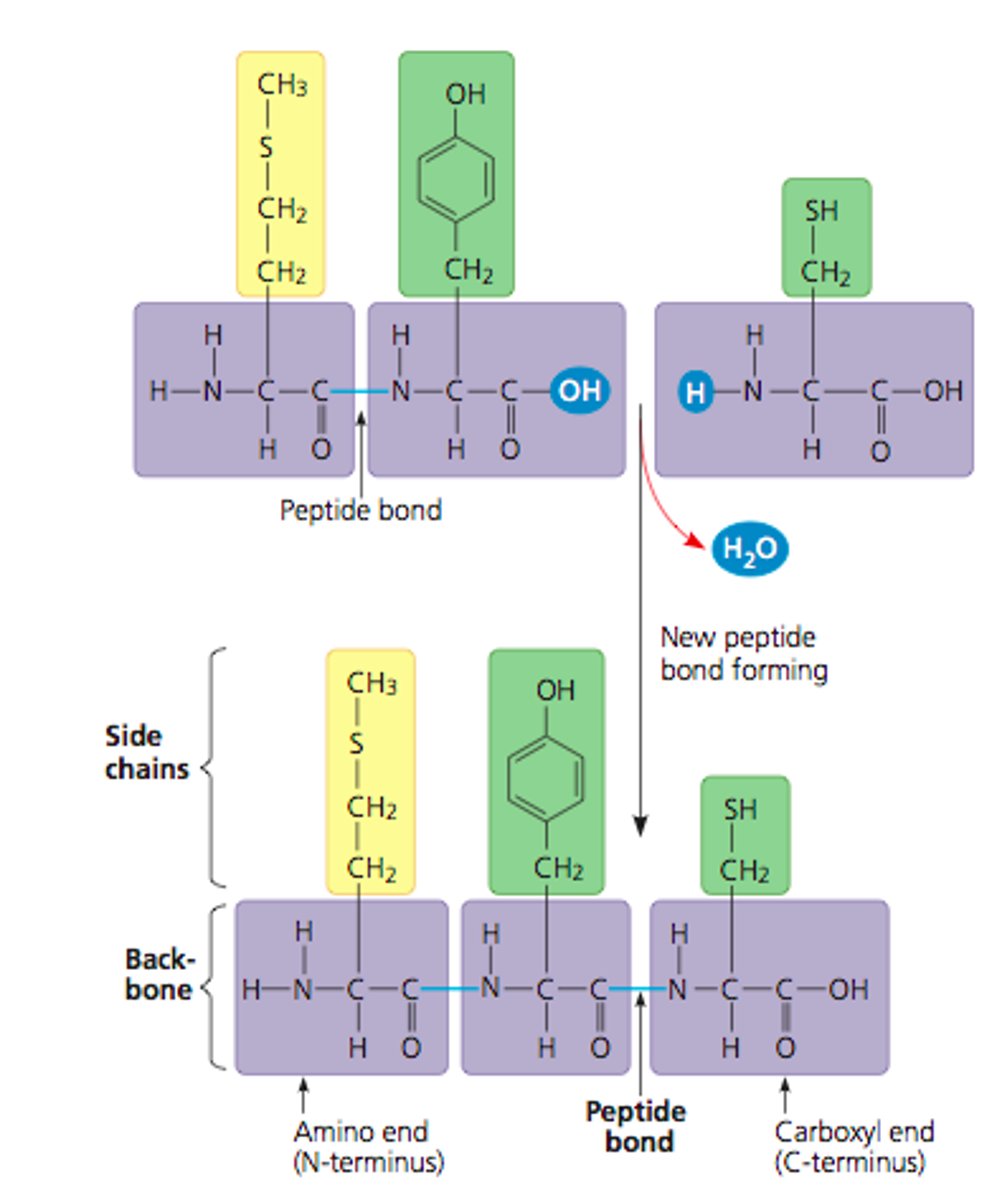
Polypeptide
A polymer (chain) of many amino acids linked together by peptide bonds.
- dehydration reaction links carboxyl of one amino to amino grp of next amino acid
- Starts from amino end (N-terminus), backbone is repetitive and wear amino acid side chains added to
-> can range in length from a few to >1000s monomers
- each polypeptide has unique linear seq of amino acids
Proteins - folding
- consist of 1 or more polypeptide twisted, folded, or coiled into unique shape
- diff to draw
Ribbon - show folds; Space-filling - shows shape
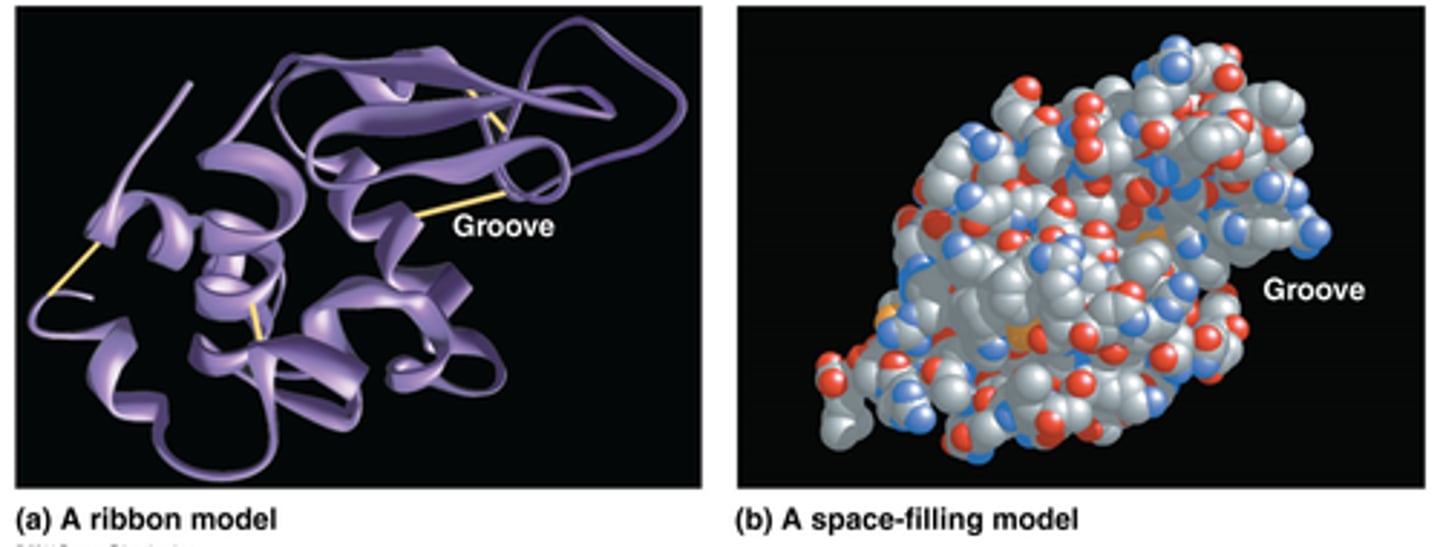
Primary structure
of a protein is amino acid sequence
- draw from amino (N) terminus end to carboxyl terminal end, numbered starting for N terminus
- Proteins - polymers of L-amino acids held by peptide bonds
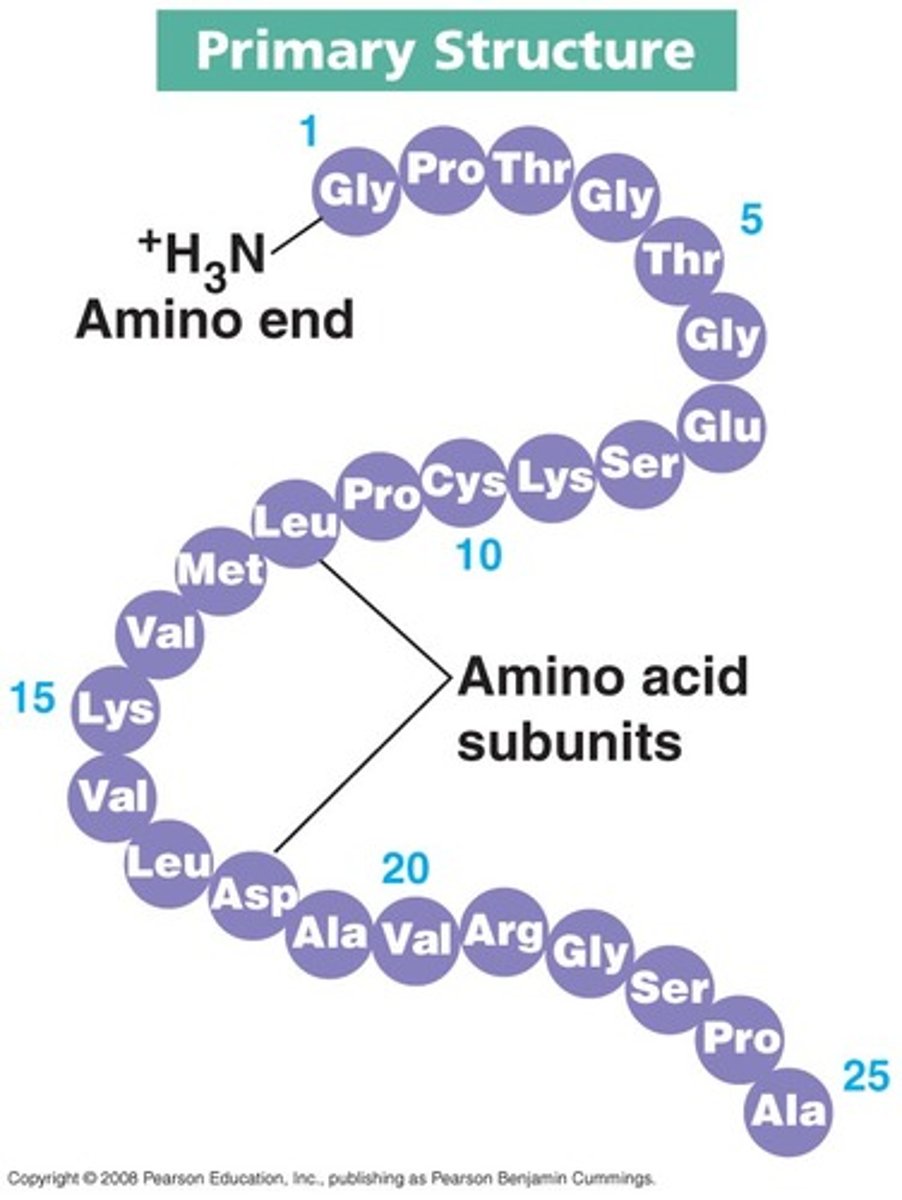
Secondary structure
is the local spatial arrangement of polypeptide chain; regular repeating structure held together by hydrogen bonds
2 regular common arrangements:
- α-helix: H bonds stabilise, between nearby residues
- β-sheet: H bonds stabilise between adjacent segments (mightn't be nearby)
Random coil: irregular arrangement of polypeptide chain
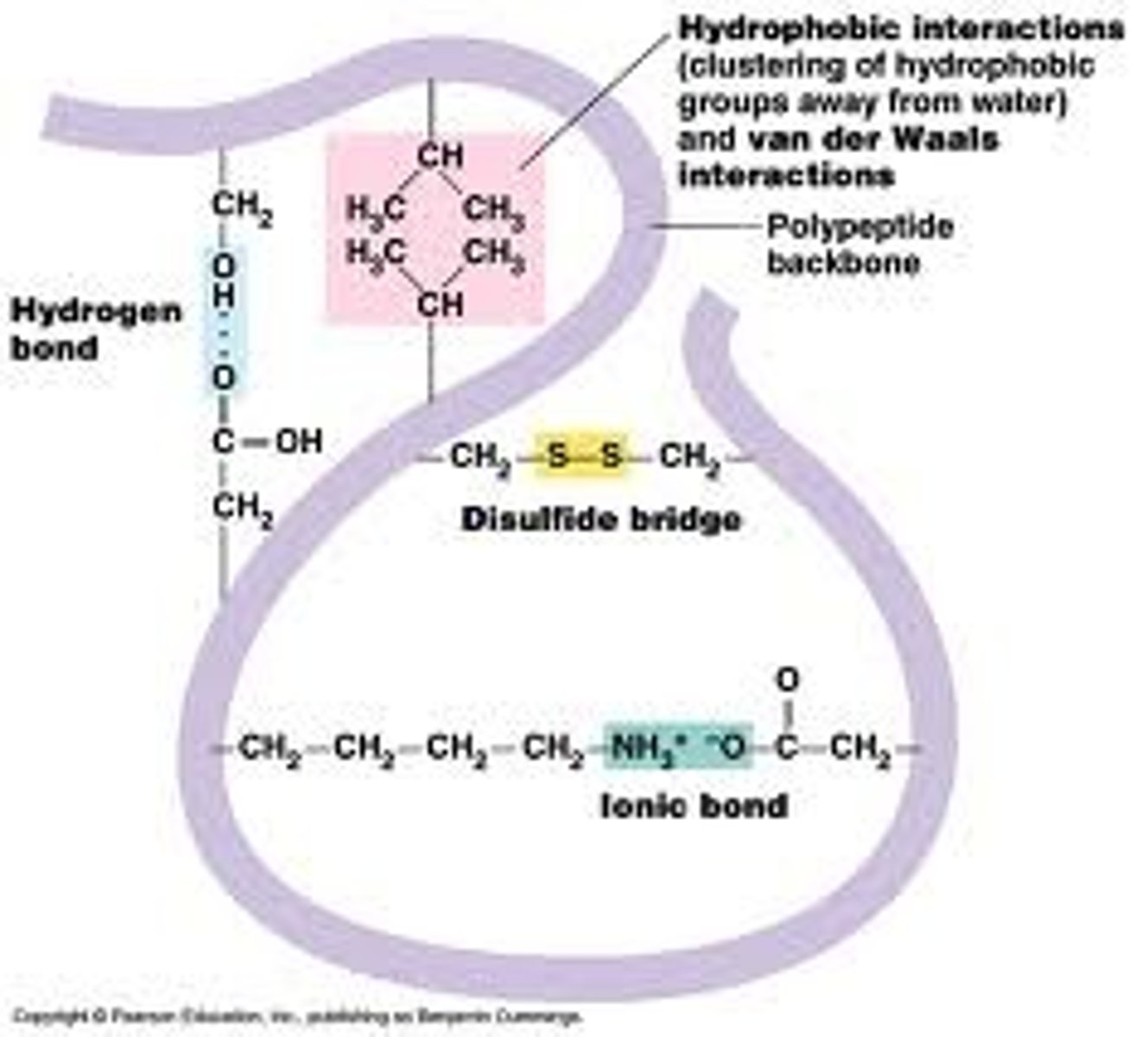
Tertiary Structure
is the overall, three-dimensional shape of a polypeptide due to non-covalent interactions between amino acids + R grps
Quaternary structure
the shape resulting from the association of two or more polypeptide subunits (assembly of polypeptide chains)
- can be identical or different inseq
α/β can represent that polypeptides have diff primary structurse
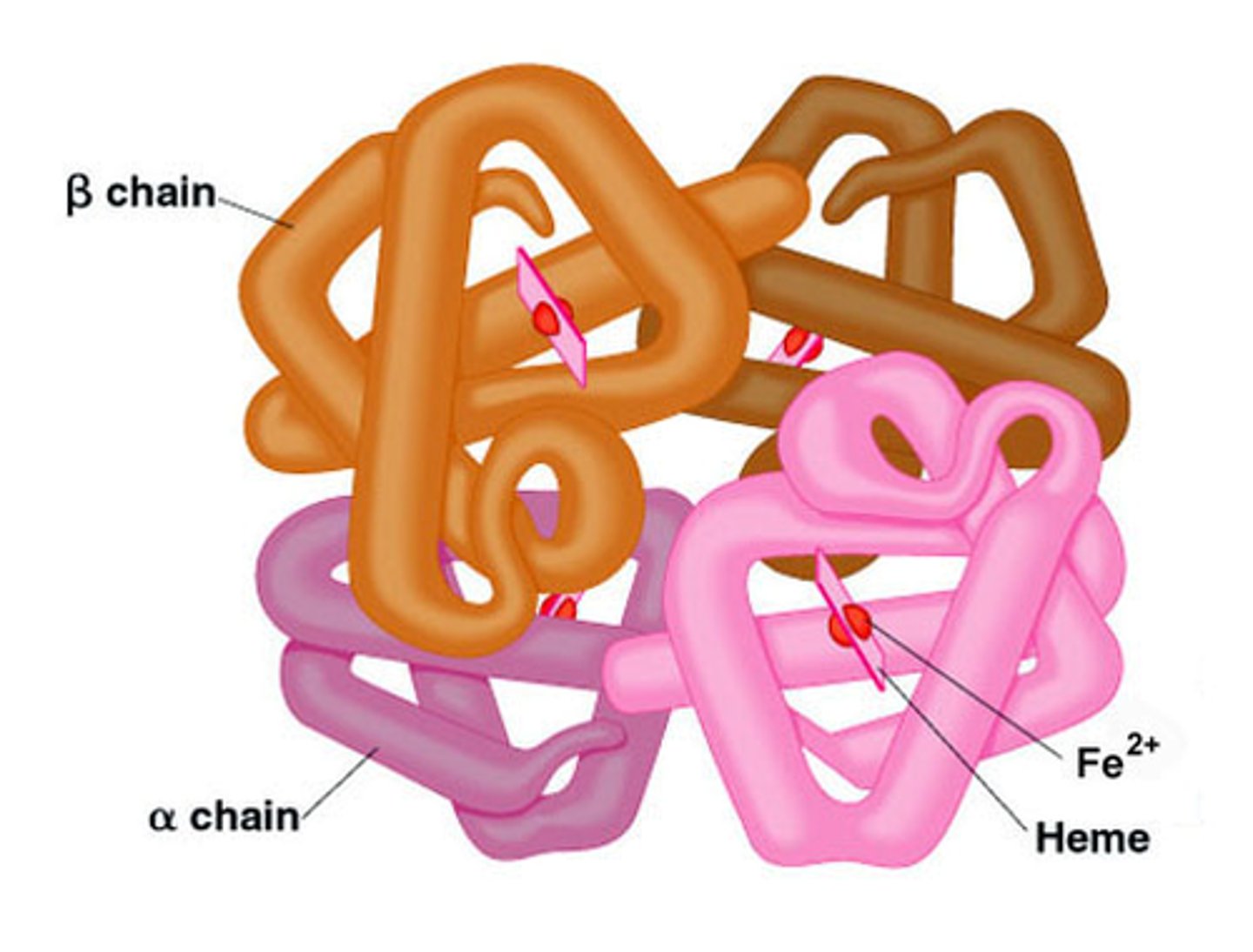
Haemoglobin is a tetramer - made of 4 polypeptides
Myoglobin and Haemoglobin - Functions
Myoglobin: stores + delivers O2 to muscle s=cells - imp in deep diving aquatic animals, allows them to store 'O' underwater
Haemoglobin: transports + delivers O2 through veins/arteries and is the major protein of red blood cells.
- are homologous proteins (similar primary, secondary & tertiary structures) and thus, have similar functions
Haemoglobin
- heme grp (co-factor) attached to polypeptide via non-covalent bonds
- Fe^2+ ion coordinated to heme and to histidine side chain
- 1 site around Fe2+ available for O2 binding
- tetramer w 4 subunits, which allows 'communication' between subunits
Myoglobin
is a single polypeptide monomer
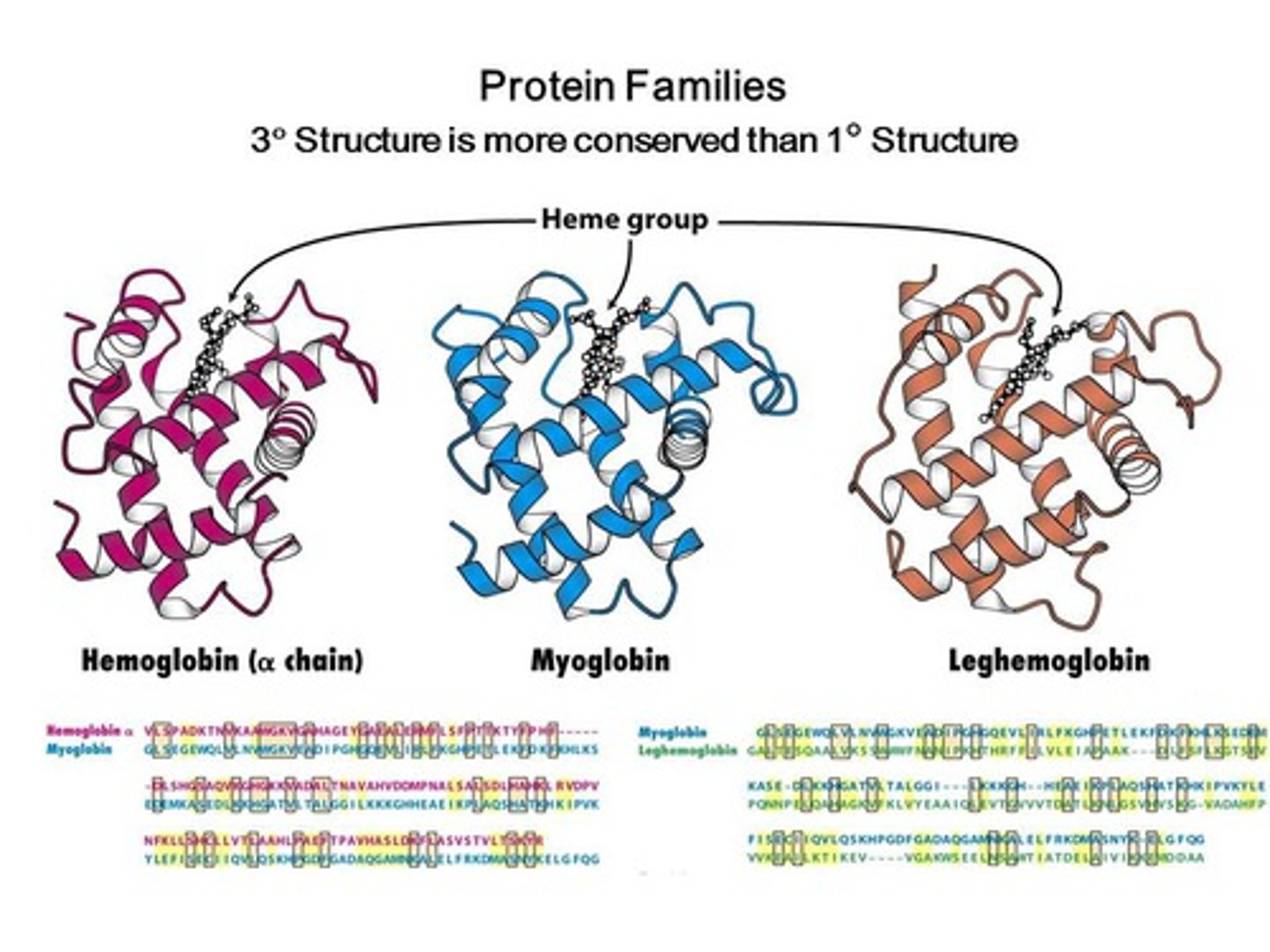
Sequence Homology
similarities in the order of nucleotides of DNA and RNA or in the order of amino acids of proteins that indicate common ancestry between organisms (note the same histidine between alpha, beta and myglobin (2nd row, HGKKV and HG)
Myo and haemoglobin function - by reverse binding O2
Myoglobin + O2 ⇌ Myoglobin-O2
Haemoglobin + 4O2 ⇌ Haemoglobin-4O2
Myoglobin: single polypeptide monomer
Haemoglobin: tetramer (allows subunit communication)
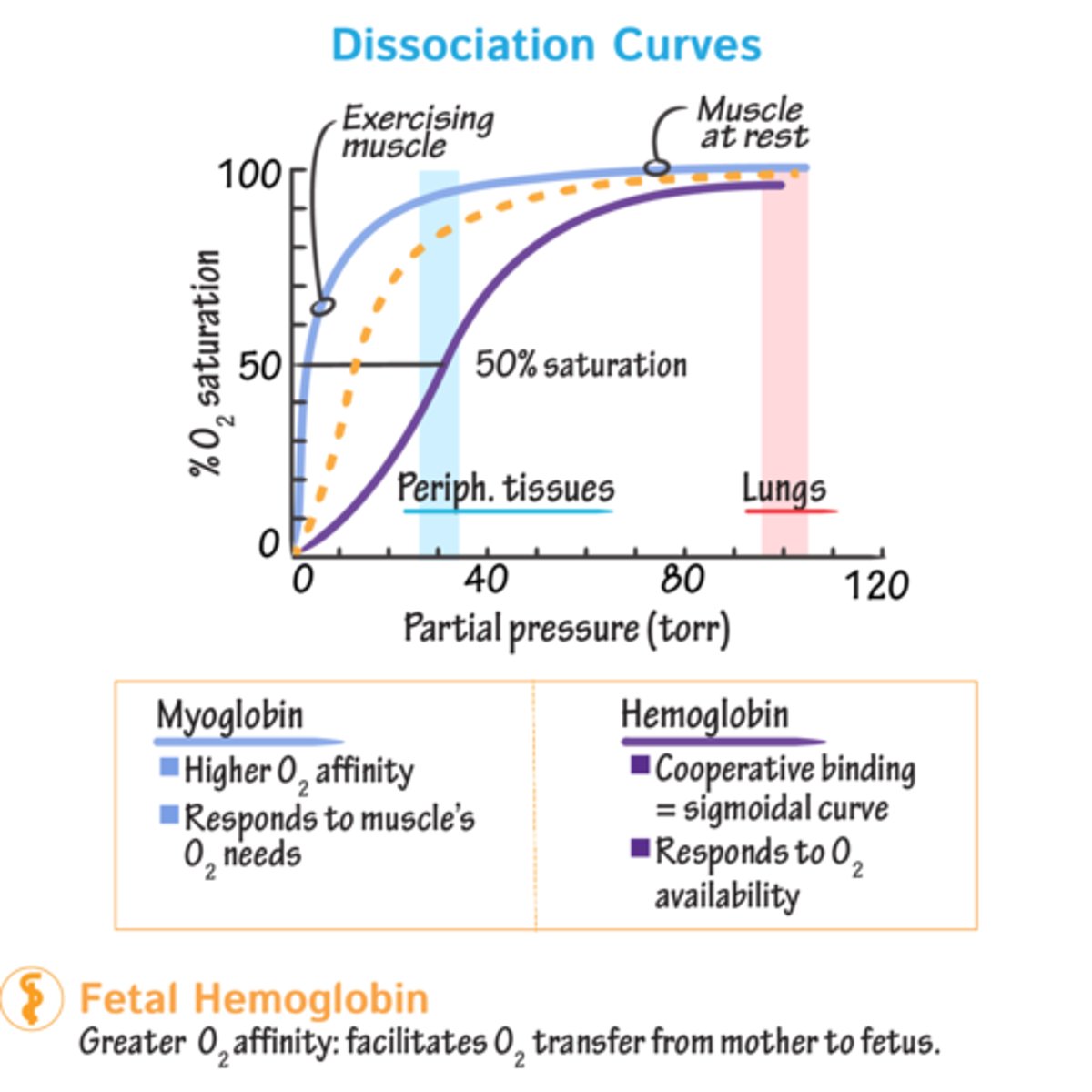
Saturation curves
Tell us % of protein mol that have 'O' mol bound at any one time. Amount varies depending on partial pressure exerted by O2 (concentration (partial pressure) of O2 - varies depending on body part).
Sat. curves of harm/myoglobin
@ high pO2 (100) of lungs, both fully loaded (sat=1)
Active tissues pO2=20
- Myo can't release O2 at this pressure; haem can release approx. 75% of O here.
- pO2=5 Myo releases 'O'. Because last stores of O2 have been depleted.
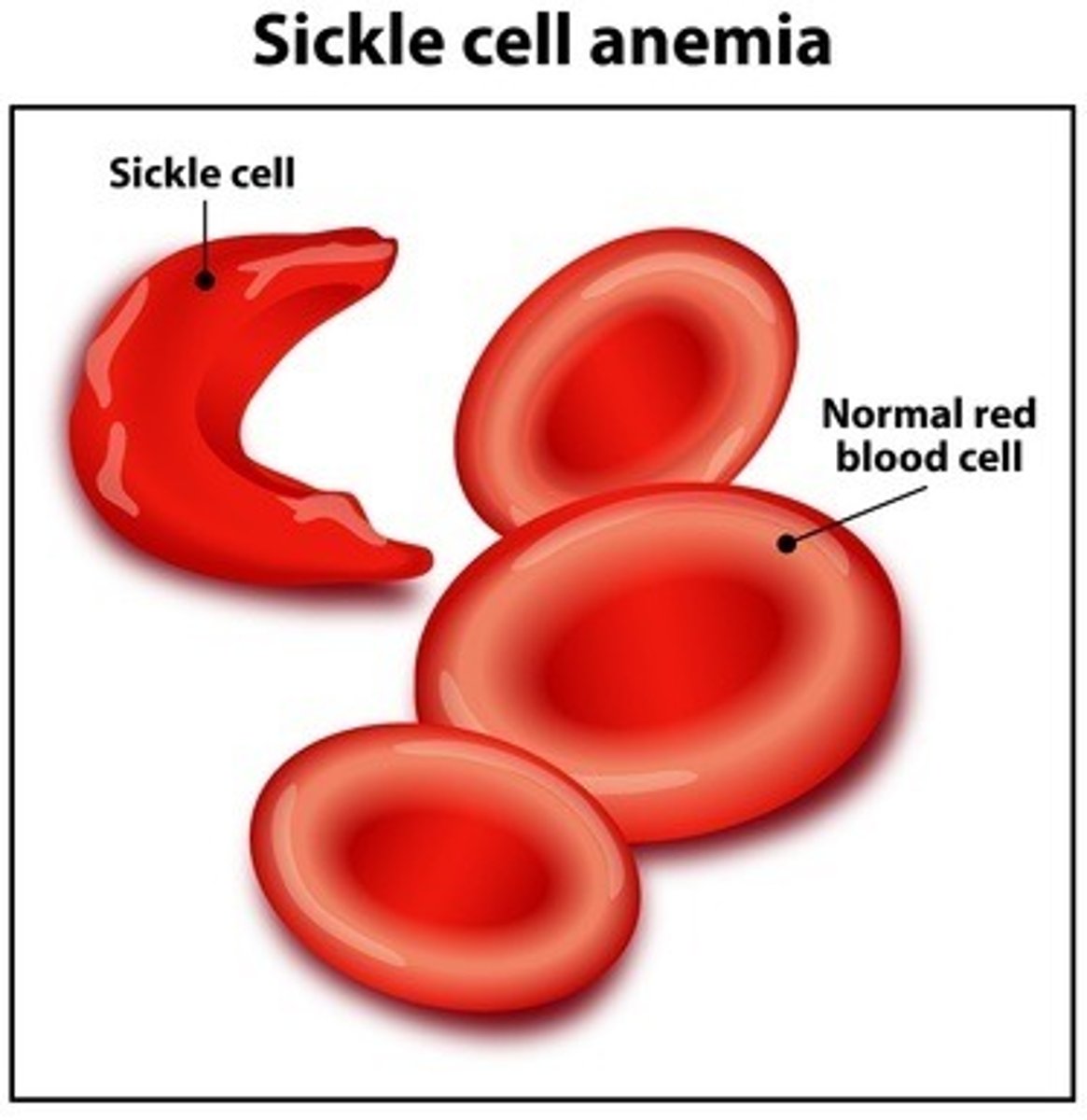
Sickle cell disease
Condition where not enough healthy red blood cells to carry 'O' throughout body; red blood calls have abnormal shapes + textures - normally uniform + cup-shaped
- accumulation can damage spleen - making pts vulnerable to infections
Sickle cell haemoglobin (HbS)
Has a single amino acid 'mutation' compared to norm. haemoglobin (HbA), which occurs in β-subunit.
- Glu to Val
- is a non-conservative amino acid subsititution
Where mutation?
Glu (-)ly side chain on norm. ham surface.
Val non-polar - substitution places hydrophobic residue on protein surface, greatly reducing solubility of deoxygenated haem. form, forming a hydrophobic patch on mutant protein surface.
Aggregation of deoxy-haemoglobin
- Val B6 forms -phobic patch on mutant protein surface
- B-chain of deoxy. haem. has another -phobic patch by Phe 85 & Leu 88
- mutant haem. mol. aggregate to hide the sticky patches
Interactions between Val 6, Phe 85 & Leu 88 from adjacent mol. (join together)
Sickle-cell disease is due to the association of multiple haemoglobin molecules
Normal: singular - do not associate w one another, each carries 'O'
Sickle: Mol interact with another and crystallise into a fiber; 'O' carrying capacity greatly reduced.
QUATENARY STRUCTURES of haem and myo- are thus different.
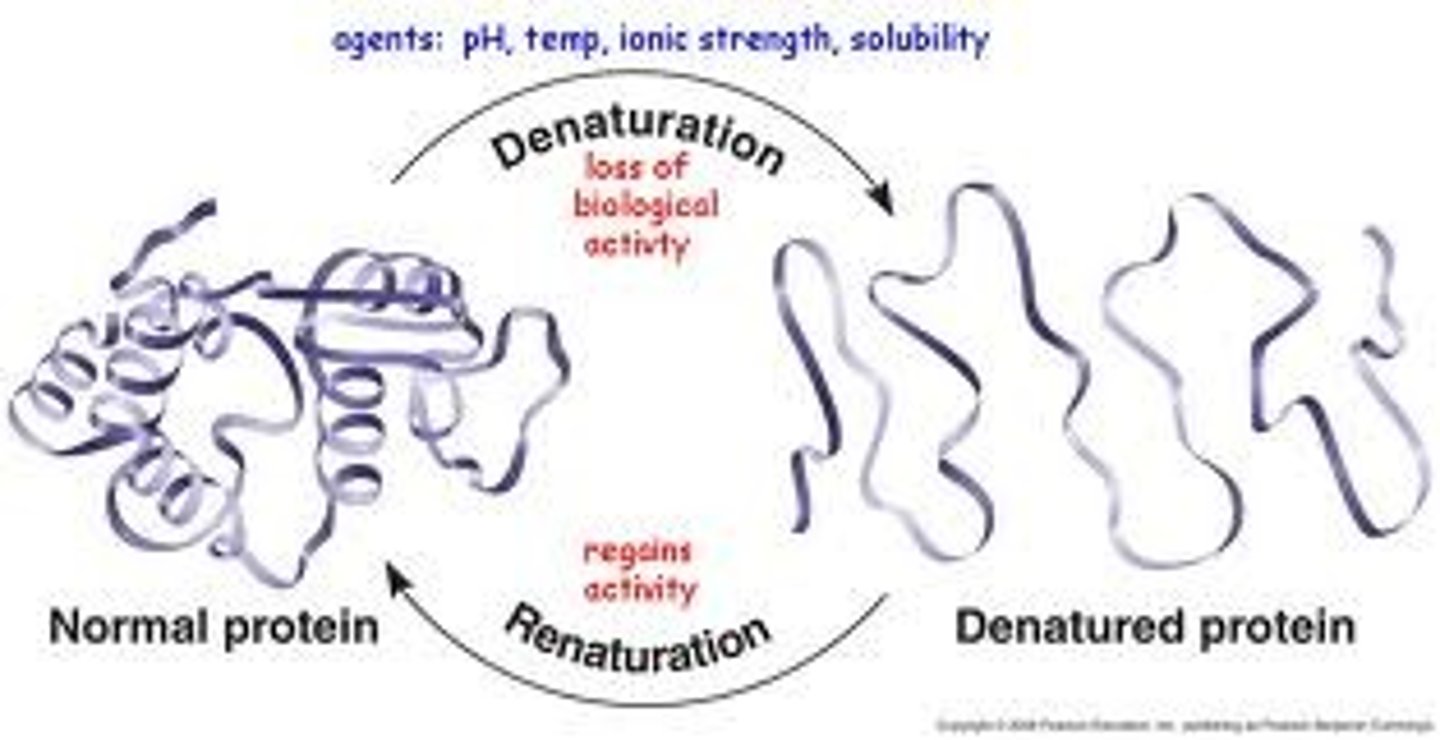
Structure - factors that affect it
Physical + chemical conditions can affect structure
e.g. pH alteration, salt concentration, temp or detergents can protein unraveling
Denaturation: loss of the native protein structure; denatured protein is biologically inactive bc shape lost, hence function
Globular proteins - Characteristics
Hydrophobic amino acid side-chain: usually located inside protein core
Charged side chains: usually outside
H bonding side chains: can be in core or located on surface
When these are disrupted, globular proteins unravel + loose activity.
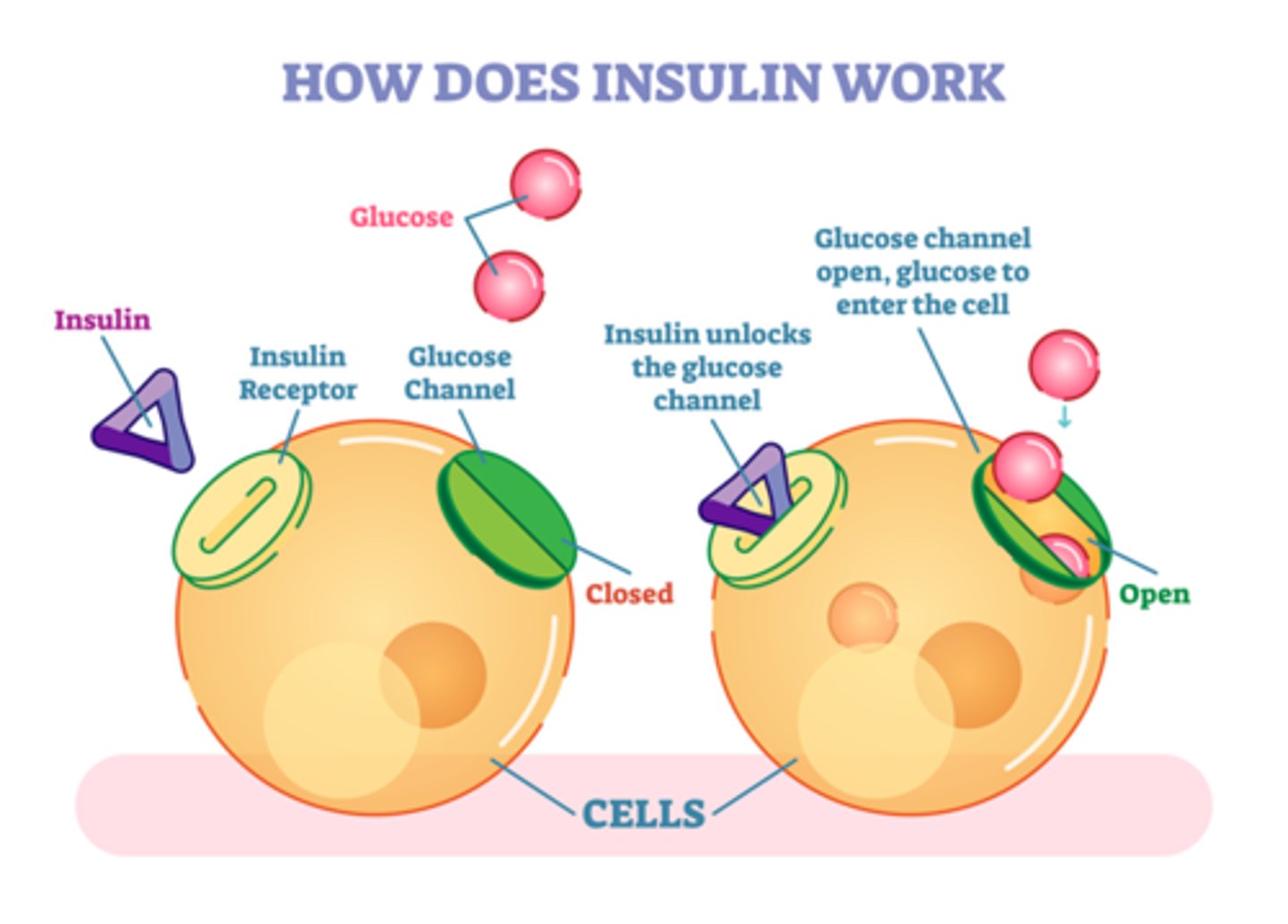
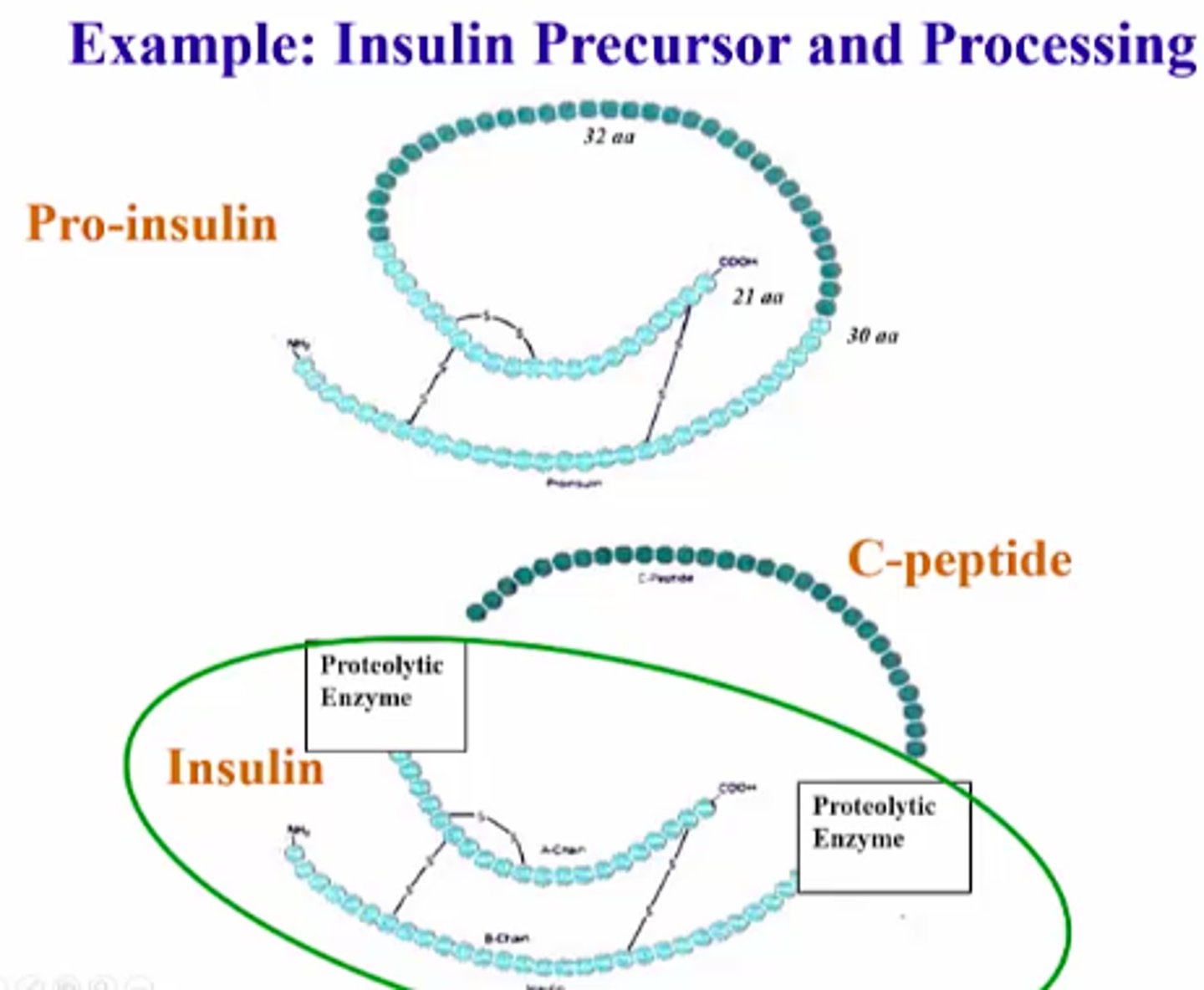
Insulin
The polypeptide hormone that controls blood glucose levels. Diabetes - disease where insulin mol no longer carry out regulation of glucose levels
Proinsulin
the inactive precursor form of insulin that is made of the A and B chains of insulin along with C-peptide.
It has a connecting piece of peptide which must be cleaved before becoming active
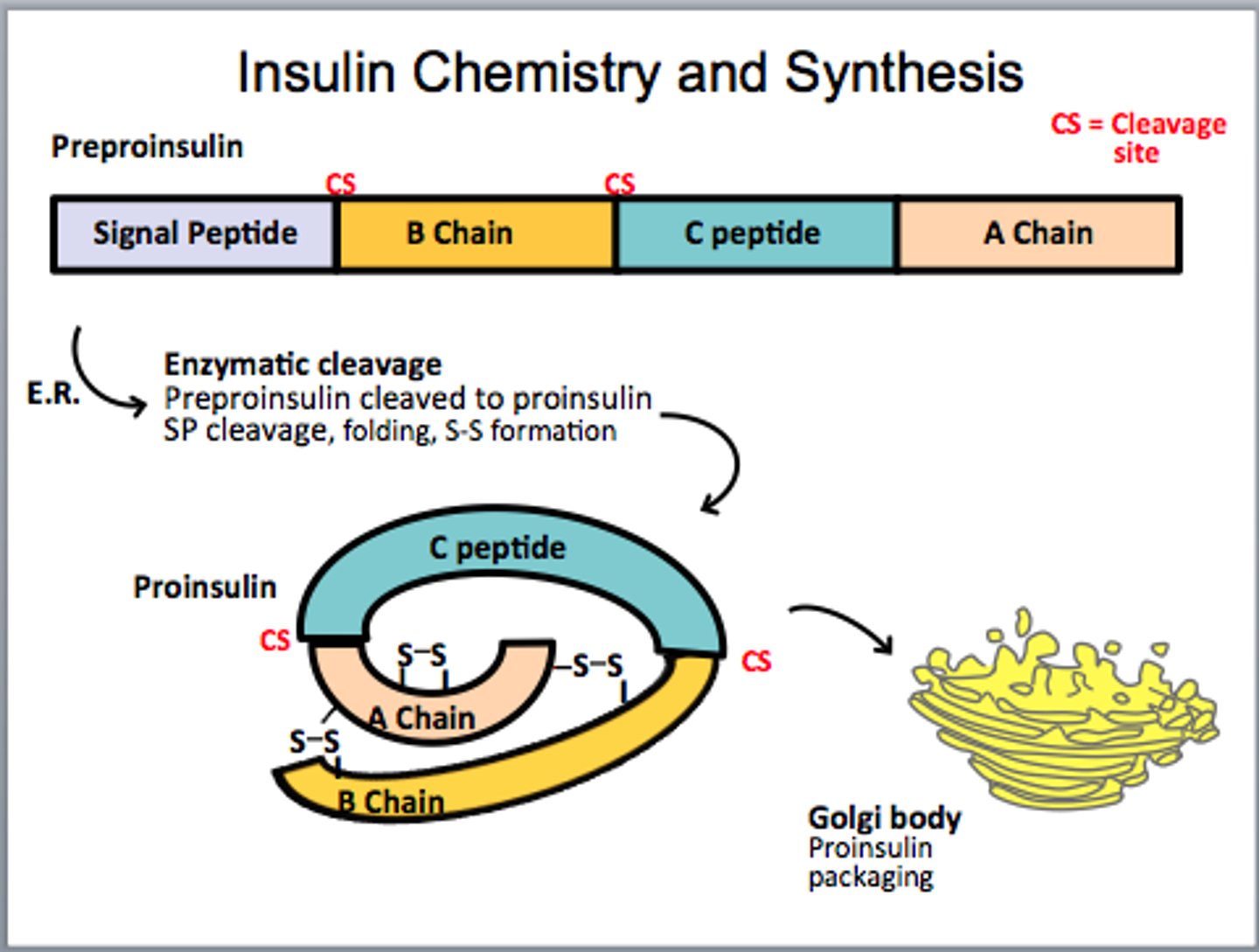
Active insulin
Insulin becomes active after proteolytic (enzyme) cleavage
- Disulfide bonds hold 2 separated chains (A & B) together
- insulin can nod bind w receptors on cell surface; regulating glucose/sugar levels
Collagen
- most abundant protein in vertebrates
- 30% of total body protein
- provides mechanical strength to connective tissues (incl. skin, bones, tendon, cartilage + blood vessels)
- fibrous and not soluble in water
Five types of collagen
28 diff collagen (amino seq simmer, but distinctly diff)
- 90% in body in collagen I
Collagen I: skin, tendon, vascular ligature, organs, bones
Collagen II: cartilage's (main component)
Collagen III: liver's surrounding connective tissue
Synthesis of tropocollagen from procollagen
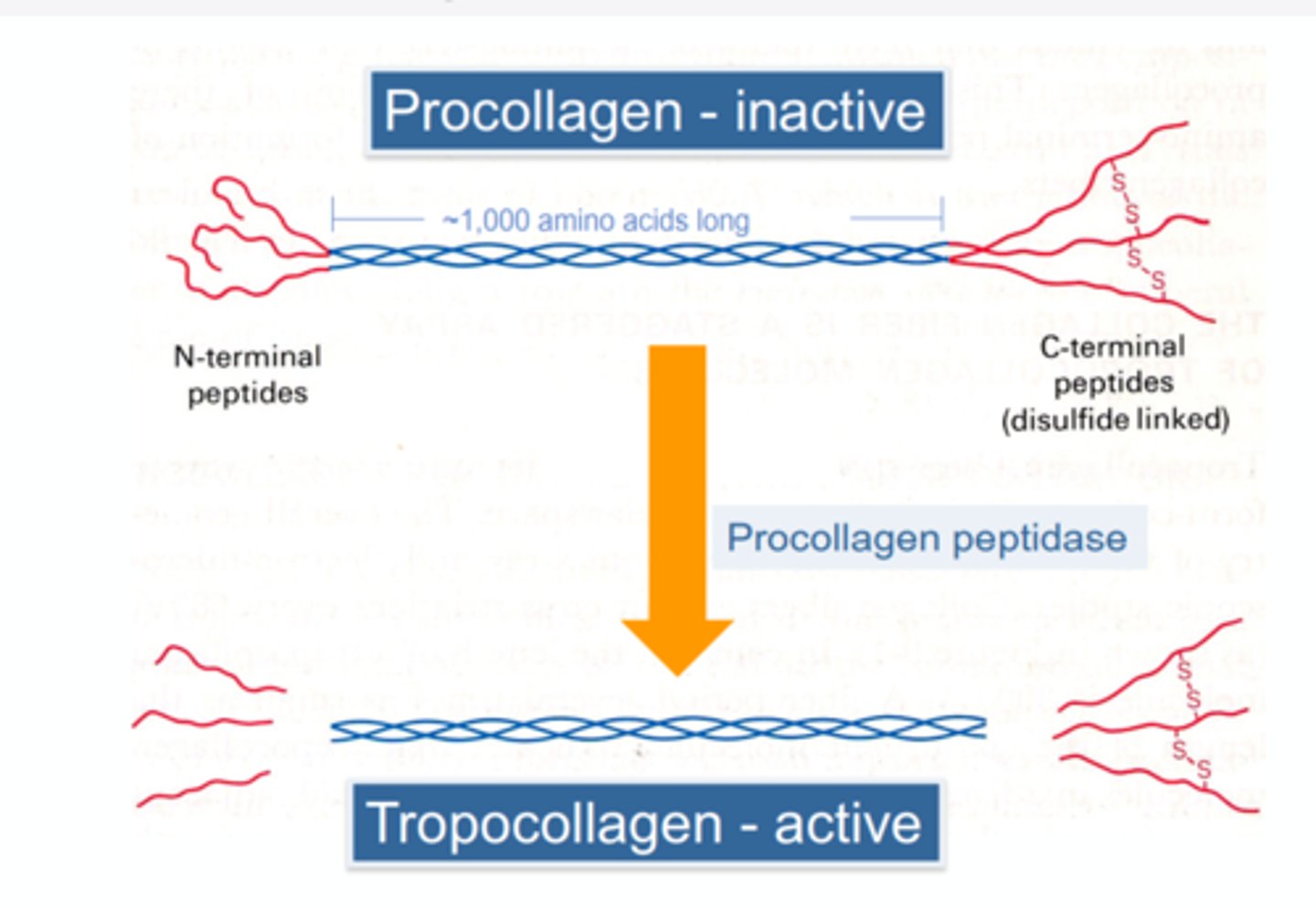
Collagen fibrils
rod-like structures, 300nM long + 1.5nM thick; are made from collagen units aligned in a staggered fashion (heads of collagen mol); units are cross-linked (covalent bonds) for strength.
- ~ 1000 amino acids per each chain
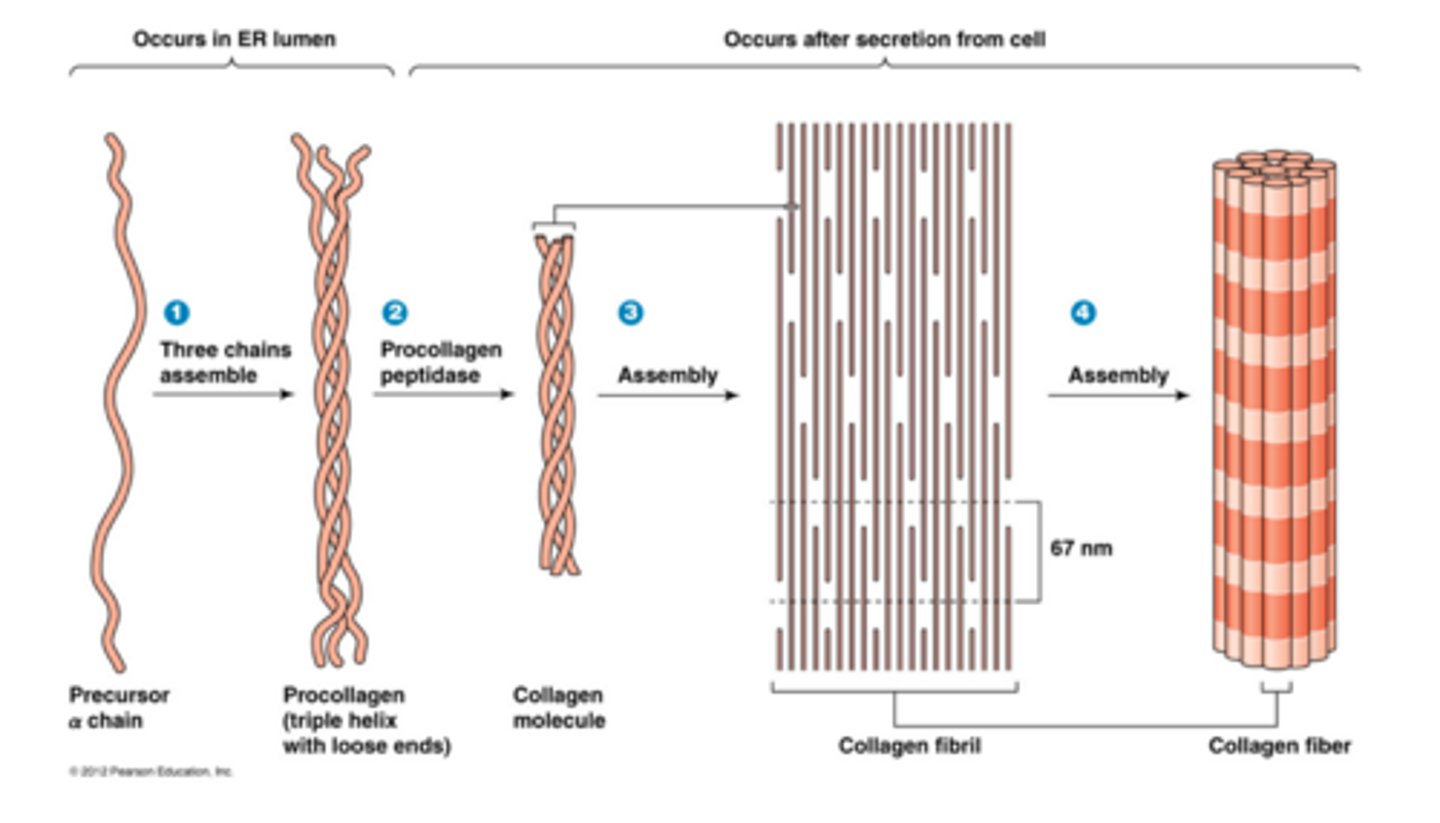
3D structure - Collagen
Single chain: helical structure; atoms drawn as spheres
Collagen mol: 3 stranded "triple-helix" of collagen - each peptide chain coloured differently. Triple-helix provides strength + flexibility to skin + cartilage
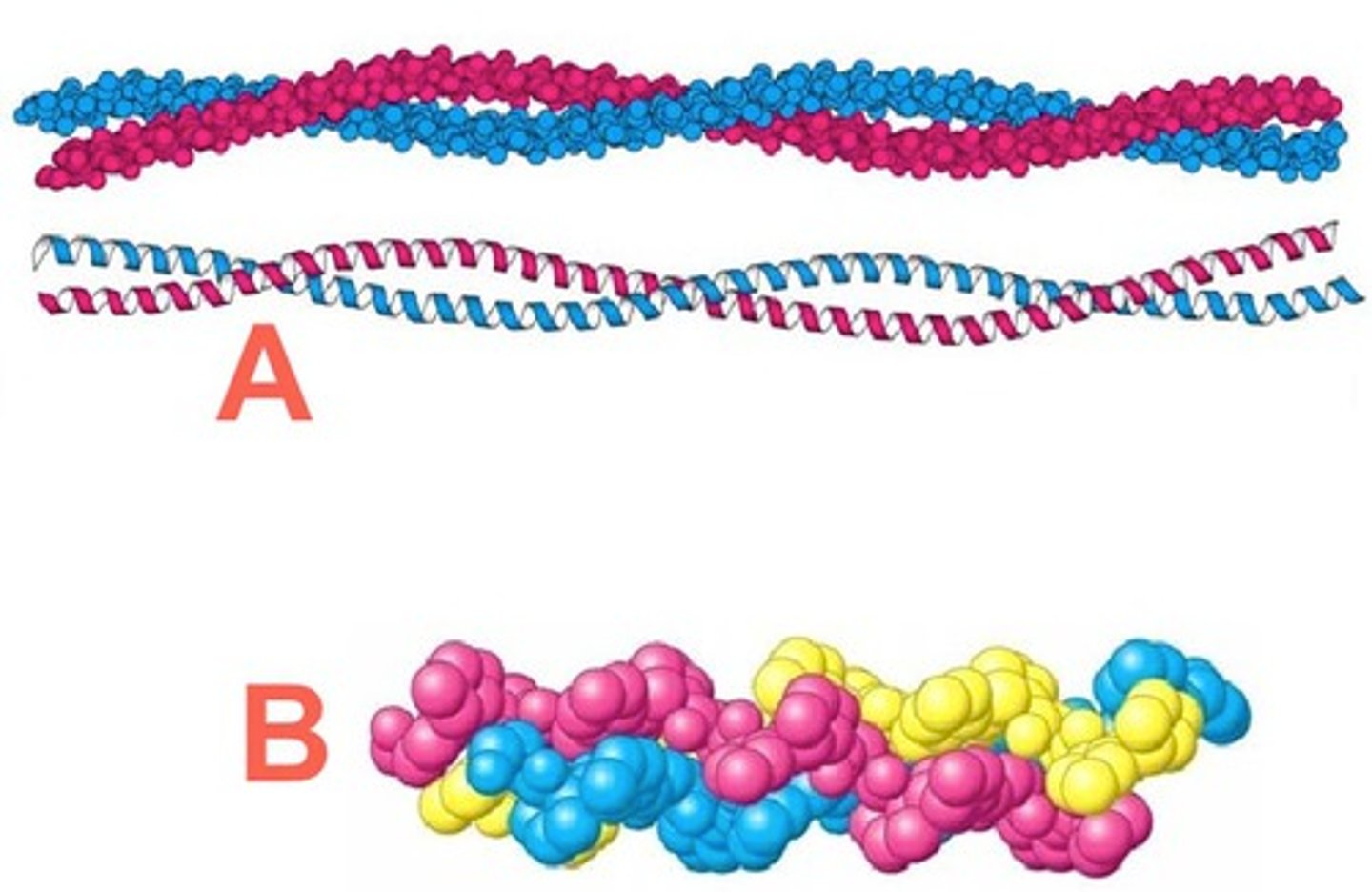
Collagen 3D structure
is not soluble in water and has an elongated shape. It *assembles tightly together into fibres" w extensive intermolecular interaction to build up macroscopic structures.
- Long axis - inside is v crowded w atoms
Inside collagen triple helix
Since all other side chains are too big, Glycine side-chains (only a H atom) points inside triple-helix
Genetic disorder - collagen
Osteogensis imperfecta or brittle bone disease: genetic disorder where glycine side chain had mutated to cysteine - which has too many atoms to find inside the helix (cannot fit), making it unstable (H bond and helical arrangement disrupted). This mutation is lethal.
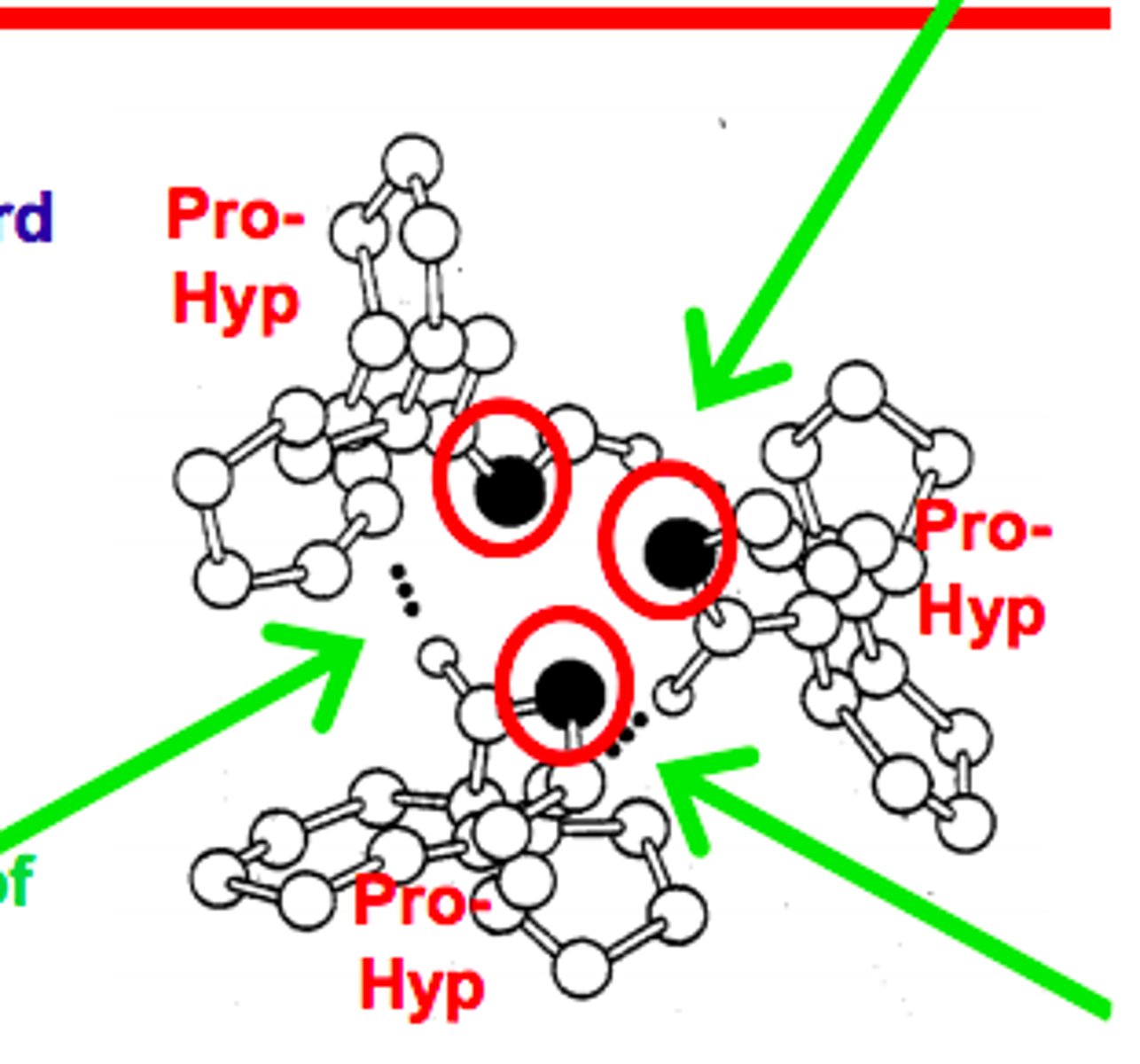
Collagen amino acid sequence
Typical of fibrous proteins, its polypeptide has repeated amino acids.
Hyp= hydroxyproline is not 1 of 20 standard amino acids
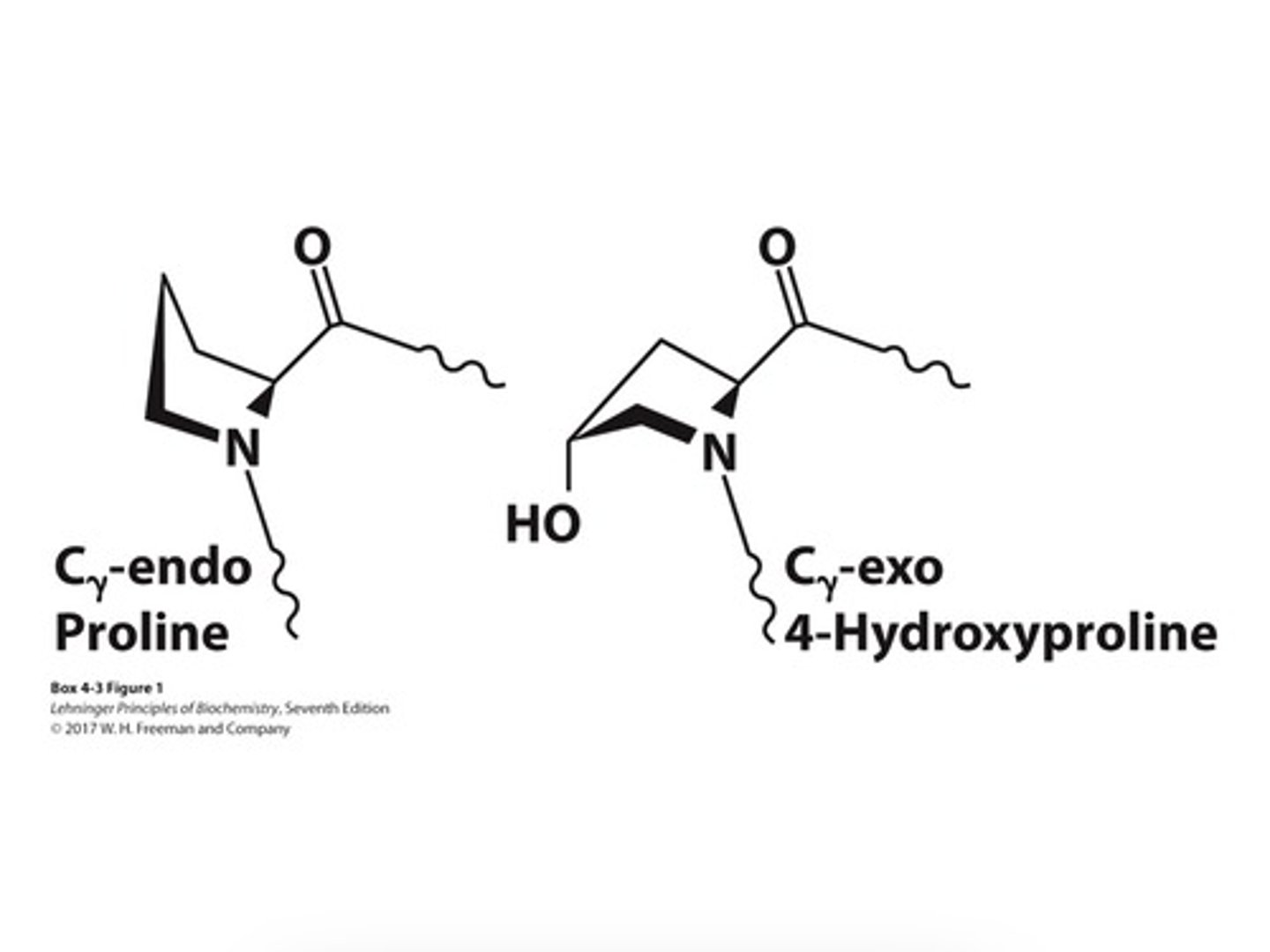
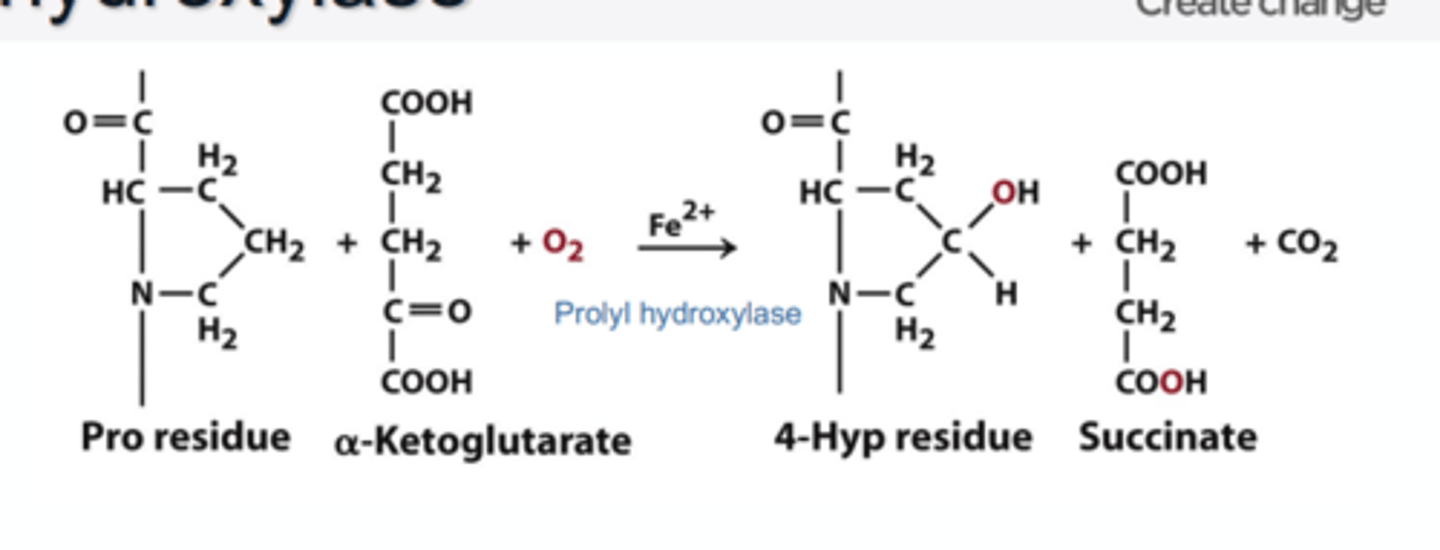
Hydroxyproline
- amino acid created not by the genetic code, but modification, after polypeptide formation of proline (by enzyme prolyl hydroxylase)
(formed by chemical modification to amino acid's existing structure)
4-hydroxyproline
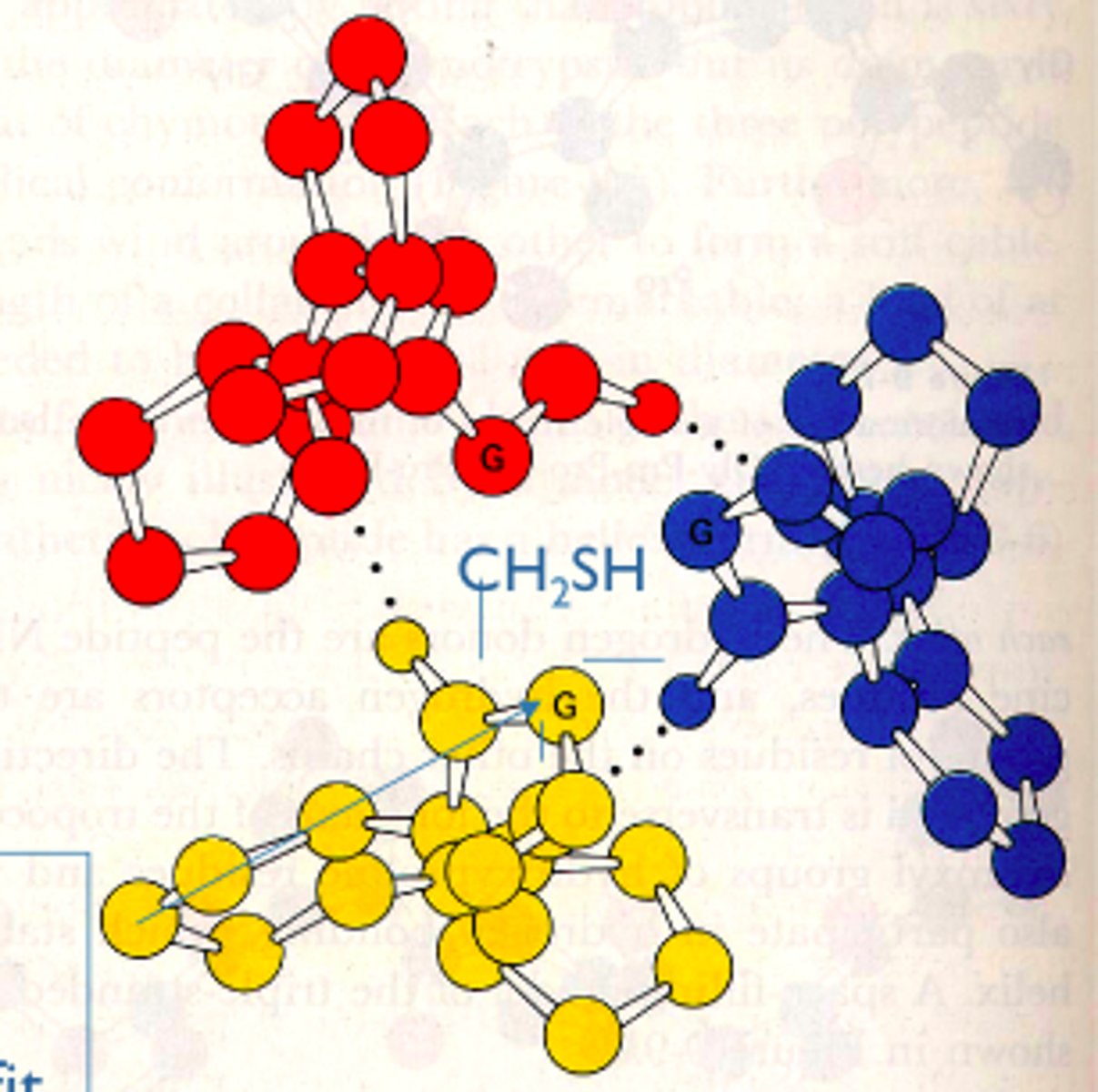
• Forces the proline ring into a favorable pucker
• Offers more hydrogen bonds between the three strands of collagen
• Catalysed by propel hydroxylate in the presence of ascorbate (vitamin C)
Vitamins
• an organic compound req as a vital nutrient by an organism
• cannot be synthesised by the organism (e.g. human). Thus, must be obtained in the diet.
• Vitamin C strengthens collagen
Vitamin C activates prolyl 4-hydroxylase
• Prolyl-4-hydroxylase (P4H) is a metalloenzyme which needs Fe2+ to be fully active.
• Vitamin C adds an electron to Fe3+ = Fe2+; activating P4H
• W/o vitamin C, proline in collagen will have less of a tendency to become hydroxylated. Collagen will be weaker.
Scurvy
• is a disease caused by a lack of vitamin C (thereby weakening collagen). Symptoms include: loss of teeth, pale skin, sunken eyes.
• Cpt Cook fed crew sauerkraut, oranges and lemons - eliminating disease's threat
Cross linking of collagen triple-helices
E.g. Two lysine side chains replaced by CH=N bond.
• Cross linking makes collagen stronger (more covalent bonds) and less soluble

Collagen Biosynthesis Steps
1) Single strands of polypeptide are made
2) Hydroxylation of lysine and proline side chains
3) The tripe helix is formed
4) Procollagen is converted to collagen by procollagen peptidase
5) Bundles form
6) Cross links form
Lipids
diverse group of hydrophobic molecules (little/no affinity for h2o bc they mostly consist of hydrocarbons - forming non-polar covalent bond - DONT mix well with water
-1 class of biological mol DO NOT form polymers
- most important lipids: fats, phospholipids + steroids
Fat
constructed from two types of smaller molecules: glycerol and fatty acids.
Fats seperat from H2O bc H2O mol form H bonds w each other excluding fats
Glycerol + fatty acids
Glycerol: 3-C alcohol w hydroxyl grip attached to each carbon
Fatty acid: consists of carboxyl gap attached to long carbon skeleton
Triacylglycerol: (or triglyceride) is created by 3 fatty acids joined to glycerol by ester linkage, in a fat
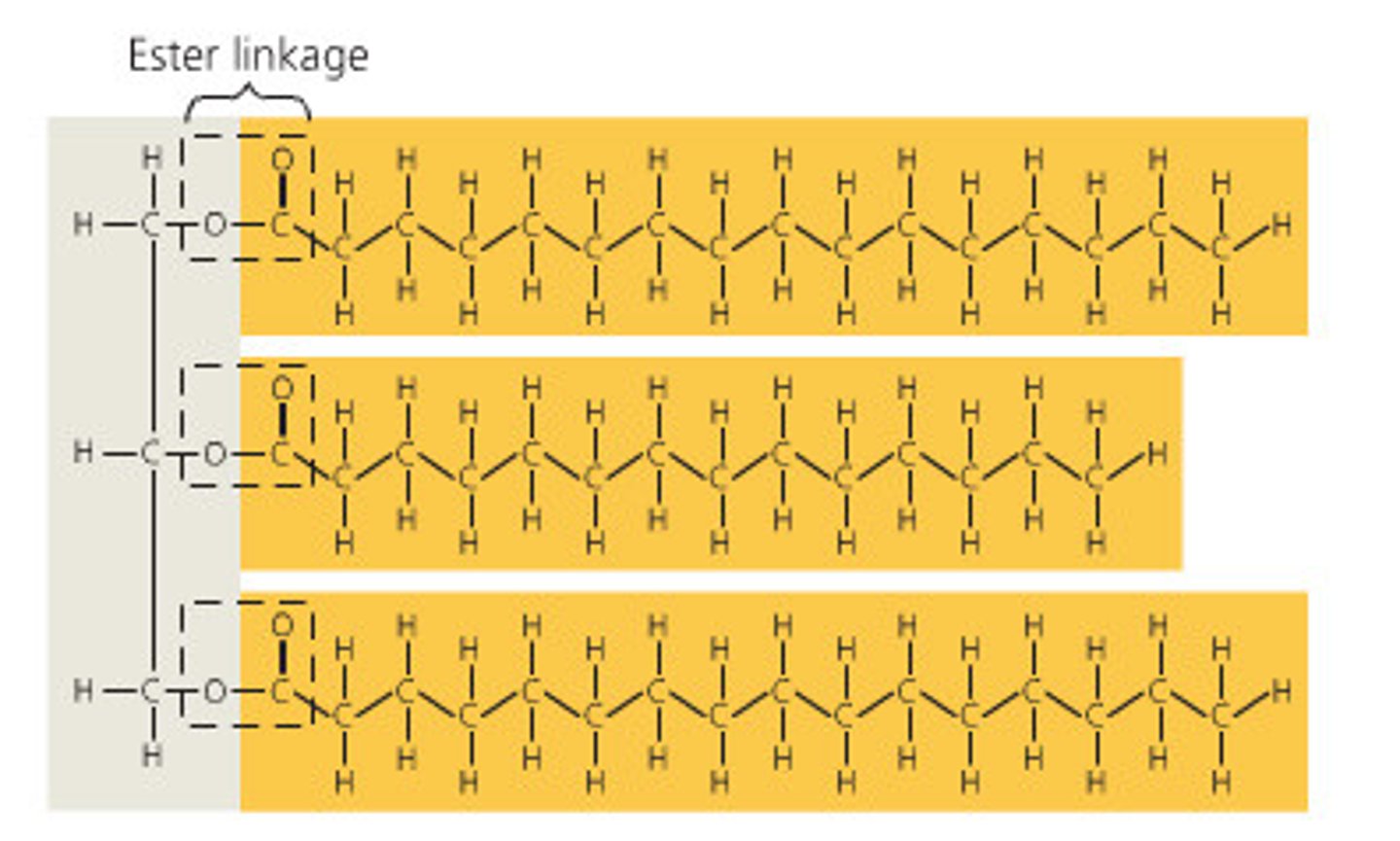
Fatty acids
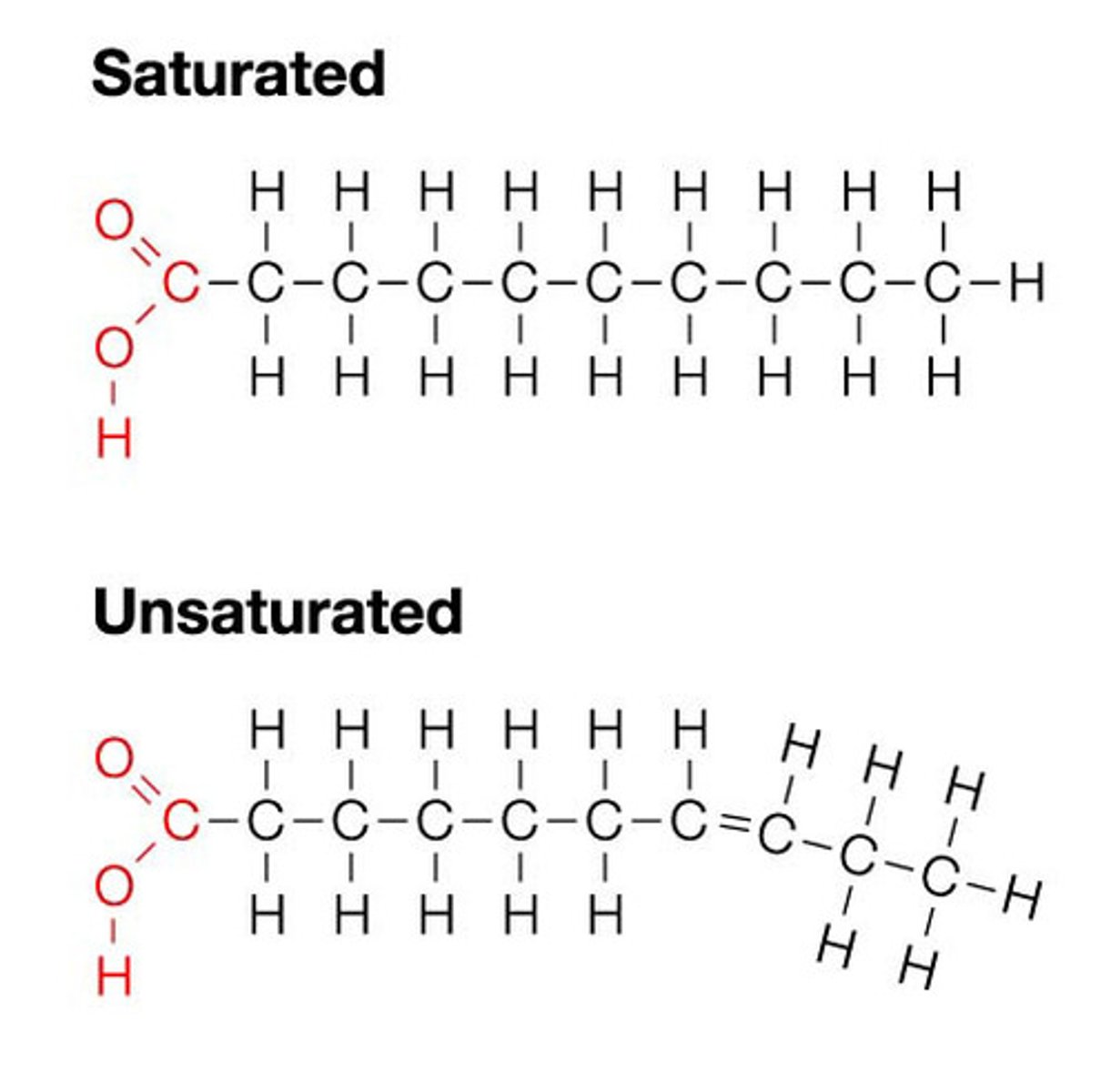
Vary in # of C and #/locations of double bonds
• Saturated fats: no double bonds e.g butter thus solid @ room temp
• Unsaturated fats: 1 or more double bonds e.g. olive oil @ room temp is liquid bc can't be packed together enough to solidify - bc of kinds in fatty acid hydrocarbon chains
Fats - disease
Diets rich in sat fats contribute to cardiovascular disease via plaque deposits
Hydrogenation
The process of converting unsaturated fats to saturated fats by adding hydrogen.
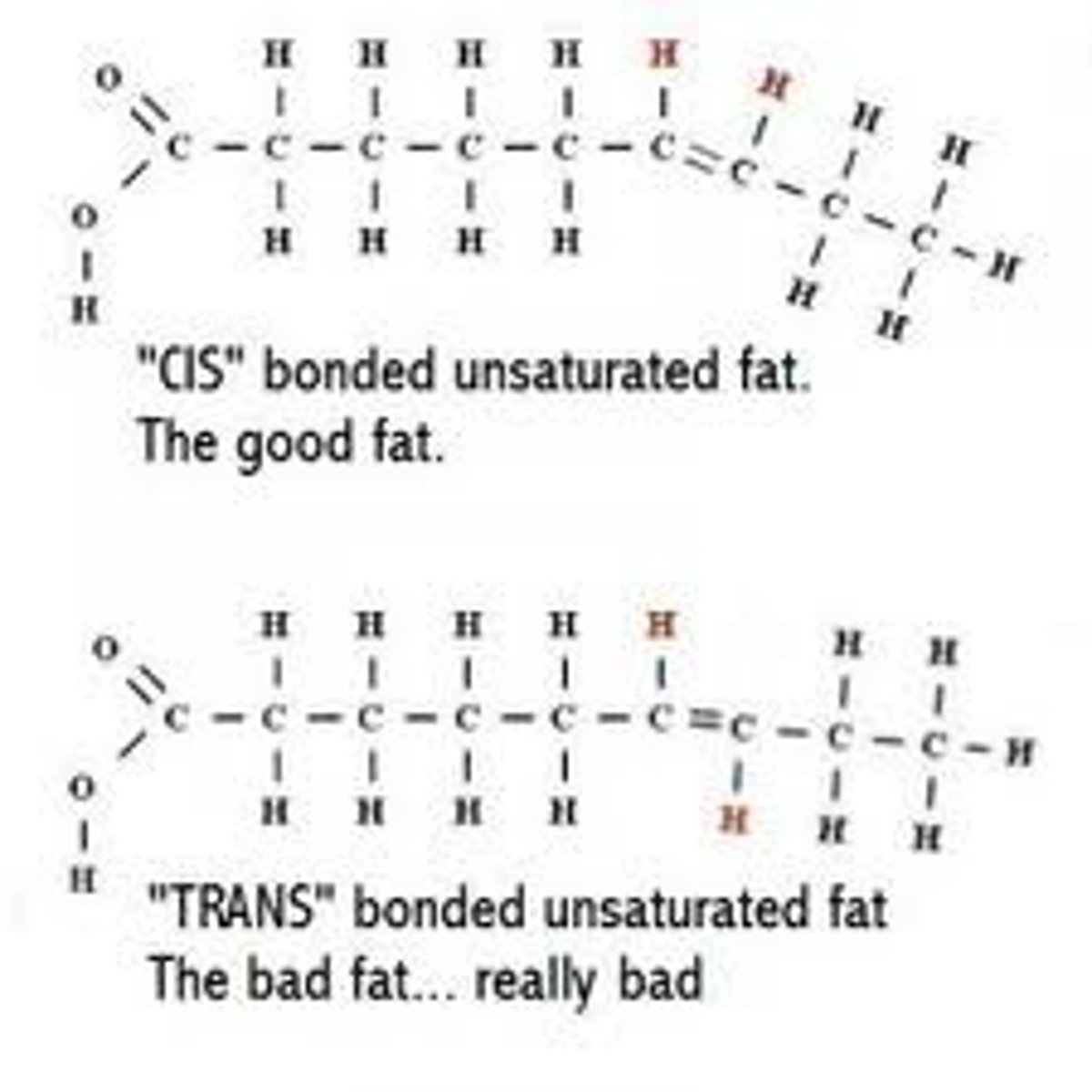
Hydrogenation veg oils created unsaturated fats w trans double bonds.
Trans fats
unsaturated fats with trans double bonds (flips)
May contribute more than sat fats to cardiovascular disease, bc they go undetected/ are harder to break down?
Adipose
Fats major function is energy storage.The tails of fats can be broken down to provide energy.
• Humans/mammals store fat in adipose cells
Adipose tissue also cushions vital organs + insulated body
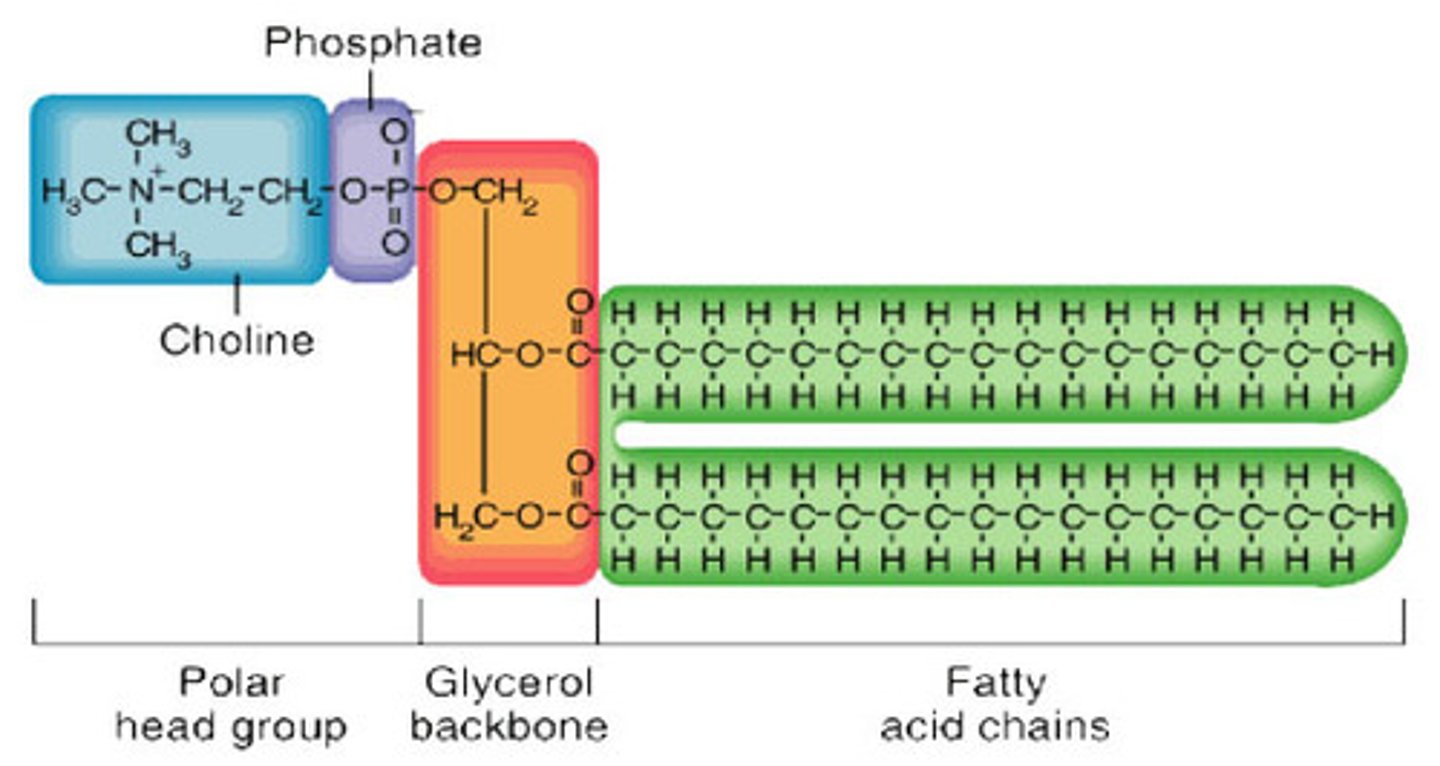
Phospholipids
a lipid of two fatty acids tails (hydrophobic) and a phosphate grp which are attached to a glycerol (forming a hydrophilic head)
• thus, when added to water, self-assemble into a bilayer - hydrophobic tails pointing interior (philic heads outside)
• the major component of cell membranes (outer layer)
Steroids
A type of lipid characterised by a carbon skeleton consisting of four rings with various functional groups attached (polar).
• mainly hydrophobic
• can be found in cell membranes (cholesterol)
• can play role as signalling mol (often)
Hormones
influence differences in gener traits (testosterone vs estrogen)
Anabolic steroids
can be natural or synthetic; interact w receptor storage stimulate muscle + bone synthesis.
• athletes take synthetic anabolic steroids to enhance performance (tech advance (mass spectrometry) allow for detection)
DNA and protein synthesis
DNA: double stranded, providing direction for replication. It directs synthesis of mRNA (messenger), controlling protein synthesis
Protein synthesis: occurs in ribosomes (molecular machines)
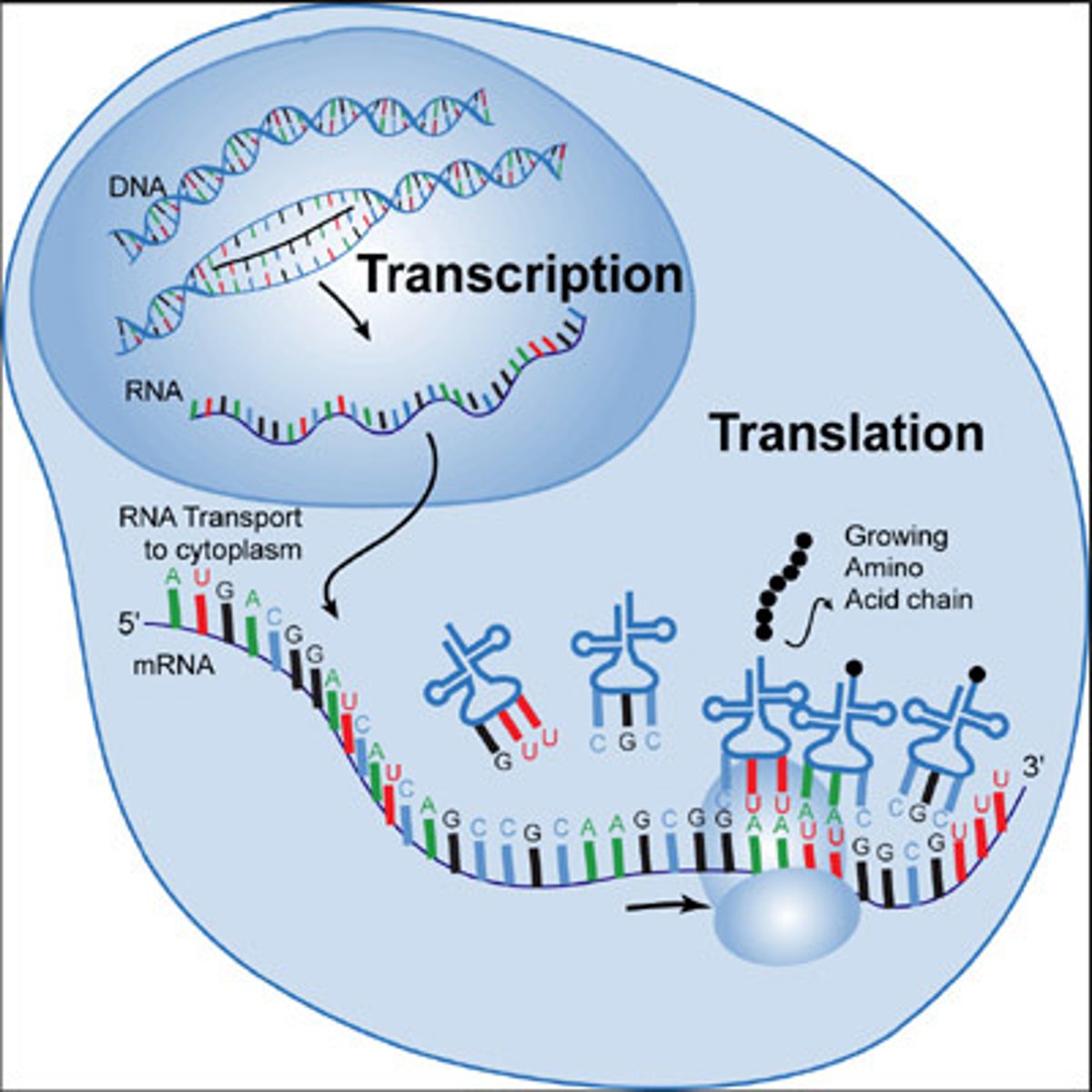
Nucleic acids (DNA, RNA)
store and transmit hereditary or genetic information
• Polymers = polynucleotides; Monomers = nucleotides
• amino acid seq of polypeptide (protein) programmed by unit of inheritance = gene (packets of info 1 gene/polypept. structure)
• Genes are found within DNA
• 1 DNA = many genes (although there are some regions which are non-coding DNA)
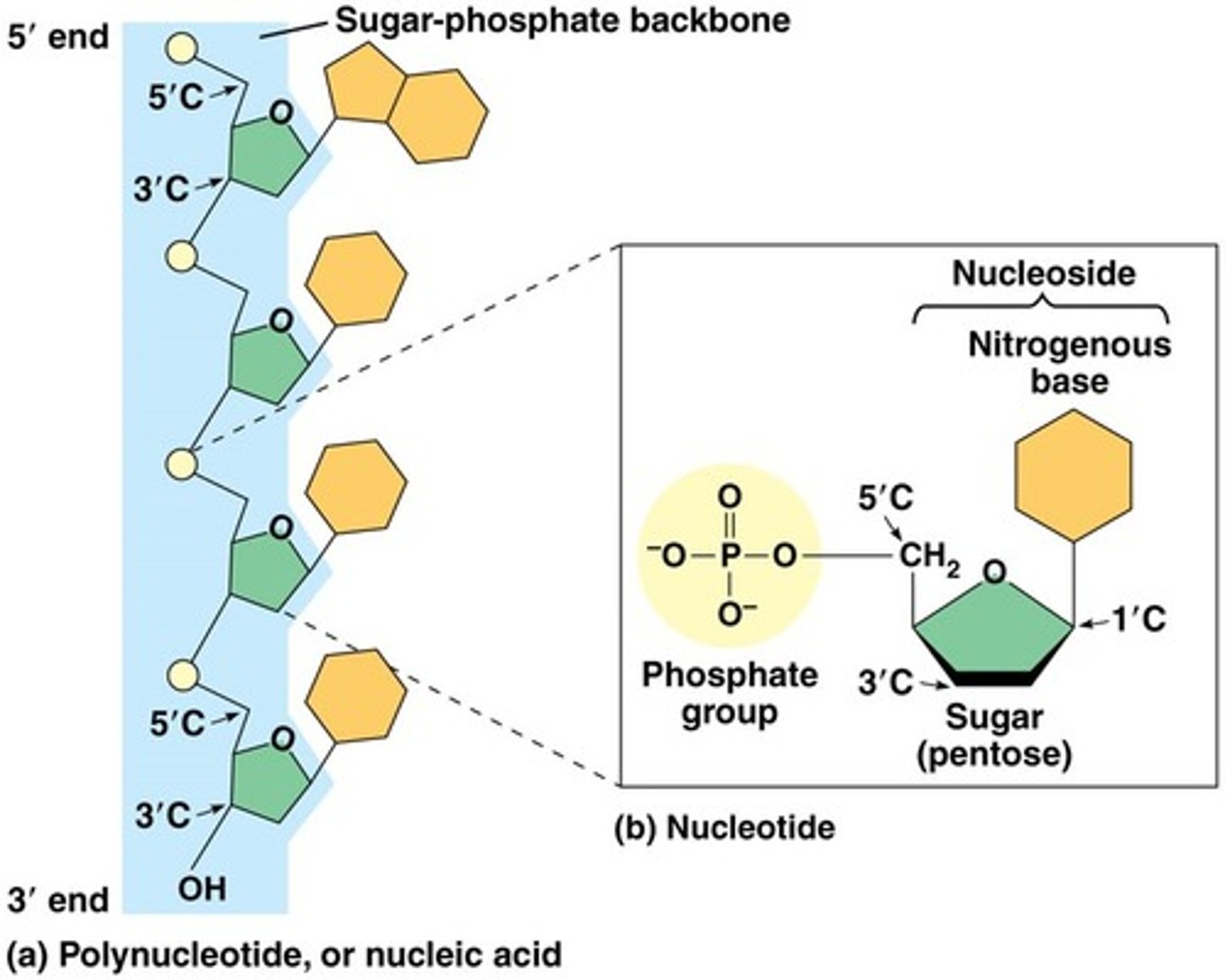
Components of nucleic acids
Nucleotides (monomer) consists of a nitrogenous base, a pentose sugar, and a phosphate
• 5' to 3' end; joined by covalent bonds between OH group on 3' C and the phosphate on the 5' of next (3' ending in OH waiting to accept next phosphate grp)
• thus making backbone: made of phosphate and sugar
• nitrogenous bases as appendages
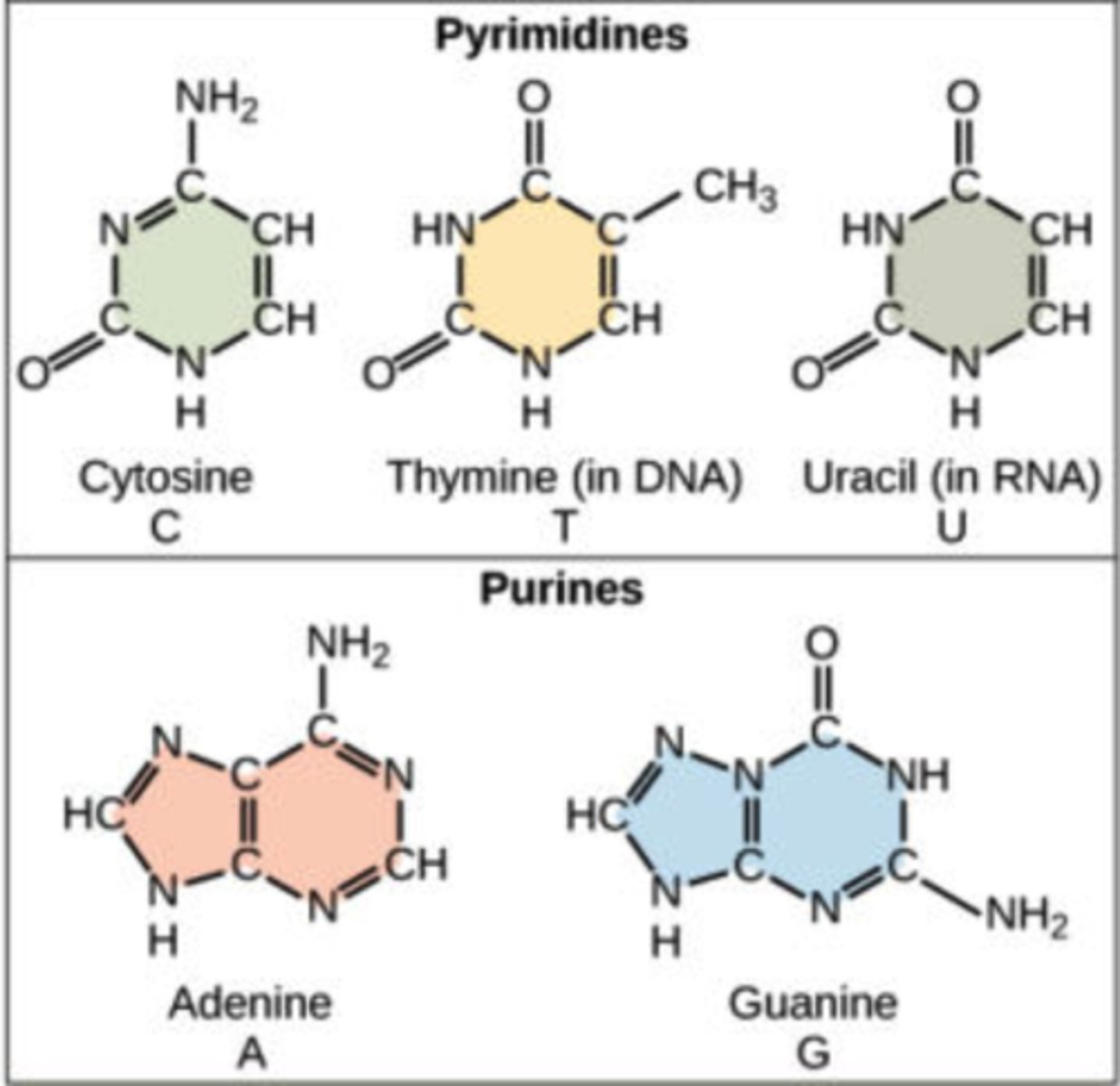
Nitrogenous bases
adenine, guanine, cytosine, thymine, uracil
Pyrimidines (6 membered rings): Cytosine, Thymine (DNA), & Uracil (RNA) (notice how U doesn't have methyl compared to T)
Purines (5+6 membered rings): Adenine, and Guanine
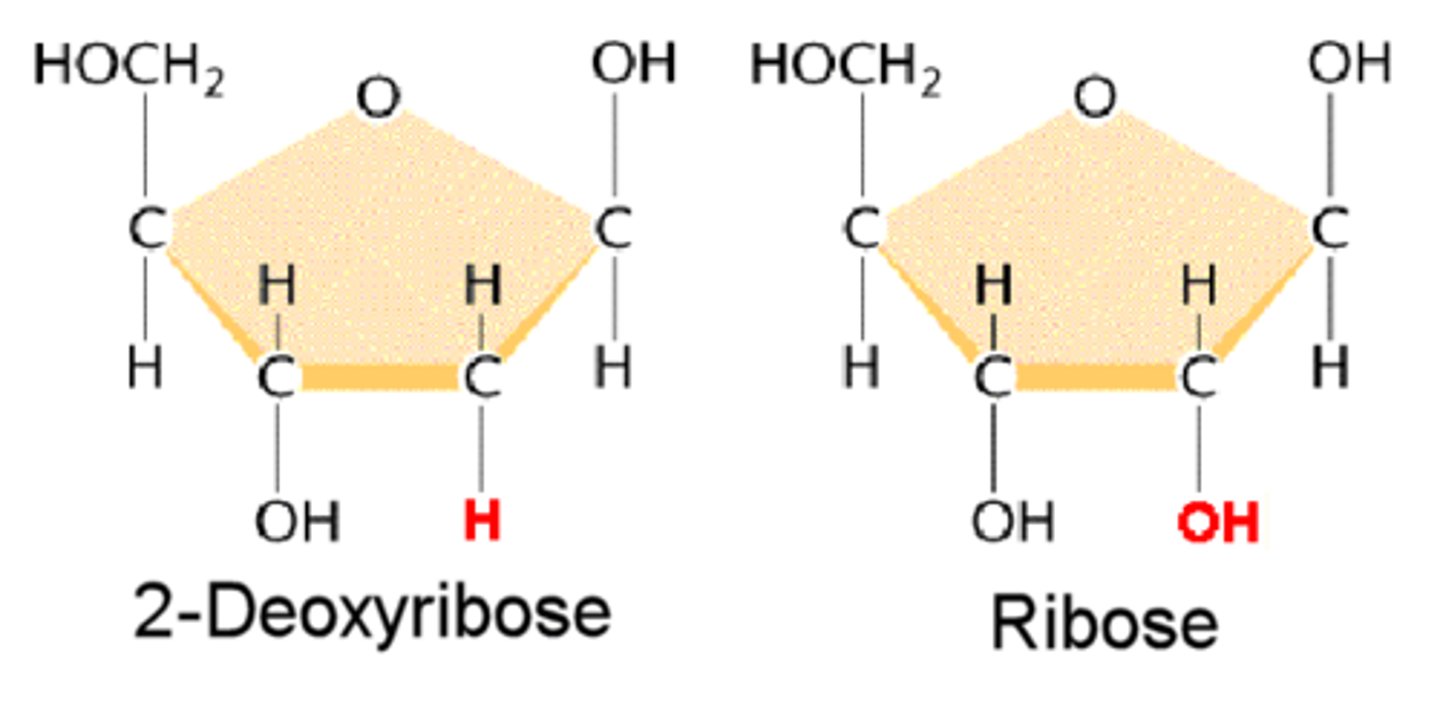
Sugar (pentose) in nucleotides
- DNA (deoxyribose) bc it lacks the 2nd C hydroxyl.
- RNA (ribose) doesn't lack/has OH. This makes it more soluble, good for cytoplasm, bad bc it can break down - bond moves susceptible
Assembly of DNA
• 2 polynucleotides spiral forming a Double Helix
• Two backbones run in opposite 5' to 3' direction, thus are antiparallel
• AT and GC (GC are stronger bc they have 3 'H' bonds, whilst AT only have 2; consequently bc they have diff # of H bonding GT and AC dont work)
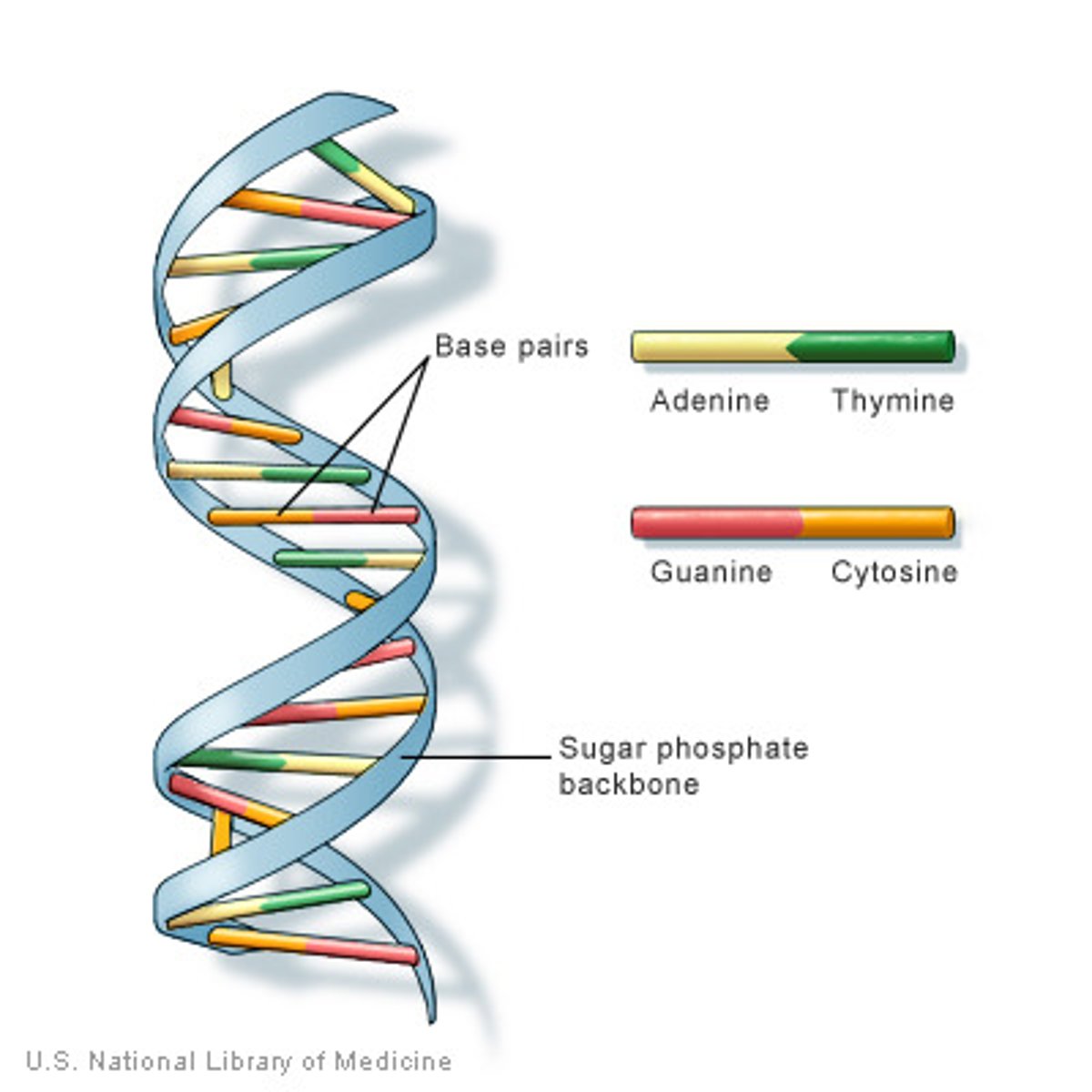
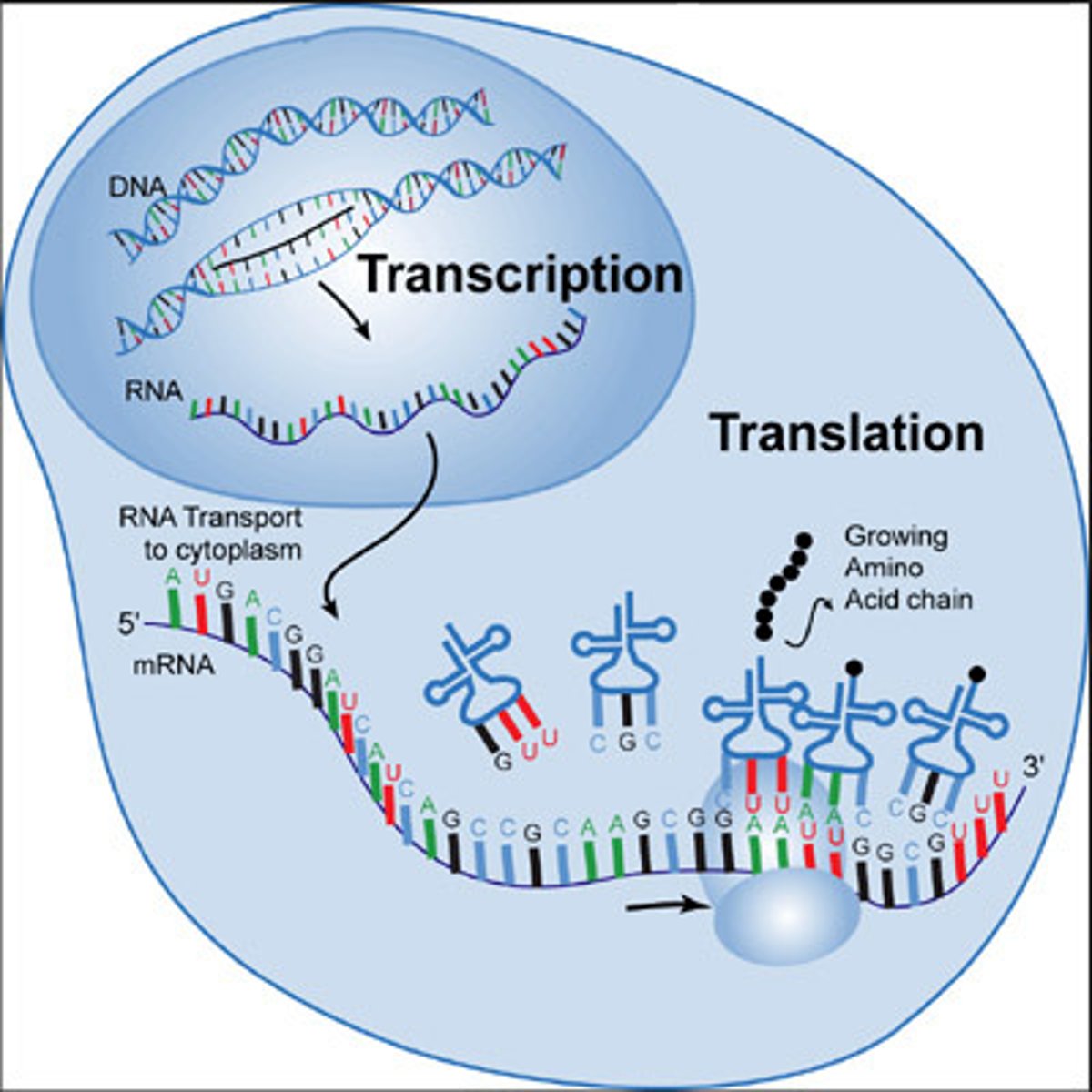
Gene to Protein
Nucleotide work together in packet of three (codons) to code for specific amino acids
• template strand of DNA may have AGT (transcription to) on mRNA it would have AGU which is seen as a Condon and is translated a protein - the amino acid serine.
Energy
Potential energy stored in food (bonds chemicals mol) (due to energy in chemical bonds) is converted in kinetic energy.
Law of thermodynamics energy can be transferred + transformed, but no created nor destroyed
Gibbs free energy
is the change in free energy ΔG
• ΔH: enthalpy or bonding energy
• ΔS: change in entropy (order/disorder - chaos favoured)
• T: temp in Kelvin
ΔG = ΔH - TΔS (or enthalpy - entropy)
• non-chaotic to chaotic favourable (-) ΔG = spontaneous (process which can be harnessed to preform work)
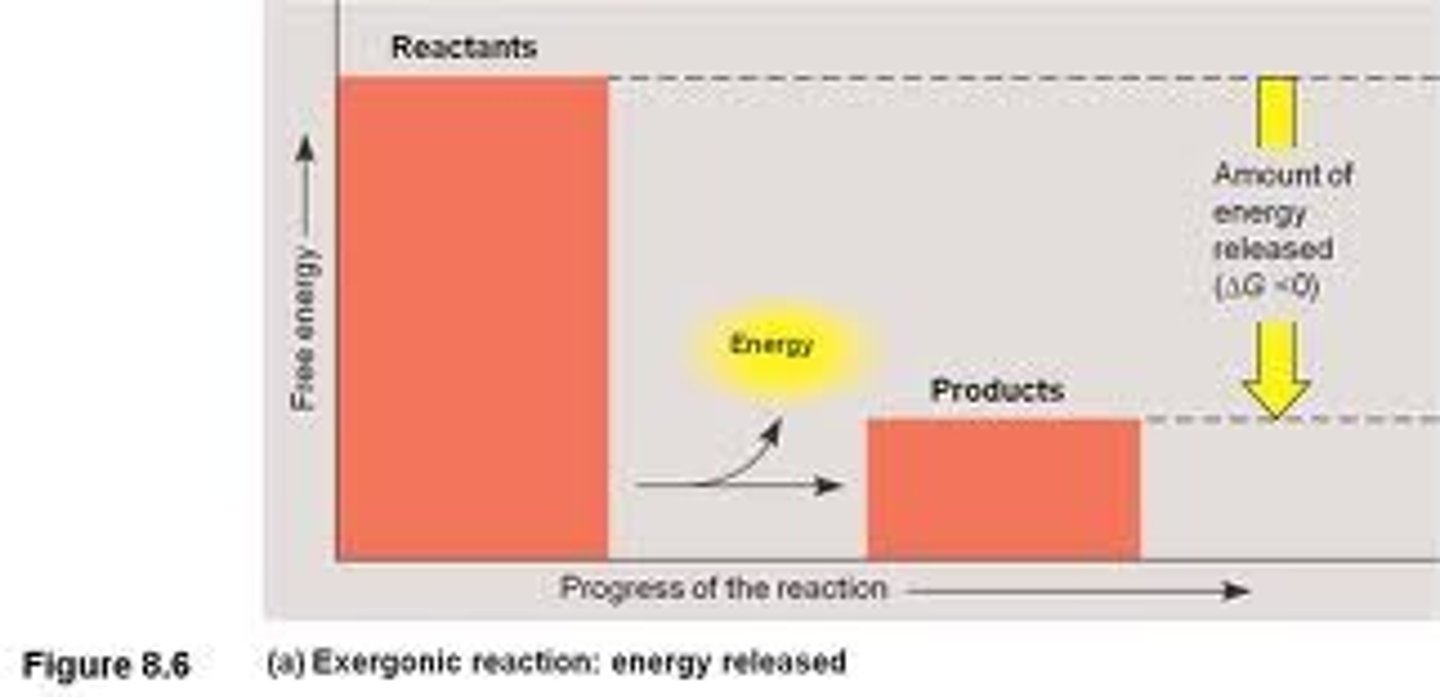
Exergonic reaction
proceeds with a net release of free energy and is spontaneous
• - ΔG
• Exer= exerts energy
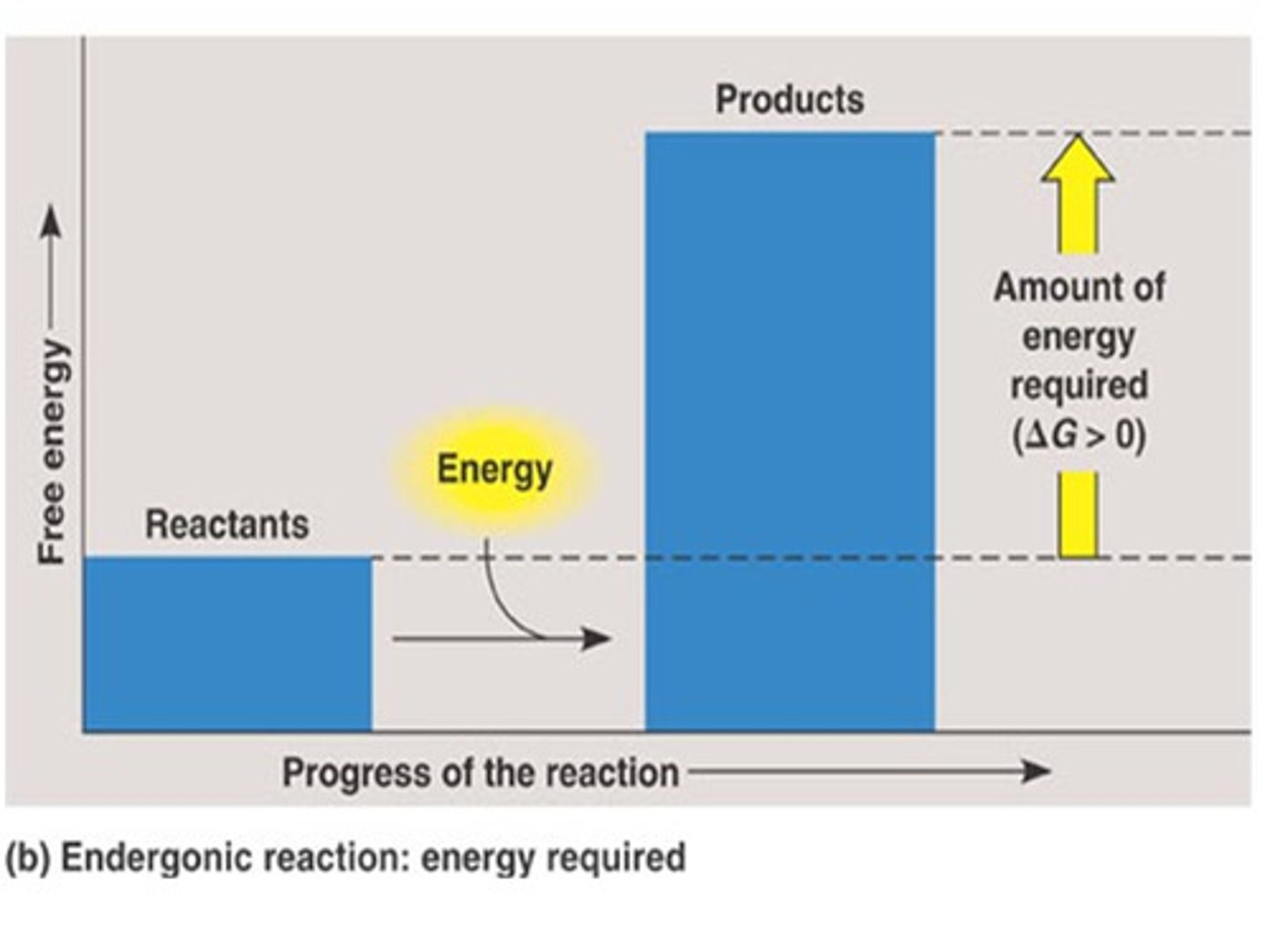
Endergonic reaction
absorbs free energy from its surroundings and is non-spontaneous
• + ΔG
• en = in/takes energy
ATP
(adenosine triphosphate) energy shuttle or bank of cell
• composed of ribose (sugar), adenine (nitrogenous base) and 3 phosphate groups
• powers cellular work by coupling exer. and endergonic reactions
• utilised to allow non-spontaneous reactions to occur
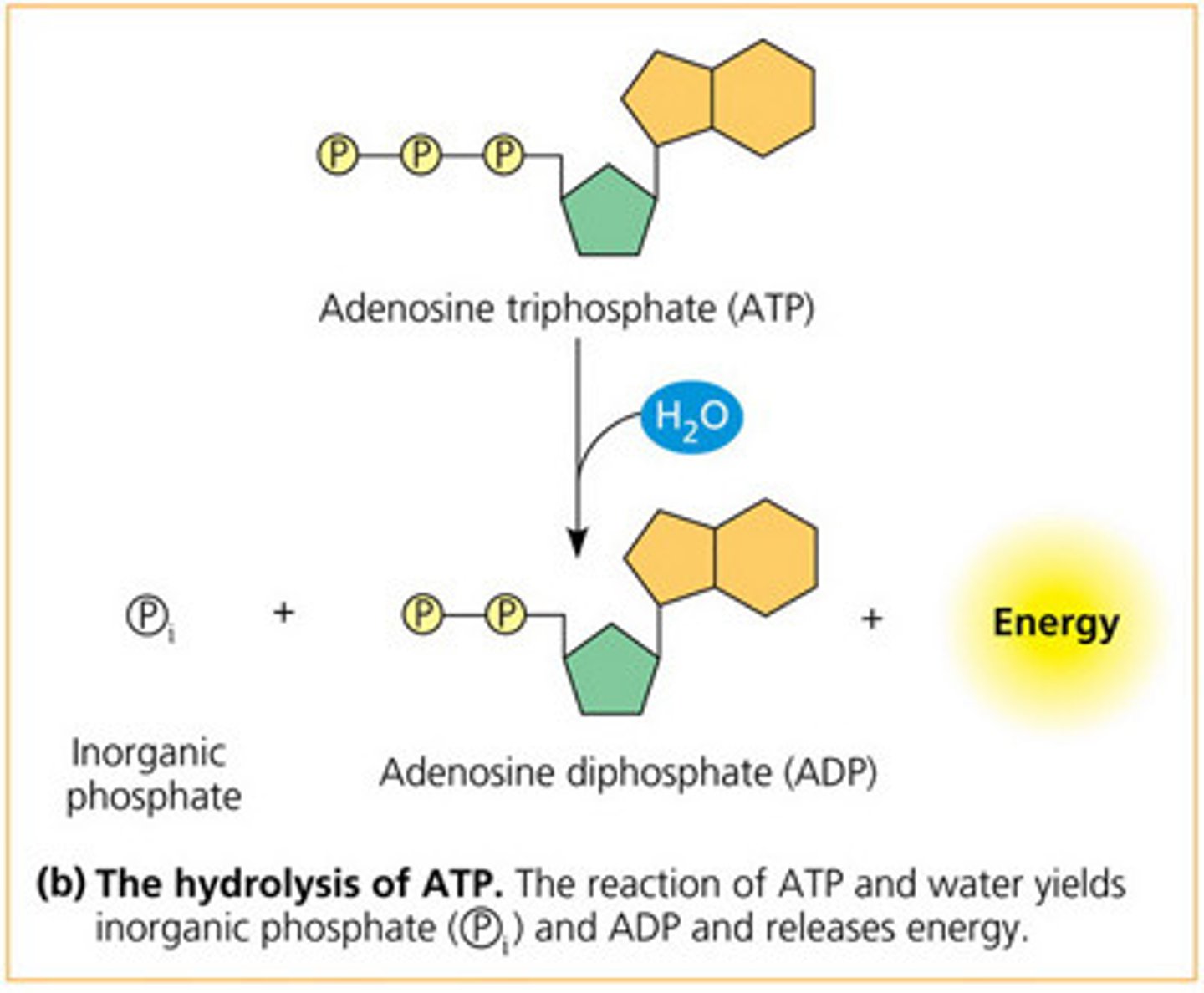
ATP hydrolysis
ATP is readily 'hydrolysed' (+H2O) breaking bond between phosphate, resulting in an inorganic phosphate + ADP (diphosphate) + energy.
• releases energy, thus is - ΔG
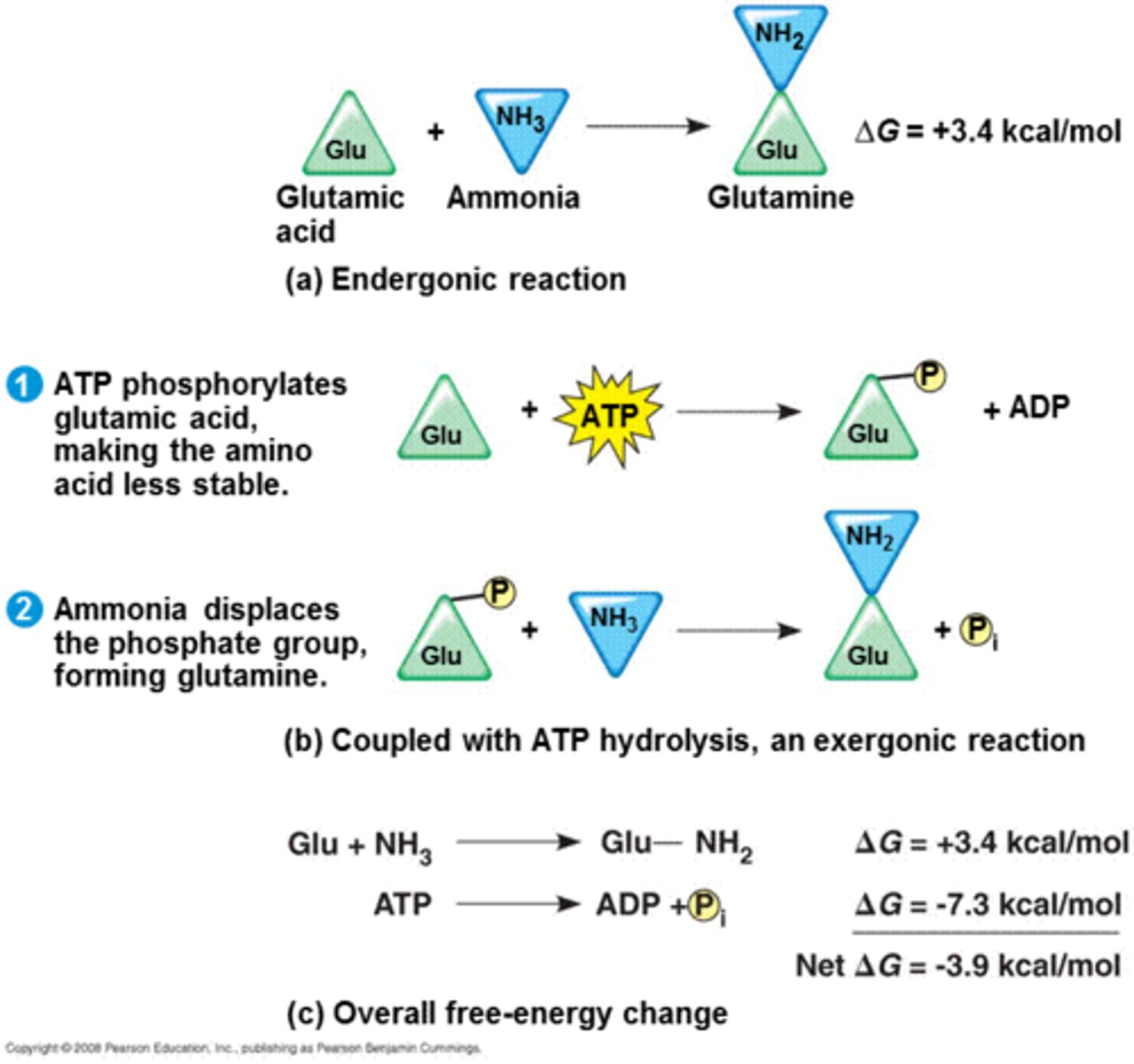
ATP drives chemical work
a) endergonic (+) ΔG; not spontaneous
b) coupled with ATP hydrolysis gets reaction to occur spontaneously, Glu has a new covalent bond formed - this is less stable; this 'P' is displaced forming glutamine - lots of energy released.
c) the coupled reaction ends up being overall spontaneous (-) ΔG
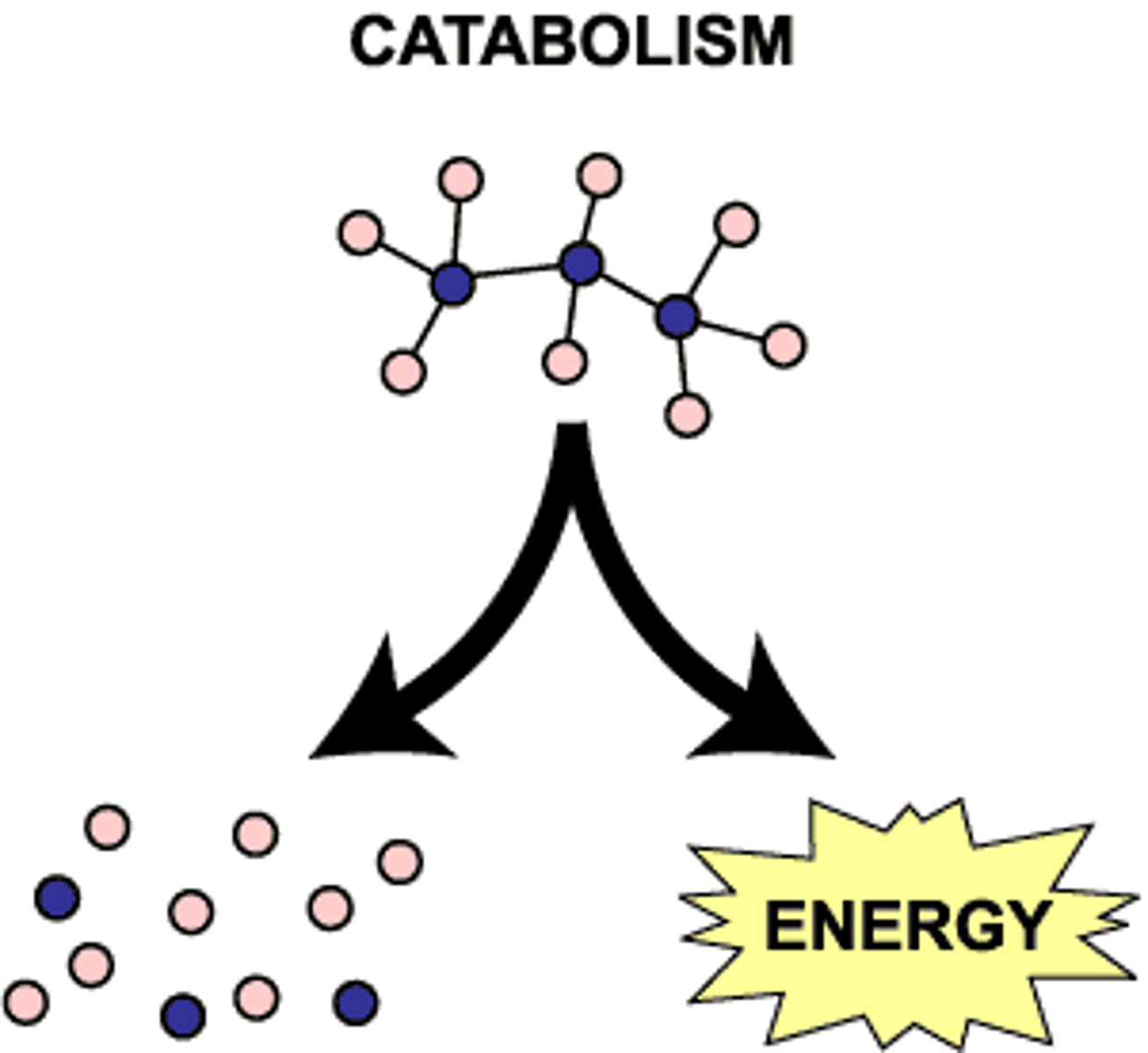
Catabolism and anabolism
Catabolism: (or catabolic pathways) release energy by breaking down complex mol into simpler compounds; e.g. cellular respiration - breakdown of glucose in presence of O, releases energy in body (catabolic pathway)
Anabolism: use of energy to build up/repair substances
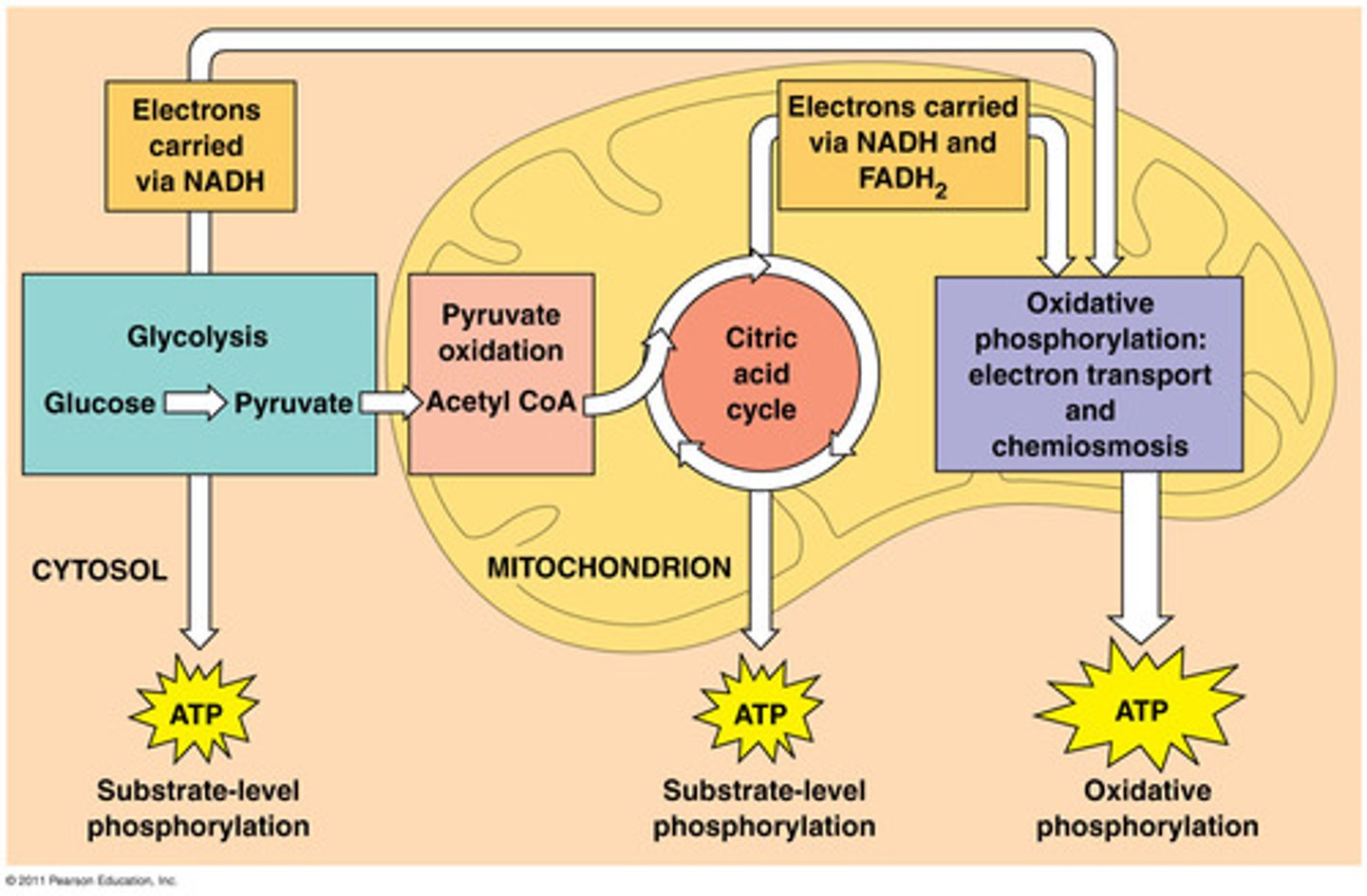
Cellular respiration
banks energy in the form of ATP mol (releases energy)

Stages of Cellular Respiration
1.Glycolysis 2. Citric acid cycle 3. Oxidative phosphorylation
approx. 38 ATP for every 1 glucose mol. that enters cellular respiration.
1. Glycolysis (outside)
breaks down glucose into two molecules of pyruvate, harvesting chemical energy.
Glucose (6 C) -(2NAD+ electrons bind, converting to NADH and 2ADP collects phosphates, becoming 2ATP)-> 3 C's (2 pyruvate produced)
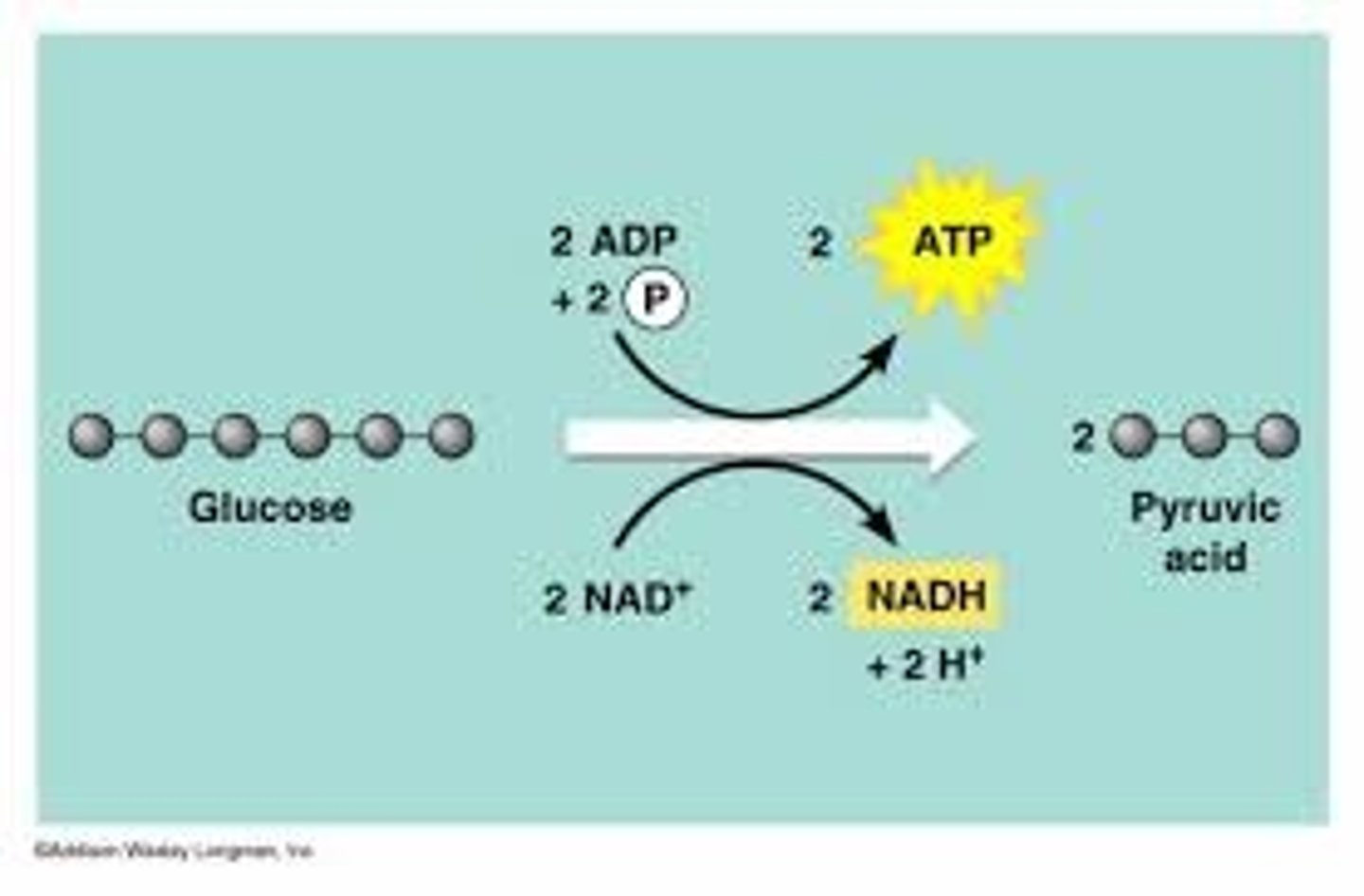
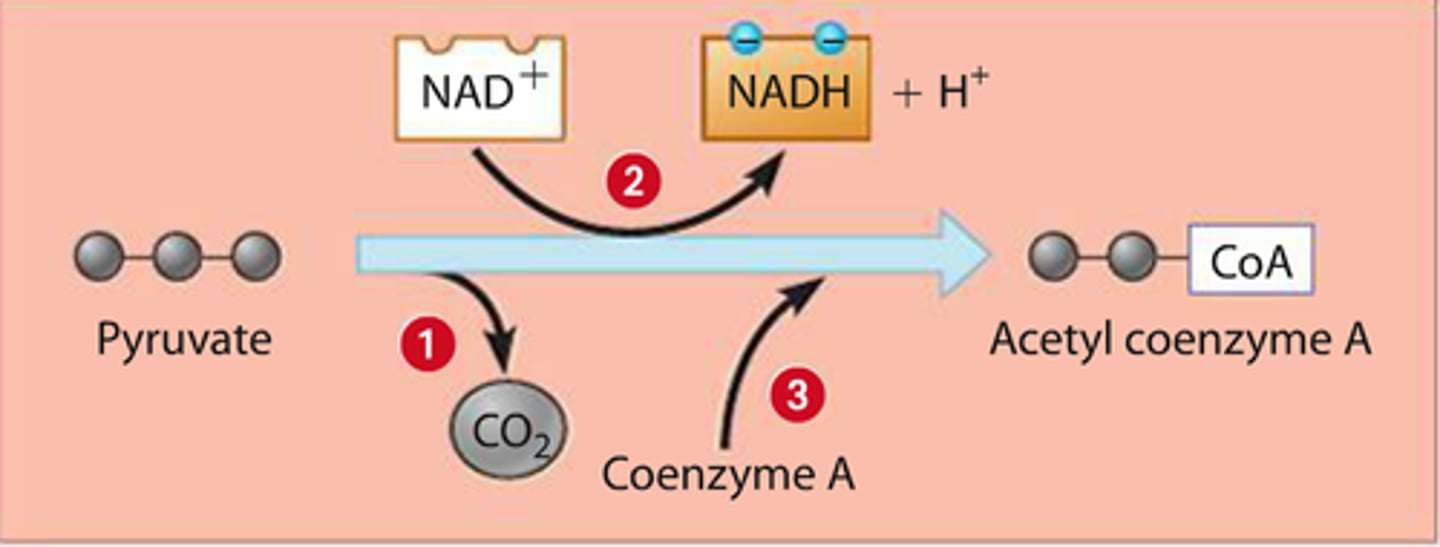
2. a) Pyruvate oxidation
pyruvate converted to acetyl CoA, linking cycle to glycolysis
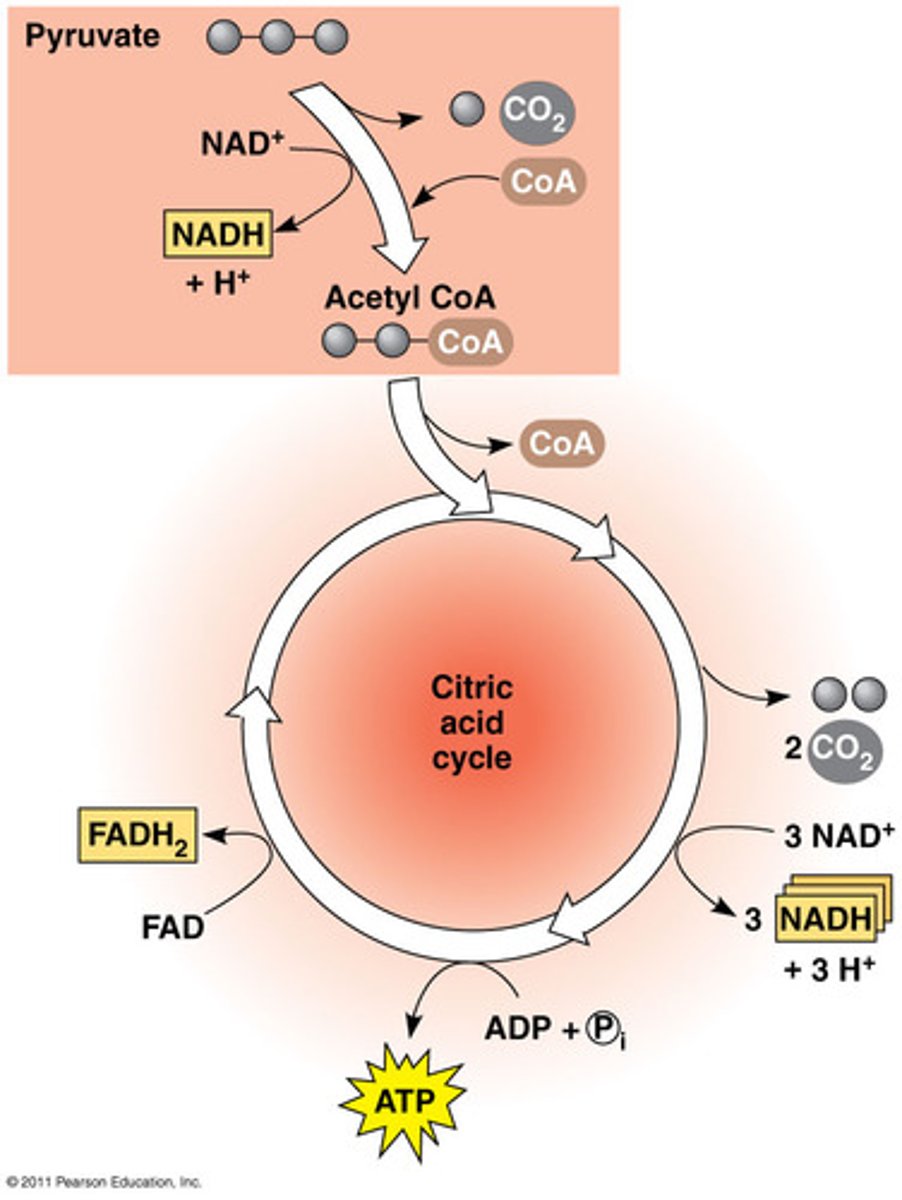
2. b) Citric acid cycle
2 C of acetyl CoA oxides and more NADH (2e; 1proton) and FADH2 (2e; 2proton) produced. This is done twice, for each pyruvate.
• completes breakdown of glucose producing a small amount of ATP, mainly supplying next stage w electrons
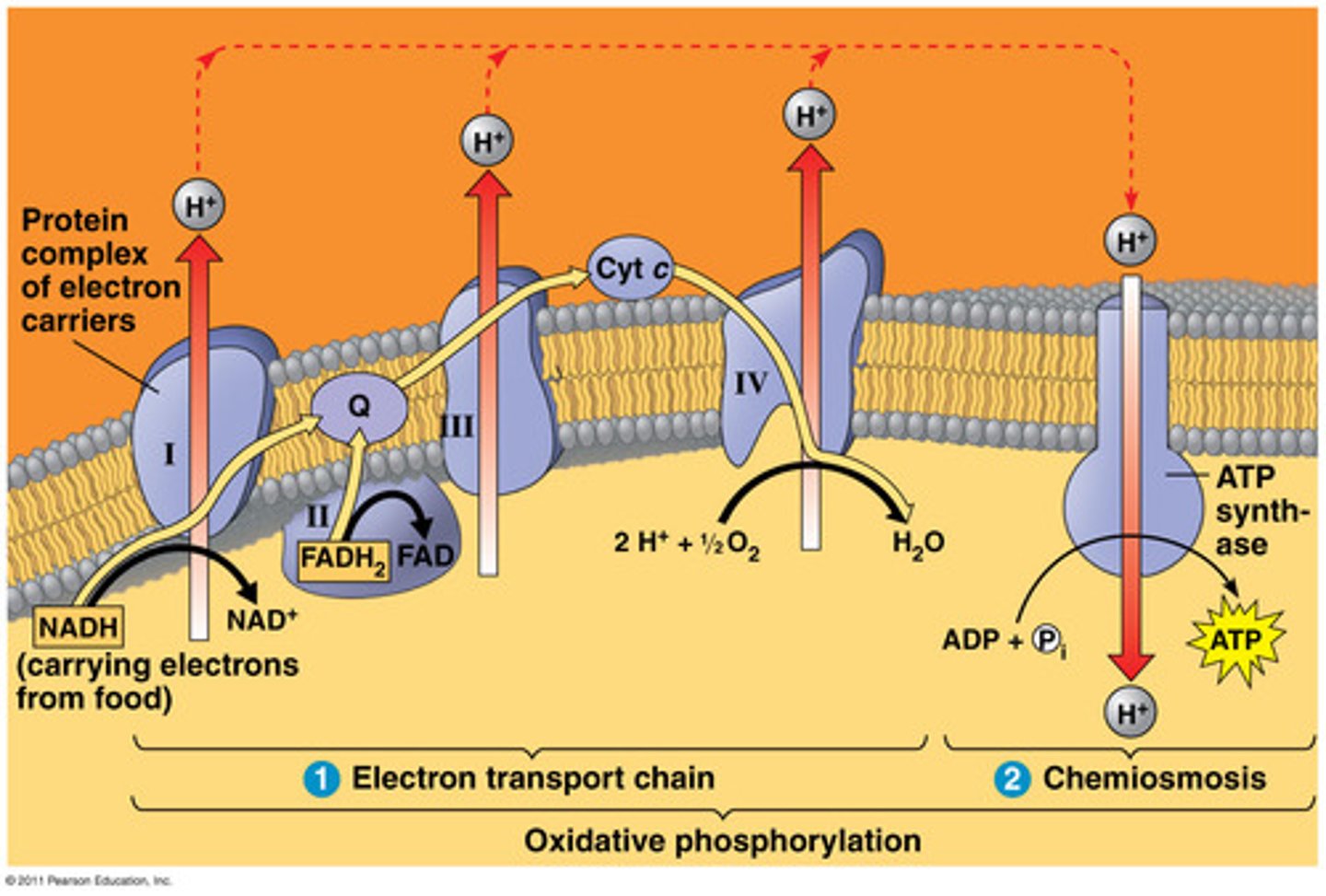
3. Oxidative phosphorylation
accounts for most of the ATP synthesis. Uses energy released by falling electrons to pump H+ across membrane. Harness energy to produce ATP.
• Electron transport chain: 4 protein complexes which they go through, creating a proton gradient enabling:
• Chemiosmosis; H+ diffuses back through inner membrane through ATP synthase complex driving synthesis of ATP (rotor -> builds charge bc outside (+) inside less, wants to come inside, potential energy)
approx. 34 ATP
Cell. resp and carbs, fats & proteins
Carbohydrates, fats and proteins can fuel cellular respiration, as they are converted to mol that enter glycolysis or citric acid cycle.
• Carbs as sugars (glycolysis)
• Fats as Glycerol fatty acids (Glycolysis, pyruvate oxidation)
• Proteins as amino acids (glycolysis, pyruvate oxidation & citric acid cycle)