BIOL121: Lecture 7-10
1/95
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
96 Terms
4 levels gene expression is regulated:
transcription
production of transcript
post-transcription
stability or function of transcript
translation
production of protein
post-translation
stability or function of protein
() proteins are proteins are always required for growth
housekeeping
() alter gene expression to help cell respond to conditions within or outside cell
regulatory proteins/transcription factors
() is a group of related genes with a single promotor and a () is a group of genes/ () that are controlled by single regulatory protein
operon
regulon
operon
Regulatory proteins come in 2 forms:
() to bind to regulatory sequences in DNA and prevent transcription of target genes
() to bind to regulatory sequences in DNA and stimulate transcription of target genes
Most () must first bind to small ligand
repressors
activators
activators
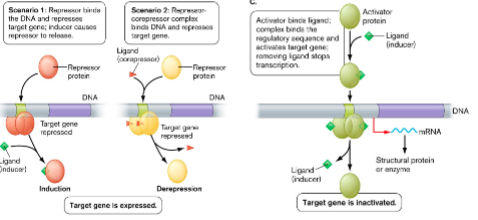
2 types of repressors:
Bind operator DNA ()
() binds to repressor causing it to release from () also called ()
Bind operator DNA ()
() disappears, repressor releases from () and target gene can be expressed also called ()
by themselves
inducer, operator, induction
only if corepressor is bound
corepressor, operator, derepression
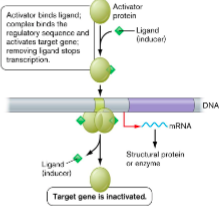
() are proteins that bind to promoters and can interact with () that’s stuck nearby to initiate ().
Some () don’t bind DNA activator sequences well unless () is present.
activators, RNA polymerase, transcription
inducer
The E. coli lac operon was discovered by () and ()
Jacques Monod
Francois Jacob
E. coli uses () for food but it can’t pass through plasma membrane () allows it entry and () is used to bring lactose inside cell.
lactose
lactose permease
proton motive force
T/F: E. coli prefers glucose over lactose
true
() encodes Beta-galactosidase
() encodes lactose permease
lacZ
lacY
T/F: Either lacZ or lacY is needed to digest lactose
false
both needed
T/F: Lactose digesting lacZYA operon of E. coli was the first gene regulatory system described
true
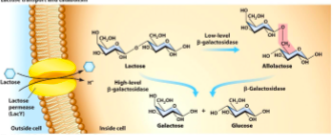
How lactose is transported and metabolized
Cells use () made by () to transport lactose into cell
Cells use () made by () to either cleave lactose into () and () or alter lactose to produce ()
lactose permease, lacY
Beta-galactosidase, lacZ, galactose and glucose, allolactose
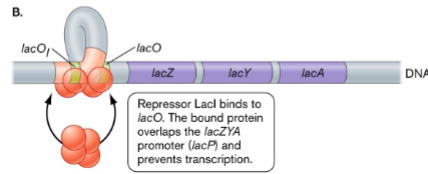
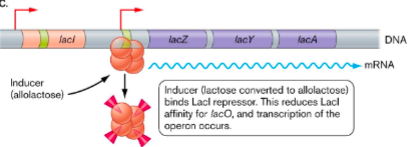
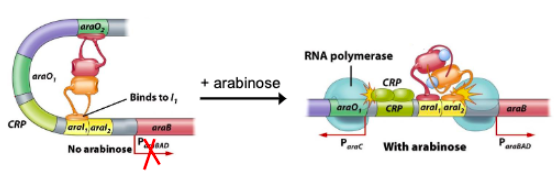
In the absence of lactose, () binds to () region and a DNA sequence bound by repressor () represses lac operon by preventing RNA polymerase from working well
lacI
operator
lacO
In the presence of lactose, () made by () when at low concentrations rearranges lactose to make inducer () which binds to () reducing its affinity to the operator allowing operon to turn on
beta-galactosidase
lacZ
allolactose
lacI
In (), an operon enabling the break down of one nutrient is repressed by presence of more favorable nutrient
catabolite repression
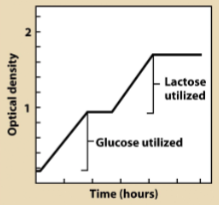
Maximum expression of lac operon requires presence of () and ()
() binds to promoter and interacts with RNA polymerase to increase rate of transcription ()
cyclic AMP (cAMP), cAMP receptor protein (CRP)
cAMP CRP, initiation
() is a sensor for glucose levels and glucose inhibits () production
phosphotransferase system (PTS)
cAMP
When glucose is present () are transferred to glucose making ().
IIA isn’t () so it inhibits cAMP production via ()
phosphates, glucose-6-P
phosphorylated, adenylate cyclase
When there is no glucose, PTS members are () meaning IIA doesn’t inhibit (), () is produced, and can activate lac operon
phosphorylated
adenylate cyclase
cAMP
Glucose transport by PTS causes () by inhibiting LacY permease activity ()
catabolite repression
inducer exclusion
Genes encoding biosynthetic enzymes are regulated by repressors called () which bind the end product of the pathway, the ()
inactive aporepressors
corepressor
The aporepressor-corepressor complex, (), can bind to operator sequence upstream of target gene or operon.
Blocks () and so transcription is mostly off
holorepressor
RNA polymerase
The () is an operon that codes for many enzymes involved in tryptophan production
tryptophan operon
When internal tryptophan levels exceed cellular needs, the excess tryptophan or () will bind to an inactive (), ().
The () then binds to an operator DNA sequence upstream of trp operon repressing the expression of structural genes by blocking ()
corepressor, aporepressor, TrpR
holorepressor, RNA polymerase
() is a mechanism to terminate transcription after it’s already started and is important in pathways where end products are ()
attenuation
low
If the concentration of tryptophan is high, the need for tryptophan biosynthesis genes is () and transcription and attenuation ().
If the concentration of tryptophan is low, the need for tryptophan biosynthesis genes is () and transcription and attenuation ().
If the concentration of tryptophan is extremely low, the need for tryptophan biosynthesis genes is () and transcription and attenuation ().
low, transcription repressed, attenuation not mentioned
moderate, transcription enabled, attenuation active
high, transcription enabled, little attenuation
Leader sequences () control biosynthesis of tryptophan and () determine if RNA polymerase can transcribe into stuctural genes
doesn’t
does
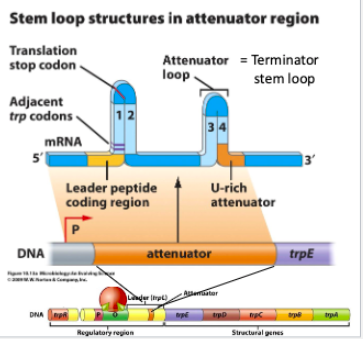
Regulation of trp operon through attenuation is based on ().
There are 4 regions of ().
() stem loops are critical
Region 3:4 () forms when trp levels are () and RNA polymerase falls off RNA before it transcribes structural genes
Region 2:3 () forms when trp levels are ()
RNA secondary structures
mRNA
2
attenuator loop, high
anti-attenuator, low
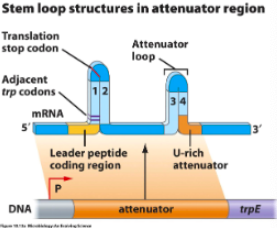
Low tryptophan levels
Tryptophan levels dictate how many () present
Ribosome pauses at () because there’s () tryptophan
Allows region () loop to form
() loop can’t form so RNA polymerase transcribes trp genes
trp-tRNAs
leader codons, no
2:3 anti-attenuator
attenuator
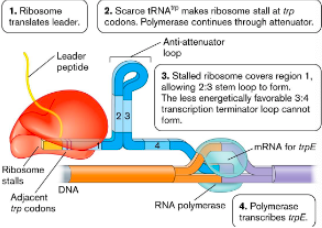
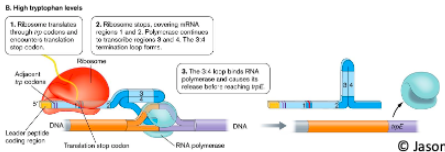
High tryptophan levels
With lots of tryptophan present, ribosome () trp codons then a stop codon
Ribosome stops and physically covers region ()
This allows () loop to form
() loop releases RNA polymerase before it can transcribe ()
translates
1:2
3:4 attenuator
attenuator, trpE
() converts arabinose into () an intermediate in a biosynthetic pathway
arabinose operon
xylulose-P
T/F: Major regulator AraC represses gene expression
false
Major regulator AraC can repress or activate gene expression depending on whether substrate is available
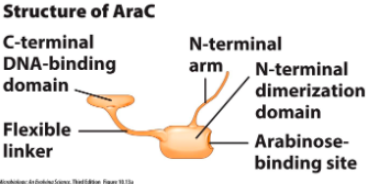
When arabinose is (), AraC shape is (). It represses expression of genes that break down arabinose.
When arabinose is (), AraC shape is (). It stimulates binding of RNA polymerase to transcribe genes.
absent, rigid and elongated
present, compact
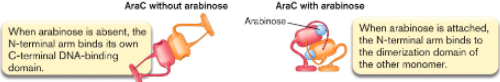
When arabinose is absent, the N-terminal arm binds to ()
When arabinose is attached, the N-terminal arm binds to ().
its own C-terminal DNA binding domain
dimerization domain of other monomer
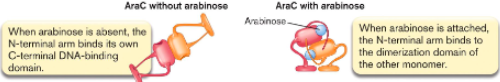
If no arabinose present, AraC acts as () and blocks ().
If arabinose is present, AraC acts as () and binds ().
repressor, transcription
activator, RNA polymerase
The () directs the expression of genes, operons, and regulons.
alternative sigma factors
Sigma Factor Regulation: Heat-shock response
() encodes heat-response sigma factor ()
() controls expression of heat-shock response genes
Secondary structures at () end of rpoH mRNA blocks ribosome, reducing () translation
rpoH, Sigma H
Sigma H
5’
Sigma Factor Regulation: Heat-shock response
Under normal conditions, () is bound by chaperones and taken for () by DnaJ, DnaK, GrpE, also known as ()
Chaperones bind () proteins to try to fix denatured proteins
Sigma H, degradation, anti-sigma factors
heat-denatured
Sigma Factor Regulation: Heat-shock response
() controls levels of Sigma H
() melts RNA secondary structure and increases translation of ()
() causes proteins to misfold which draws chaperones away from () and allows it to promot transcription of ()
heat
heat, Sigma H
heat, Sigma H, heat shock regulon
() are found within bacterial intergenic regions and regulate transcription or stability of mRNAs
small regulatory RNAs
() nature of sRNA allow these molecules to bind mRNA which can either () mRNA or make it susceptible to ()
antisense
target
degradation
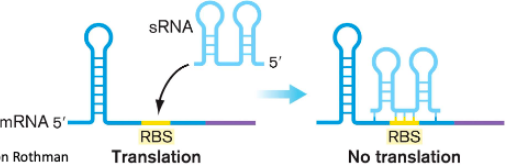
Some () bind to () and block the ribosome which () translation or () translation
sRNA
ribosome-binding site
turns off
inhibits
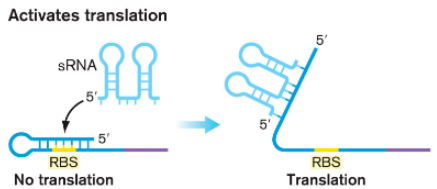
Some () bind to () and free () to () translation or () translation
sRNAs
mRNA sequence
Ribosome-binding site
allow
activate
Some () bind to an () and cause () to form. Cells don’t like () and this will turn translation () and promote ()
sRNAs
mRNA sequence
dsRNA
dsRNA
off
mRNA degradation
() refers to process where bacterial cells work together at high density and was discovered in () a bioluminescent bacterium that colonizes the light organ of Hawaiian squid
quorum sensing
vibrio fischeri
Induction of quorum sensing gene system requires accumulation of secreted small molecule called ()
autoinducer
At a certain extracellular concentration, secreted autoinducer reenters cell and binds to () which in case of Vibrio fisheri is () and activates transcription of luciferase target genes that confer bioluminescence
regulatory protein
LuxR
What triggers burst in luminescence in V. fisheri?
() synthesizes () which diffuses out of cell
When a critical level is reached, () reenters cell and binds to ()
() complex activates transcription of luciferase genes that make luminescence
LuxI, autoinducer
autoinducer, LuxR regulatory protein
LucR autoinducer
4 techniques to extract microbial DNA:
cells lysed with lysozyme to degrade cell wall, treated with detergents to dissolve membrane
proteins removed in high-salt solution
cleared lysate containing DNA passed through column containing silica resin that specifically binds DNA
extracted DNA examined with variety of analytic tools
PCR is used to () DNA sequences using () to target DNA sequence of interest and apply () and ()
copy
specific primers
cyclical heat
Taq, Thermus aquaticus DNA polymerase
() separates PCR products by size and lets you see them on agarose gel
gel electrophoresis
() is an early sequencing method still in use today and uses a mixture of deoxy- and dideoxynucleotides to sequence DNA
sanger sequencing
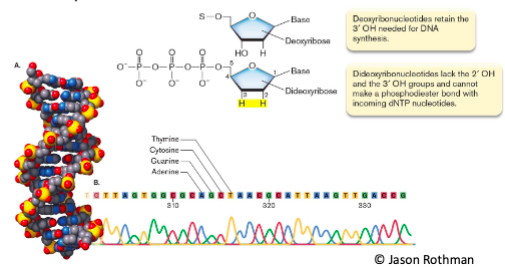
() sequencing builds chains of DNA and detects which base is added next
illumina
() sequencing is part of () sequencing and sequences target PCR fragments
amplicon
illumina
() sequencing is part of () sequencing and sequences random fragments of DNA
shotgun
illumina
4 types of genetic manipulation of microbes:
mutagenesis
random
site-directed
targeted gene editing (CRISPR/Cas9)
restriction endonucleases
gene cloning
() is used to identify genes involved in microbial processes of interest.
A population of cells is exposes to a mutagen that alters genetic code at random locations within genome
random mutagenesis
() is used when gene of interest is known.
The bacteria’s gene (allele) is replaced by plasmid containing different gene or allele.
targeted (site-directed) mutagenesis
() is useful when you want to find genes involved in some processes
isolating mutants
If a gene involved in acid resistance is knocked out then it () grow at low pH
can not
() usually involves using antibiotic resistance genes in sequences that will interrupt the gene of interest creating ()
generating mutants
gene knockout
Isolating mutants
Randomly allow () to integrate
screen mutants for desired phenotype
Specifically target a suspected gene of interest ()
screen for correct recombination event
transposon
site directed mutagenesis
3 reasons why transposons should be easy to identify:
contains antibiotic resistance genes
cells with transposons grow with antibiotics present
create large insertion mutations
knock out gene function
easy to identify sequence
mutated gene next to transposon
Create mutants by interrupting gene with ().
Identify mutated gene by using sequence DNA interrupted by (). Use known sequence of () to identify gene it interrupts.
transposon x3
Targeted gene mutagenesis using () can knockout gene of interest with another gene
homologous recombination
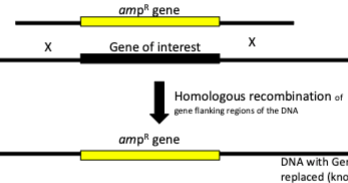
Introducing genes/DNA in bacteria on plasmid
Put gene of interest on () and transform () into () cell.
Plasmid will () once inside bacterial cell
plasmid vector, vector into competent cell
replicate itself
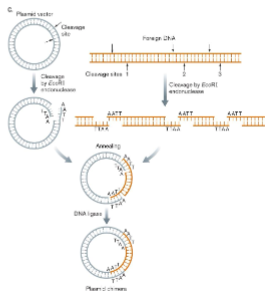
Regulation of gene can be determined by fusing () of gene of interest to () and it encodes an easily assayed protein like glowing fluorescent protein.
promoter
reporter gene
2 types of reporter fusions:
operon fusion/transcriptional fusion
shows transcriptional control of gene
gene fusion/translational fusion
shows transcriptional and translational control of gene
What is a virus?
noncellular particle that must infect host to replicate
T/F: All viruses are obligate intracellular parasites which means they depend on host metabolism
true
() is the virus particle and consists of nucleic acid and protein coat
virion
T/F: Viruses sometimes have a protective protein coat called a capsid
false
always
Virus genomes only contain information for () and ()
taking over host cell
making viral proteins
The viral () packages the genome and delivers it into host cell.
The viral () is composed of repeated protein subunits
capsid x2
Capsid + genome = ()
nucleocapsid
Different viruses make 2 different capsid forms:
symmetrical and asymmetrical
Viruses can be classified by 4 characteristics:
shape
structure
genome composition
replication mechanism
() have a structure that exhibits rotational symmetry and is a polyhedral with 20 identical triangular faces
icosahedral viruses
The () is sometimes enclosed in an () formed from the host cell’s membrane
capsid
envelope
Between the envelope and capsid you’ll find () proteins
tegument
() have a capsid that consists of a long tube of protein with genome coiled inside and vary in length depending on genome
filamentous viruses
T/F: Icosahedral viruses include bacteriophages as well as animal and plant viruses
false
filamentous viruses?
In () capsid monomers form tube around genome
filamentous viruses
() have complex multipart structures and include Tf bacteriophages that have () head and () neck
asymmetrical viruses
icosahedral
helical
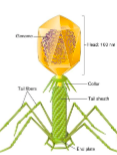
() virus has a genome that is surrounded by several layers including a core envelope studded with spike proteins and an outer membrane
pox
Viral Life Cycle:
attach to host cell
get viral genome into host cell
replicate genome
make viral proteins
assemble capsids
release progeny viruses from host cell
Viral genomes
DNA/RNA can be:
Include genes encoding viral proteins:
ss/ds, linear/circular
capsid, envelope proteins, any polymerase not in hose
What Baltimore family is herpes and small pox?
1 dsDNA
What Baltimore family is parvovirus and geminiviruses?
2 ssDNA
What Baltimore family is rotavirus and reoviruses?
3 dsRNA
What Baltimore family is COVID, HepC, cold virus?
4 +ssRNA
What Baltimore family is flu, rabies, ebola?
5 -ssRNA
What Baltimore family is HIV?
6 RNA retrovirus, reverse transcriptase
What Baltimore family is HepB?
7 DNA pararetrovirus