CHEM 114A Midterm 2 Flashcards
1/14
Earn XP
Description and Tags
Lecture 5 Slide 41 to Lecture 11 Slide 10, Chapters 5-8
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
15 Terms
Rapid pace of discovery dependent on several essential techniques:
Restriction enzymes
Blotting techniques
DNA sequencing
Solid-phase synthesis of nucleic acids
Polymerase chain reaction (PCR)
Restriction enzymes
Act as precise scissors for cutting specific DNA sequences
Recognize specific sequence in double-stranded DNA and cleave both strands
Cleavage possess 2-fold rotational symmetry
Recognized sequence is palindromic (same when read in forward and reverse direction)
CC GC / GG
GG / CG CC
Enzyme names consist of 3-letter abbreviation of host org, followed by strain designation and Roman numeral
Used to cleave large DNA molecules into smaller sizes that are more amenable to analysis
Pattern of fragments produced can also serve as fingerprint of that specific DNA molecule
Visualizing restriction enzyme cleavages
Gel electrophoresis used to separate different sized fragments
DNA visualized by staining with ethidium bromide and using UV light, which causes ethidium bromide to fluoresce orange
Every DNA sequence has characteristic cleavage pattern
Blotting techniques
Allows separation and identification of specific proteins and nucleic acids using gel electrophoresis
Southern blotting
Used to identify restriction fragments on a gel
Radioactively labeled complementary single-stranded DNA can be bound to gel with this technique
1) Add DNA fragments to agarose gel and complete electrophoresis
2) Transfer DNA by blotting to nitrocellulose sheet
3) Add 32^P-labeled DNA probe
4) Autoradiography
5) DNA probe is revealed on autoradiogram
Northern blotting
Identifying an RNA sequence on a gel using complementary radiolabeled DNA probe
Western blotting
Use radiolabeled antibody to identify protein on a SDS-page gel
DNA sequencing
Genome sequences of entire organisms can be determined
Frederick Sanger invented Sanger dideoxy method
Based on generation of DNA fragments whose length is determined by identity of last base on the sequence
Collection of fragments obtained through controlled termination of replication
Uses dideoxy analogs of each NTP to stop replication
4 separate reaction mixtures each contain small amount of 1 dideoxy analog (these are randomly incorporated)
DNA polymerase used to make complement of particular DNA which has been denatured to single strands
Incorporation of analog prevents further chain elongation
Flouresence more commonly used now instead of radioactive reagents
Modern DNA sequencing can sequence >10^6 bases per day
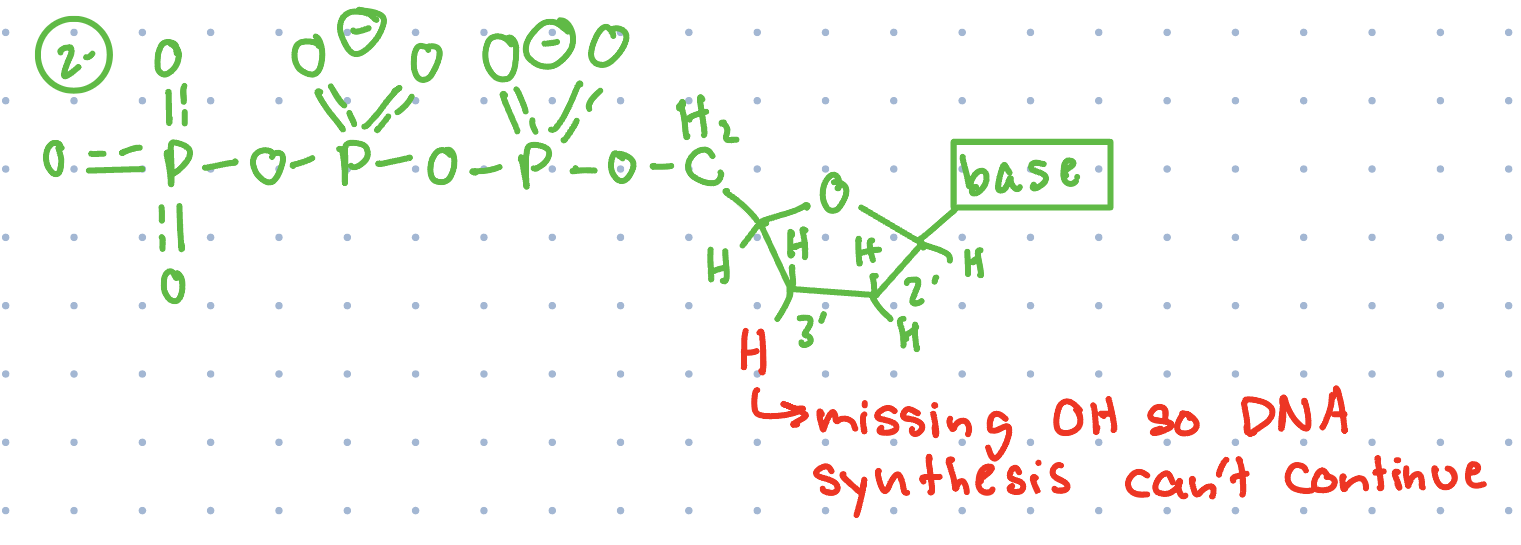
DNA sequencing example (with adenosine)
1) Have 3’ end DNA to be sequenced and 5’ end complementary primer
2) Add DNA polymerase I + labeled dATP, TTP, dCTP, dGTP + dideoxy analog of dATP
3) Add nucleotides to primer that match DNA
4) At points where there is T in DNA (sometimes skipped and instead add in normal dATP), sequenced strand adds in dideoxy analog of dATP, ending replication and making a fragment (the dATP analog will only be inserted where a T was located in DNA being sequenced)
5) This is done through the whole sequence so that you get a variety of random fragments
6) Radiolabeled dideoxy sequencing reactions are run on a denaturing polyacrylamide gel (8%) that has lanes for T, A, G, and C
7) Different sized fragments are separated
8) Sequence is read from the pattern of chain termination (longer chains closer to 3’ end will be longer and run slower on gel so will be near top, shorter chains closer to 5’ end will be near bottom)
Solid-phase synthesis of nucleic acids
Specific sequences of nucleic acids can be synthesized and used to identify or amplify other nucleic acids
DNA strands can be chemically synthesized by sequential addition of activated monomers
Occurs in 3’ to 5’ direction
Size limitation of about 100 bases
Allows generation of short DNAs that can be used to amplify genes using PCR
Can be used to make DNA probes for blotting
Recent: multiple DNAs synthesized corresponding to much larger sequence and joined to form new tailor-made genes
PCR
Polymerase chain reaction
Allows billion-fold amplification of small quantities of specific DNA to obtain sufficient quantities for further characterization
Kary Mullis developed in 1984
Flanking sequences f DNA target must be known
Short DNA must be synthesized that is complementary to these flanking sequences - primers
Components needed for PCR
Pair of primers that hybridize and are complementary to flanking sequences of DNA target
All four dNTPS (A, C, T, G)
Heat-stable DNA polymerase (rom thermophilic bacteria)
Thermal cycler (machine that cycles between different temperatures)
PCR cycle
1) Strand separation - 2 strands of target DNA molecule are separated by heating at 95 C
2) Hybridization of primers - solution is quickly cooled to 54 C to allow primers to hybridize to 5’ and 3’ ends of target DNA
3) DNA synthesis - solution heated to 72 C = optimal temp for DNA synthesis by Taq DNA polymerase (heat stable polymerase from thermophilic bacterium Thermus aquaticus)
Cycle repeated about 20-35 times
After n cycles, sequnce should be amplified 2^n fold
Amplification is a millionfold after 20 cycles and billionfold after 30 cycles
Where PCR is used
Medical diagnostics - very small amount of bacterial and viral DNA detected using PCR (even if can’t sequence from this small amount, can amplify so that can sequence), can detect HIV at early stage
Forensics - DNA from crime scene can be greatly amplified for further identification
Molecular archeology - DNA from extinct orgs (bones) can be amplified for evolutionary studies (Neanderthal genome)