4 - PCR Detection Methods
1/21
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
22 Terms
Types of PCR
• Allele specific PCR
• Conventional PCR
• Digital
• Multiplex
• Nested
• Real time
• Reverse transcriptase
Main principles of PCR
• New chain requires primer as polymerase can only extend existing double strand, oligonucleotides used, approx 20
• Extension only proceeds in 5’ to 3’ direction (polymerase only adds to 3’)
• Two strands are anti-parallel
Requirements for conventional PCR
• Target DNA (template)
• Buffer, magnesium (co-factor)
• Thermostable DNA polymerase
• Deoxynucleotide triphosphates (dATP, dTTP, dCTP, dGTP)
• Forward and reverse primers complementary to target sequence
Allele specific PCR
• Detect specific mutations e.g. cystic fibrosis CFTR gene
• DNA polymerase distinguishes between match and mismatch at 3’ end, primer designed with mismatch base corresponding to mutation
• Sickle cell caused by mutation in HBB gene which codes for B-globin (glutamine to valine)
• To detect, one common primer is used and other specific for one allele (A WT, or T mutant), two separate reactions set up
Nested PCR
• Variation of standard PCR that enhances specificity and yield of amplicons
• Two primers used, outer primers flank region of DNA containing amplicon of interest, nested primer corresponds to the precise region within the DNA to be amplified
• Outer primers used in first round, product serves as template in second round with nested primers
Multiplex PCR
• Allows for concurrent amplification of multiple targets with different primers
• Requires two or more probes that can be distinguished from each other
Real time PCR (q-PCR)
• Allows for analysis of PCR products in real time, allows for quantification in exponential phase, very sensitive
• LightCycler Pro
• Two types of flourescent technologies can be used - intercalating dyes or 5’ nuclease probes
Intercalating dyes
• Incorporate into minor grooves of dsDNA and only then do they fluoresce
• Emission will increase in each cycle during the extension phase, low or absent during denaturation
• Advantages - can be used with any pair of primers, cheaper and requires less knowledge than 5’ nuclease probes
• Disadvantages - specificity diminished due to risk of amplifying non specific products or primer dimers, melt curve analysis must be performed to distinguish between specific and non specific products
Melt curve analysis
• Temperature gradually increased from 60 to 95, fluorescence monitored continuously
• High emission at low temperatures when PCR products are double stranded
• Products of different lengths or GC content will have different melting temperatures creating distinct peaks
• Primer dimers have low melting temperature (peak at lower temp)
5’ Nuclease probes
• Double labelled oligonucleotide probes with two reporter and quencher fluorochromes which emit a signal upon cleavage
• Based on principle of fluorescence resonance energy transfer (FRET), energy transfer between two light sensitive molecules
• Non-extendable at 3’ end, designed to anneal to target internally of the primers
• Upon binding, probe is degraded by exonuclease activity of polymerase
• Increase in emission directly proportional to amount of product
Data acquisition
• Point in exponential phase when amplification of product reaches detection level
• The more input DNA the less cycles it will take to make a specific amount of amplicon
• Threshold is chosen based on the variability of the baseline, 10 times the SD of baseline
• Threshold cycle (ct) values are then calculated by determining the point at which fluorescence exceeds this threshold
Multiplex PCR
• Good if there is a limited amount of input material
• Up to 6 different colours
• Limited to to existing knowledge of microorganisms genome, no distinction between live and dead organisms, unexpected mutations not detected
• Used in combination with cultures
Reverse transcription q-PCR
• Used for quantification of mRNA expression
• RNA must be good quality, consistent conditions, absence of co-amplifying genomic DNA
• Normalisation - correction for experimental variations in RT and PCR amplification efficiency, reference gene used
RT q-PCR method
• RNA converted to complementary DNA (cDNA) by enzymatic RT reaction
• Creates complementary strand of DNA based on RNA sequence, DNA primer required, ssDNA then used as template to synthesise dsDNA (cDNA)
• Entire mRNA sample can be converted to cDNA by oligo(dT) primers that anneal to poly A tails of mRNA
• Random hexamers can also be used, primers 6 nucleotides in length, randomly bind to sample at any location
Data quantification using a standard curve
• Sample of a known concentration is used to make serial dilutions
• Ideal slope is -3.3 (up to 3.6) for 10 fold dilutions, measures efficiency
• CT values are plotted against quantity, unknown amount of input can be measured
Data quantification using the comparative threshold cycle method
• Calculate relative expression levels compared to a calibrator (control)
• Value of unknown target is normalised to a reference gene
• The efficiency of the PCR amplification for the target and reference gene must be approximately equal
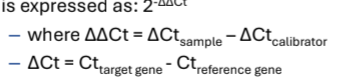
Digital PCR
• Based on limited dilution PCR
• Each individual reaction either contains a DNA molecule or not, gives yes or no signal, some may contain multiple
• At low lambda values most reactions contain no target molecules and few a single copy
• At high lambda values nearly all reactions are positive with multiple copies of the target molecule
Advantages and disadvantages of dPCR
• Advantages - absolute quantification with no standard curve, very sensitive, increased resistance to inhibitors, fast TAT (2h)
• Disadvantages - lower dynamic range, some samples may need further dilution if initial input was high
Digital PCR platforms
• Plate based systems - sample dilutes, partitioned into thousands of individual reactions, end-point PCR performed on partitions, readout and absolute quantification
• Emulsion based systems - reaction chambers consist of small water droplets separated by oil (droplet digital PCR)
Data visualisation
• 1D plots - one axis, relative fluorescence units (RFU), any dots above threshold are positive partitions
• 2D plots - multiplexing 2 reactions, RFU on both axis (different colours), negative in bottom left, double positive in upper right
Secondary analysis
• Based on results of the absolute quantification
• Mutation detection, copy number analysis, gene expression analysis
Applications of PCR assays
• Microbiology - pathogen detection, design primers in highly conserved regions due to high mutations, load can be determines
• SNV - one set of primers with two allele specific fluorescent labelled probes used e.g. FVL mutation
• Foetal RHD genotyping - cell free foetal DNA in maternal circulation, qPCR used to amplify RHD gene
• Measurable residual disease - qPCR for detection of MRD
• Chromosomal translocations - tumour specific PCR targets, primers designed to anneal to opposite sides of the breakpoint