BIO130
0.0(0)
Card Sorting
1/188
There's no tags or description
Looks like no tags are added yet.
Study Analytics
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
189 Terms
1
New cards
Extracellular matrix
specialized material outside the cell
2
New cards
lysosome
degradation of cellular components that are no longer needed. not in plants
3
New cards
Cell wall
cell shape, protection against mechanical stress. not present in animal cells, could be in bacteria
4
New cards
vacuoles
degradation- similar to animal lysosome, or storage- small molecules and proteins (depends on type)
5
New cards
Chlotoplast
site of photosynthesis
6
New cards
cytoplasm
contents of the cell outside the nucleus, includes membrane bound organelles
7
New cards
cytosol
aqueous part of the cytoplasm, does not include the organelles, does include the ribosomes and cytoskeleton
8
New cards
lumen
inside of membrane bound organelles
9
New cards
6 functions of cellular functions at membranes
compartmentalization, scaffold for biochemical activities, selectively permeable barrier, transport solutes, respond ot external signals, interactions between cells
10
New cards
Fluid mosaic model of the membrane (singer and Nicolson 1972)
includes many proteins and many lipids, most proteins and most lipids can move around (fluid)
11
New cards
membrane bilayer
two leaflets/laters, composed of lipids with hydrophyllic or polar head group, hydrophobic tails (amphipathic)
12
New cards
amphiathic
different chemical/biophysical properties on each side
13
New cards
membranes are composed of
different lipids: phospholipids, sterols, glycolipids
14
New cards
What are the 6 functions that occur at membranes?
* compartmentalization
* scaffold for biochemical activities
* selectively permeable barrier
* transport solutes
* respond to external signals
* interactions between cells
* scaffold for biochemical activities
* selectively permeable barrier
* transport solutes
* respond to external signals
* interactions between cells
15
New cards
Membrane bilayers are composed of
Hydrophillic head and hydrophobic tails that arrange as a lipid bi-layer, they are amphipathic
16
New cards
amphipathic
made up of 2 diff biochemical/biophysical properties of different side
17
New cards
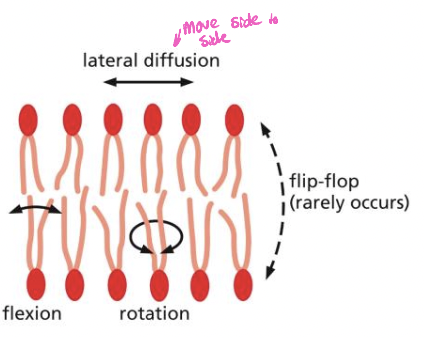
Fluid Mosaic Model of the membrane
* proposed in 1972 by singer and nicolson
* involves free movement of phospholipid in the membrane
* each can diffuse laterally, rotate, flex all rapidly
* can flip-flop but is rare (flip to other side of bi-layer)
* this movement is regulated to allow for transport, signaling etc
* involves free movement of phospholipid in the membrane
* each can diffuse laterally, rotate, flex all rapidly
* can flip-flop but is rare (flip to other side of bi-layer)
* this movement is regulated to allow for transport, signaling etc
18
New cards
Membrane lipids
phospholipids, sterols, glycolipids
19
New cards
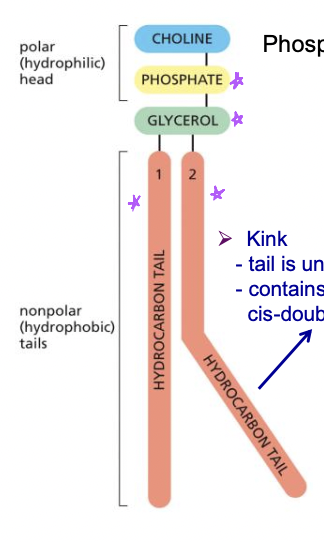
phospholipids
* most have glecerol group =phosphoglycerides
* include: group at the top-phosphate-glycerol-hydrocarbon tail
* when tail is saturated, its straight, when unsaturated has a kink meaning theres a cis-double bond
* include: group at the top-phosphate-glycerol-hydrocarbon tail
* when tail is saturated, its straight, when unsaturated has a kink meaning theres a cis-double bond
20
New cards
Membrane bi-layers
In an aqueous environment, phospholipids will spontaneously form a bi-layer
* polar heads interact with water
* hydrophobic tails interact with eachother
* polar heads interact with water
* hydrophobic tails interact with eachother
21
New cards
Phospholipid bilayers: sealed compartments
* move to create circle to get fully away from water otherwise the ends are touching water
* this is energetically favourable
* this is energetically favourable
22
New cards
Liposomes:
* artificial lipid bilayers
* used to study lipid properties, membrane protein properties, drug delivery
* used to study lipid properties, membrane protein properties, drug delivery
23
New cards
factors that effect membrane fluidity
* temperature
* lower temperatures are less fluid (freezes)
* too cold can become unsaturated
* composition
* phospholipid saturation: cis-double bonds increase fluidity at lower temperatures (reduce tight packaging)
* phospholipid tail length: shorter hydrocarbon tails increase fluidity at lower temperatures (lipid tails interact less)
* shorter tails allow for more movement cause dont hit eachother
* lipid composition: addition of cholesterol in animal cell membranes. stiffens membrane, less permeable to water
* lower temperatures are less fluid (freezes)
* too cold can become unsaturated
* composition
* phospholipid saturation: cis-double bonds increase fluidity at lower temperatures (reduce tight packaging)
* phospholipid tail length: shorter hydrocarbon tails increase fluidity at lower temperatures (lipid tails interact less)
* shorter tails allow for more movement cause dont hit eachother
* lipid composition: addition of cholesterol in animal cell membranes. stiffens membrane, less permeable to water
24
New cards
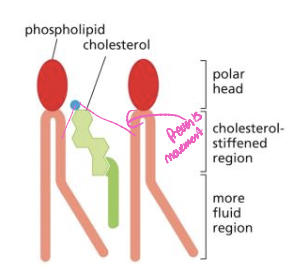
Sterols-cholesterol
* can be up to a 1:1 ratio of cholesterol and phospholipids
* decreases mobility of phospholipid tails (stiffens membrane)
* plasma membrane is less permeable to polar molecules
* goes between the phospholipids to make the membrane less permeable
* decreases mobility of phospholipid tails (stiffens membrane)
* plasma membrane is less permeable to polar molecules
* goes between the phospholipids to make the membrane less permeable
25
New cards
Lipid movement to the other leaflet (flip flopping)
* enzymes in the ER membrane flip lipids from one leaflet to other
* scramblase moves lipids to do flip flip
* catalyzes the reaction
* random phospholipids
* scramblase moves lipids to do flip flip
* catalyzes the reaction
* random phospholipids
26
New cards
Asymmetry of lipid bilayer
* the bilayer faces certain directions
* the side that faces the cytosol will always face the cytosol
* enzymes in the Golgi membrane flip lipids from one leaflet to the other
* catalyzed by flippases
* rapid flip flop of specific phospholipids to the cytosolic leaflet
* some can bind cytosolic proteins at the plasma membrane
* certain lipids have to be on specific side of the bilayer
* the side that faces the cytosol will always face the cytosol
* enzymes in the Golgi membrane flip lipids from one leaflet to the other
* catalyzed by flippases
* rapid flip flop of specific phospholipids to the cytosolic leaflet
* some can bind cytosolic proteins at the plasma membrane
* certain lipids have to be on specific side of the bilayer
27
New cards
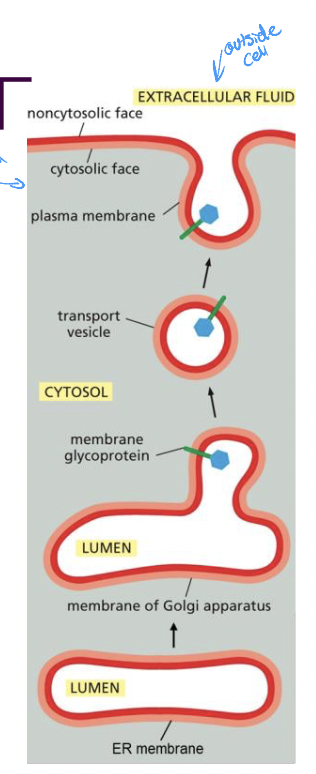
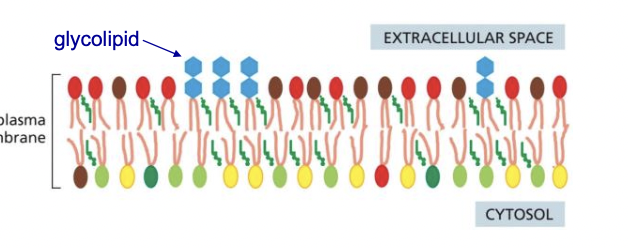
Glycolipids and Glycoproteins
* formed by adding sugar groups to lipids/proteins on luminal face of golgi
* end up on plasma membrane, inside of some organelles
* noncytosolic face
* protect the membrane from harsh environments
* end up on plasma membrane, inside of some organelles
* noncytosolic face
* protect the membrane from harsh environments
28
New cards
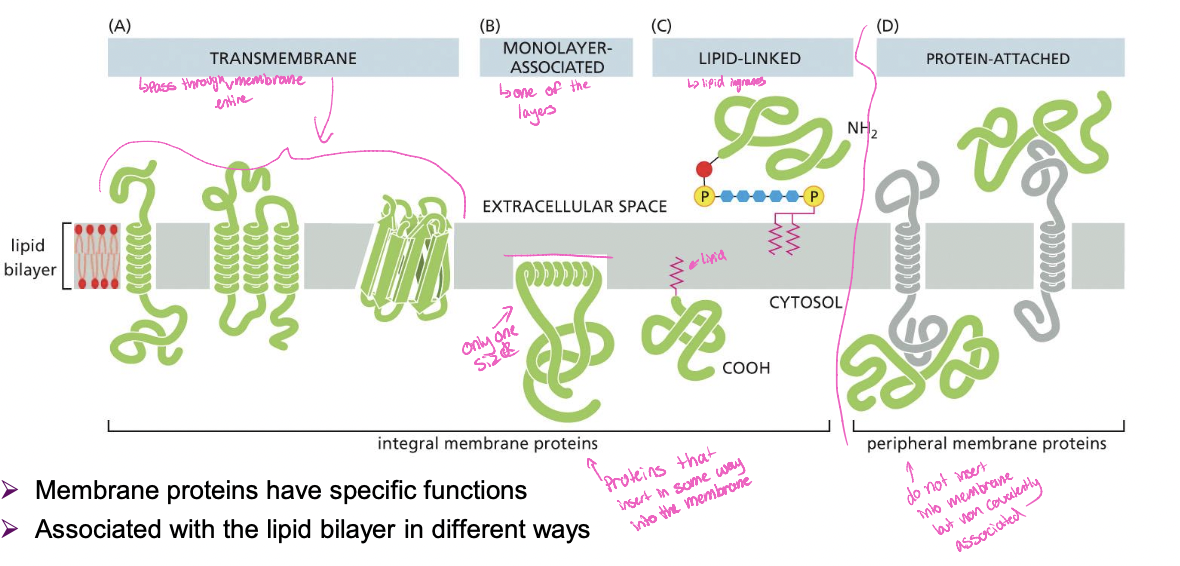
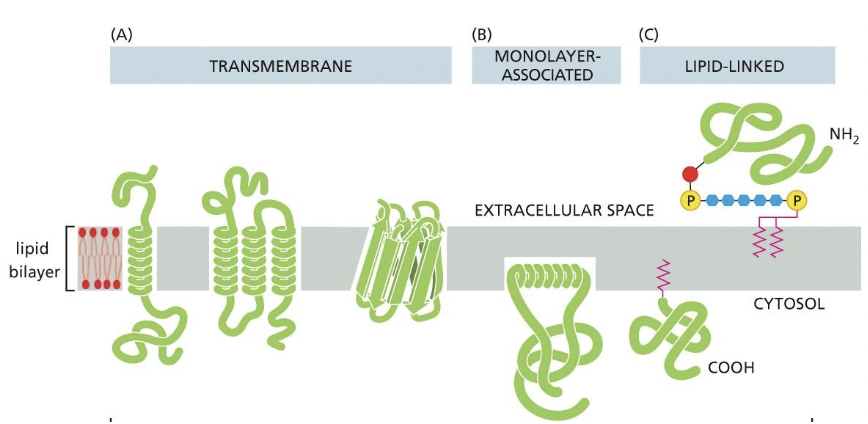
4 Membrane proteins
* transmembrane \*
* monolayer-associated \*
* lipid linked\*
* protein attached (peripheral membrane proteins)
\*integral membrane proteins
\
* monolayer-associated \*
* lipid linked\*
* protein attached (peripheral membrane proteins)
\*integral membrane proteins
\
29
New cards
Integral Membrane proteins
* proteins directly attached to lipid bilayer
* inserted into lipid bilayer transmembrane and monolayer associated
* or attached to a lipid which is inserted into lipid bilayer (lipid linked)
* extraction methods use detergents (lipid bilayer destroyed)
* inserted into lipid bilayer transmembrane and monolayer associated
* or attached to a lipid which is inserted into lipid bilayer (lipid linked)
* extraction methods use detergents (lipid bilayer destroyed)
30
New cards
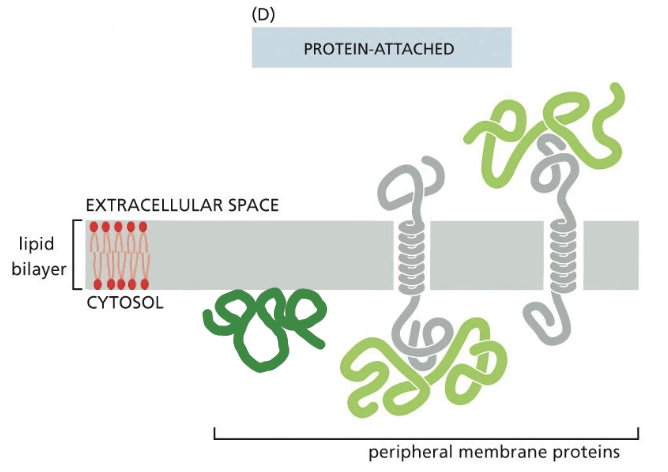
Peripheral membrane proteins
* proteins do not insert themselves in to membrane
* on either face of membrane
* bound to other proteins
* bound to lipids
* by non-covelent interactions
* gentil extraction methods used (lipid bilayer intact)
* on either face of membrane
* bound to other proteins
* bound to lipids
* by non-covelent interactions
* gentil extraction methods used (lipid bilayer intact)
31
New cards
Transmembrane proteins
Proteins are ampipathic
* hydrophillic domains -aqueous - outside membranes
* aa side chains are popar
* hydrophobic membrane-panning domains -within membrane
* aa side chains are non-polar
\*not all transmembrane proteins are alpha helixes
* hydrophillic domains -aqueous - outside membranes
* aa side chains are popar
* hydrophobic membrane-panning domains -within membrane
* aa side chains are non-polar
\*not all transmembrane proteins are alpha helixes
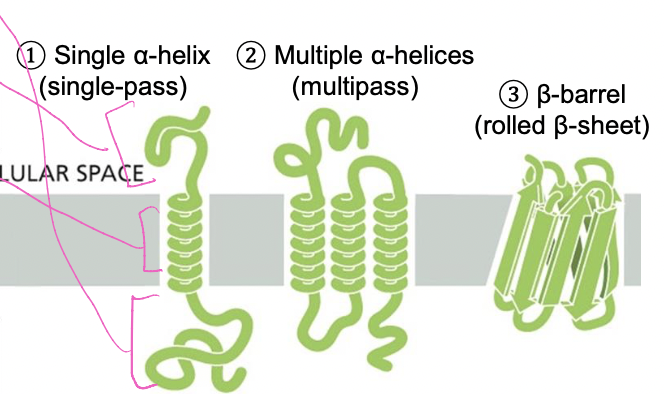
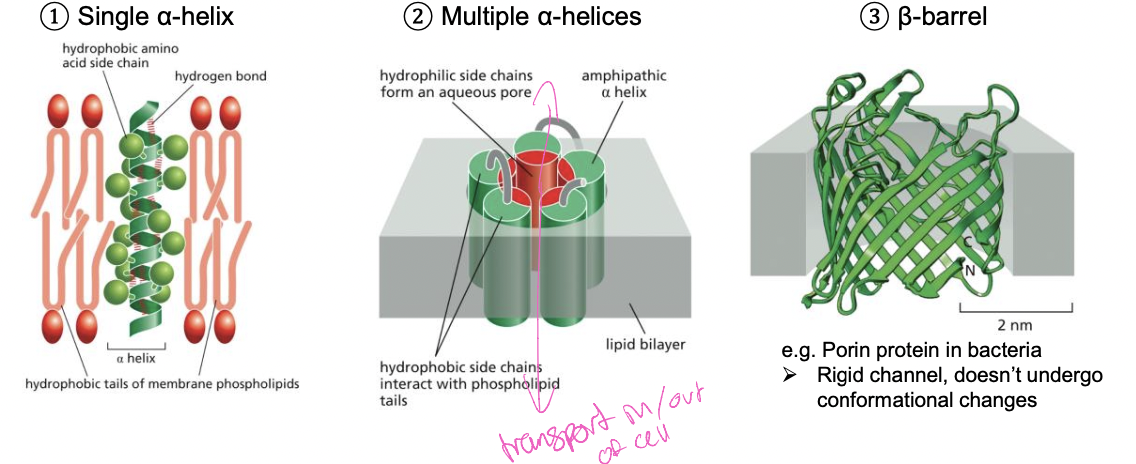
32
New cards
Examples of membrane spanning domains
1. single a-helix with
2. multiple a-helices
* create a channel to transport things in and out of the cell
* hydrophilic side chains go in middle to form aqueous pore
3. B-barrel
* rigid channel doesn’t undergo conformational changes that create large channel
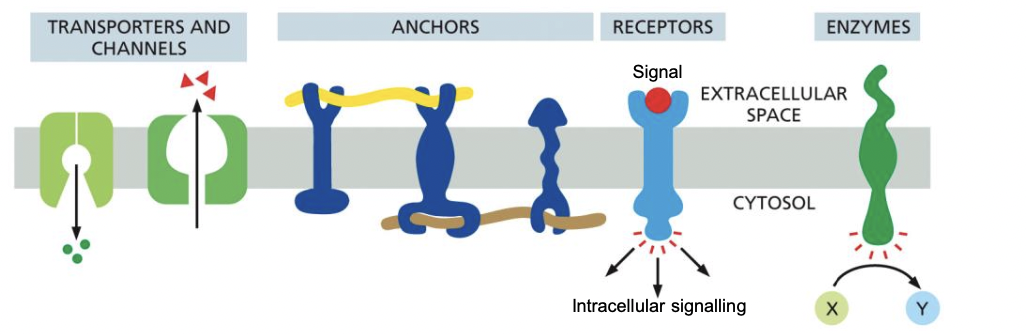
33
New cards
Transmembrane function proteins
* each transmembrane protein has a specific orientation- essential for function
* Transporters and channels
* anchors
* receptors
* enzymes
* Transporters and channels
* anchors
* receptors
* enzymes
34
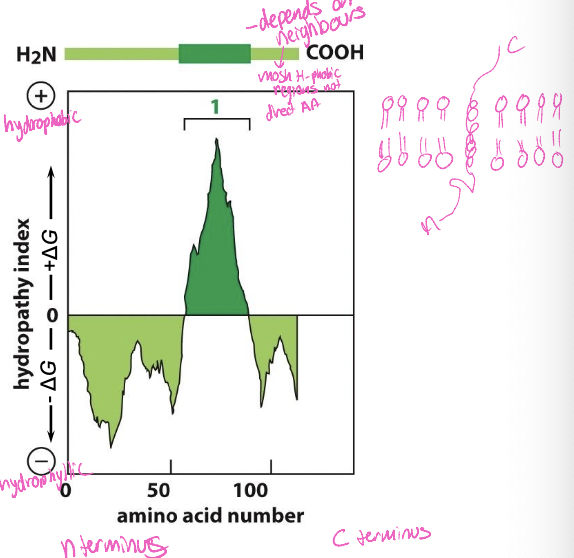
New cards
Two ways to identify structures
* x-ray crystallography
* determines 3D structure
* hydrophobicity plots
* segments of 23-30 hydrophobic amino acids can span the lipid bilayer as an a-helix
* determines 3D structure
* hydrophobicity plots
* segments of 23-30 hydrophobic amino acids can span the lipid bilayer as an a-helix
35
New cards
Hydrophobicity plots
* X-axis: n-terminus to c-terminus
* y-axis: hydrophobic to hydrophilic
* depends on neighbours’s
* most h-phobic regions not direct AA
* y-axis: hydrophobic to hydrophilic
* depends on neighbours’s
* most h-phobic regions not direct AA
36
New cards
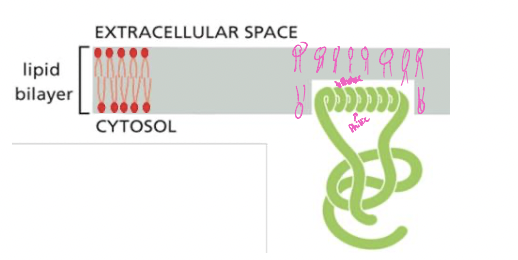
Monolayer-associated membrane proteins
* proteins anchored on cytosolic face by an amphipathic a-helix
* example: proteins in membrane bending
* vesicle budding at the ER
* only half-way embedded into the bilayer
* example: proteins in membrane bending
* vesicle budding at the ER
* only half-way embedded into the bilayer
37
New cards
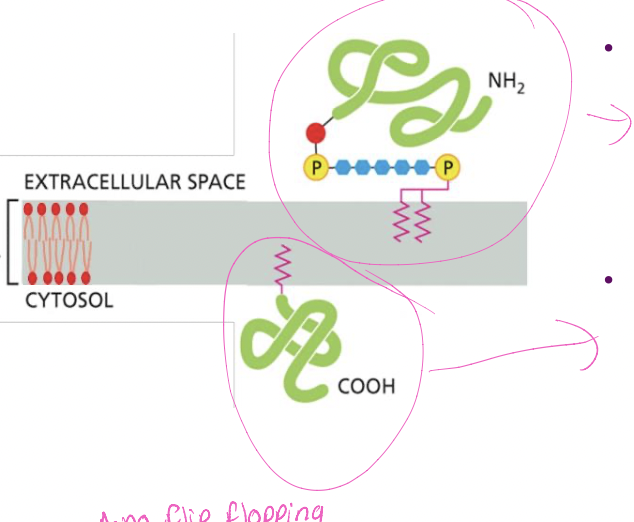
lipid-linked membrane proteins
* proteins with a GPI anchor (Phosphorous, and double pink insert)
* synthesis in ER lumen
* end up on cell surface
* non cytosolic face
* protein with other lipid anchor (fatty acid, prenyl)
* cytosolic enzymes add anchor
* directs protein on cytosolic face
* synthesis in ER lumen
* end up on cell surface
* non cytosolic face
* protein with other lipid anchor (fatty acid, prenyl)
* cytosolic enzymes add anchor
* directs protein on cytosolic face
38
New cards
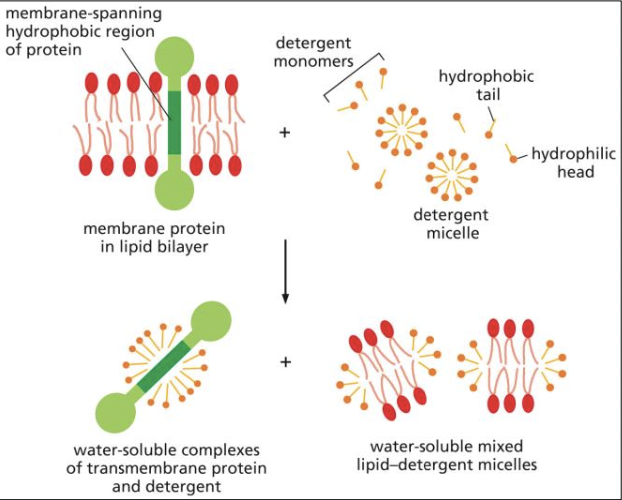
Techniques for extraction of membrane proteins
* use detergent with hydrophonic tails and hydrophillic heads to btake out the protein by surrounding the hydrophobic centre of the protein
39
New cards
Lateral diffusion of membrane proteins
* lateral diffusion within leaflet
* no flip flop
* study of protein movement: FRAP
* fluorescence recovery after photobleaching
* protein fused to GFP
* green fluorescent protein
* lateral mobility can also be restricted
* no flip flop
* study of protein movement: FRAP
* fluorescence recovery after photobleaching
* protein fused to GFP
* green fluorescent protein
* lateral mobility can also be restricted
40
New cards
Permeability of the lipid bilayer
* artificial bilayer
* impermiable to most water soluble molecules
* proteins help make membrane more permiable
* cell membrane
* membrane transport proteins to transfer specific molecules
* facilitated transport
* impermiable to most water soluble molecules
* proteins help make membrane more permiable
* cell membrane
* membrane transport proteins to transfer specific molecules
* facilitated transport
41
New cards
Permeable
* movement via simple diffusion through lipid bilayer
* high concentration to low concentration
* down the concentration gradient
* more hydrophobic or non-polar molecules
* faster diffusion across lipid bilayer
* high concentration to low concentration
* down the concentration gradient
* more hydrophobic or non-polar molecules
* faster diffusion across lipid bilayer
42
New cards
Impermeable
* these require membrane proteins for transport
* take a long time
* not readily going across
* take a long time
* not readily going across
43
New cards
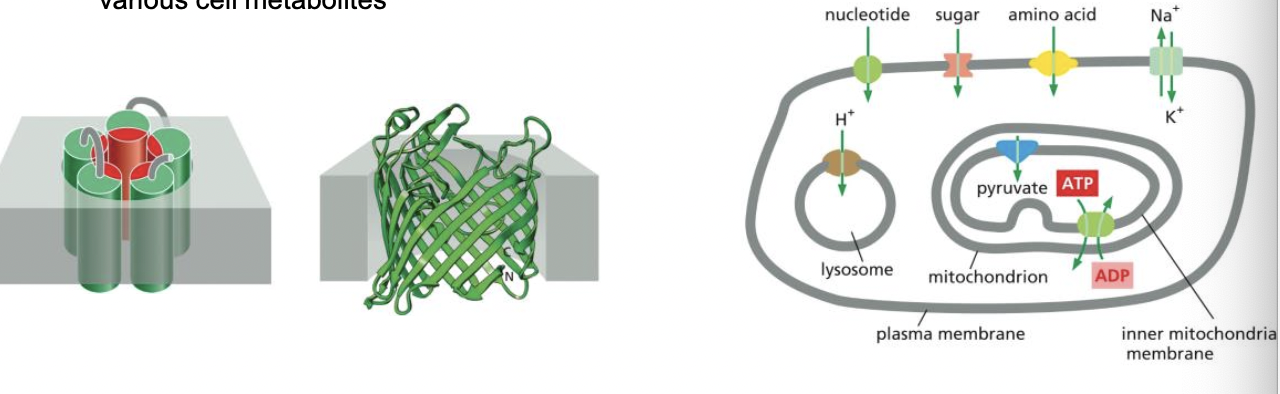
proteins involved in membrane transport
* transmembrane transport proteins go through membrane
* create a protein-lined path across cell membrane
* transport polar and charged molecules
* ions, sugars, amino acids, nucleotides, various cell metabolites
* each transport protein is selective:
* transports a specific class of molecules
* different cell membranes
* have a different complement of transport
* create a protein-lined path across cell membrane
* transport polar and charged molecules
* ions, sugars, amino acids, nucleotides, various cell metabolites
* each transport protein is selective:
* transports a specific class of molecules
* different cell membranes
* have a different complement of transport
44
New cards
Transport can happen through
Transporter and channel proteins
45
New cards
Channel proteins general
* selective
* size and electric charge of solute
* transport
* transient interactions as solute passes through
* no conformational changes for transport through an open channel
* Binds weakly to transported molecule and does not change conformation when opening an dtransporting
* size and electric charge of solute
* transport
* transient interactions as solute passes through
* no conformational changes for transport through an open channel
* Binds weakly to transported molecule and does not change conformation when opening an dtransporting
46
New cards
Transporter (AKA carrier proteins) general
* selectively
* solute fits to binding site
* transport
* specific binding of solute
* series of conformational changes for transport
* binds strong to transported molecule and changes in conformation to push the solute through
* solute fits to binding site
* transport
* specific binding of solute
* series of conformational changes for transport
* binds strong to transported molecule and changes in conformation to push the solute through
47
New cards
Passive transport happens through and requires
* simple diffusion, channel mediated, transporter mediated
* only needs the protein
* uses transporter proteins, channel proteins
* passive and go with concentration gradient
* only needs the protein
* uses transporter proteins, channel proteins
* passive and go with concentration gradient
48
New cards
Active transport happens through and requires
* transporter proteins
* using a pump
* uses energy
* goes against concentration gradient
* using a pump
* uses energy
* goes against concentration gradient
49
New cards
Concentration gradient and resting membrane potential
* electrical gradients= concentration gradient +membrane potential
* see which has more pull see which way goes
* gradients can work together or against one another
* most cells will go with concentration gradient
* passive and active transport for solutes with a net charge
* usually inside of cell is negatively charged
* see which has more pull see which way goes
* gradients can work together or against one another
* most cells will go with concentration gradient
* passive and active transport for solutes with a net charge
* usually inside of cell is negatively charged
50
New cards
Channel proteins detailed
* hydrophilic pore across membrane
* most channel proteins are selective
* ion channels transport a specific ion
* ion size and electric charge
* passive transport of solute
* transient interactions with channel wall as solute passes through
* faster transport by channels that by transporters
* most channel proteins are selective
* ion channels transport a specific ion
* ion size and electric charge
* passive transport of solute
* transient interactions with channel wall as solute passes through
* faster transport by channels that by transporters
51
New cards
Ion channels can be
Gated (signal required to open) or non gated (always open)
52
New cards
Non-gated ion channels
* K+ leak channels
* always open
* K+ moves out of cell
* major role in generating resting membrane potential in plasma membrane of animal cells
* resting potential is when + leaks to outside of cell in order to balance charges
* always open
* K+ moves out of cell
* major role in generating resting membrane potential in plasma membrane of animal cells
* resting potential is when + leaks to outside of cell in order to balance charges
53
New cards
Gated ion channels
* require a signal to open channel
* specific ions will be transported
* specific ions will be transported
54
New cards
Types of gated ion channels and signals to open
* voltage
* ligand (extracellular)
* ligand (intracellular)
* mechanically gated (mechanical stress signal)
* ligand (extracellular)
* ligand (intracellular)
* mechanically gated (mechanical stress signal)
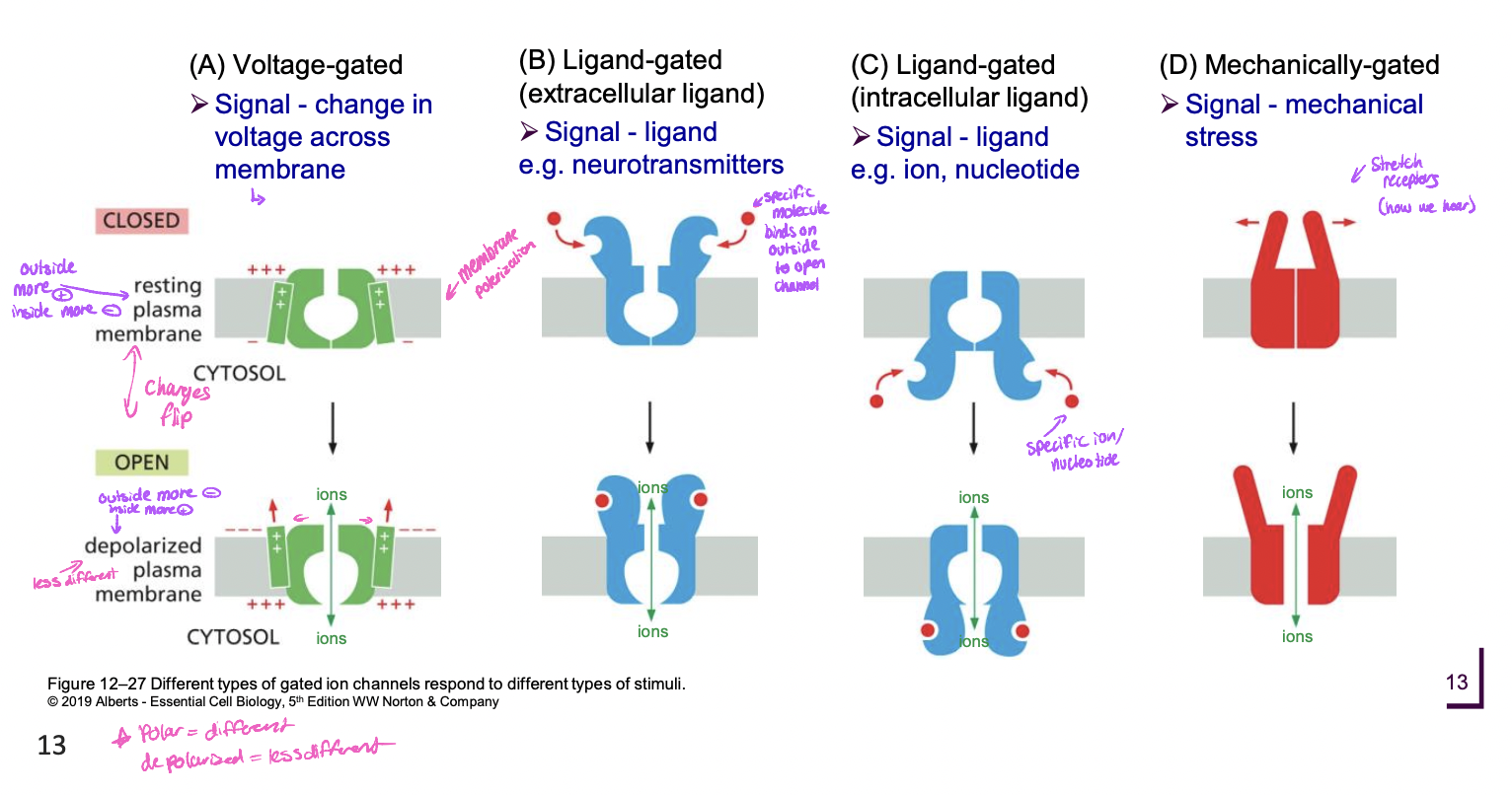
55
New cards
Transporter proteins detailed
* transporter binds to a specific solute
* goes through a conformational change
* transports solute across the membrane
* goes through a conformational change
* transports solute across the membrane
56
New cards
speed of transporter proteins
* initially has a fast speed btu can only move so fast cause has to change conformation alot has a lower max speed than channel
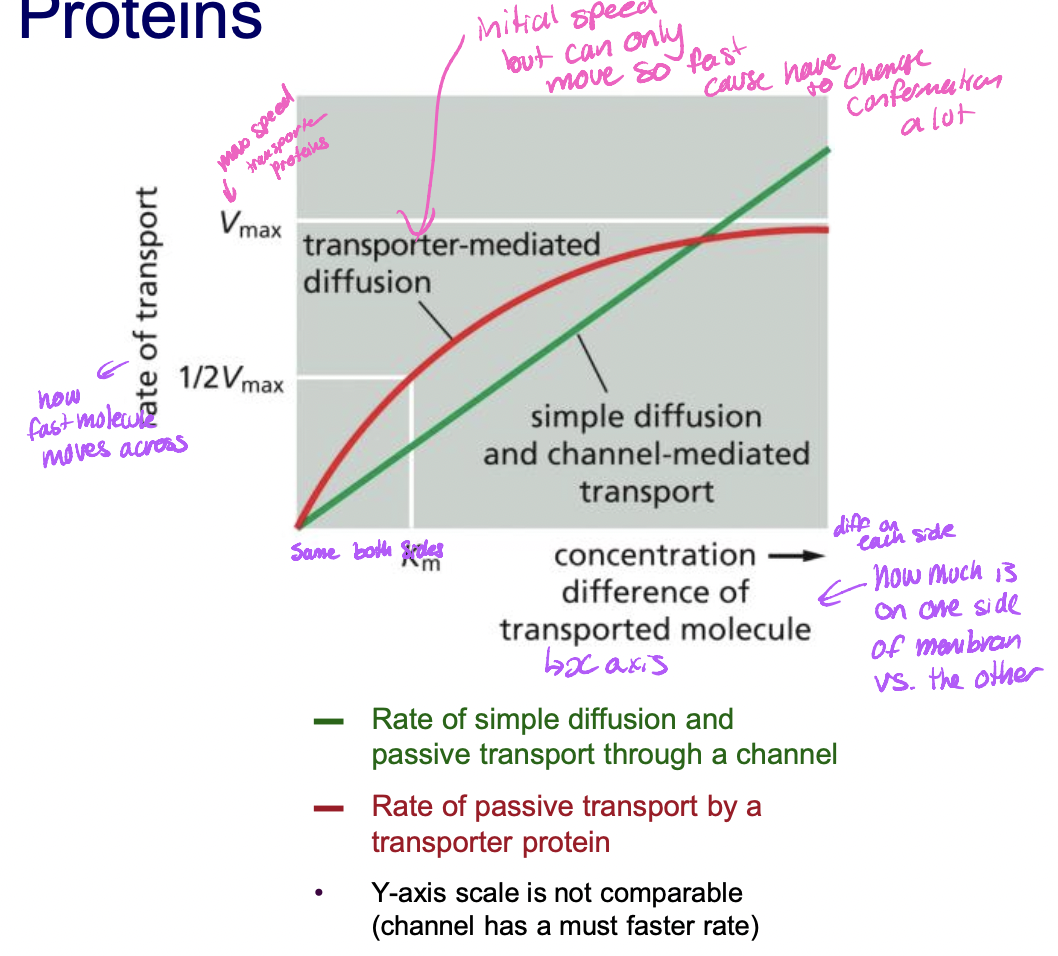
57
New cards
Passive transport by transporter proteins (uniporter)
* transports one solute down its electrochemical gradient
* direction of transport is reversible but always goes down gradient (only reverses if gradient switches)
* glucose transporter (GLUT uniporter)
* transports glucose down the concentration gradient
* can work in either direction (in or out of the cell)
* direction of transport is reversible but always goes down gradient (only reverses if gradient switches)
* glucose transporter (GLUT uniporter)
* transports glucose down the concentration gradient
* can work in either direction (in or out of the cell)
58
New cards
Types of active transport
* need energy not always in the form of ATP
* gradient driven pump
* 1st solute down its gradient, 2nd solute against its gradient
* first creates the energy needed to move the second
* ATP-driven pump (ATPases)
* atp hydrolysis (energy); moves solute against its gradient
* light driven pump (bacteria)
* light energy moves solute against gradient
* gradient driven pump
* 1st solute down its gradient, 2nd solute against its gradient
* first creates the energy needed to move the second
* ATP-driven pump (ATPases)
* atp hydrolysis (energy); moves solute against its gradient
* light driven pump (bacteria)
* light energy moves solute against gradient
59
New cards
Symport vs antiport
* symport: 2 solutes moved insame direction
* antiport: 2 solutes moved in opposite directions
* both: free energy from 1st solue moving down its electrical gradient is used to transport the 2nd solute against its chemical gradient
* antiport: 2 solutes moved in opposite directions
* both: free energy from 1st solue moving down its electrical gradient is used to transport the 2nd solute against its chemical gradient
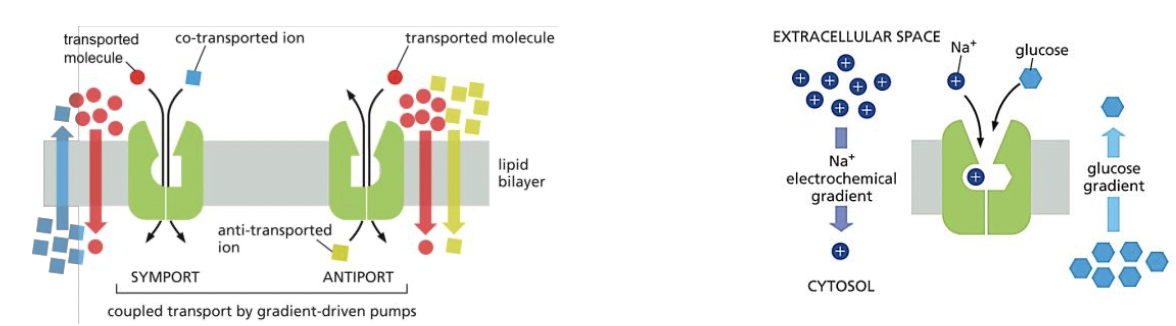
60
New cards
example of symport: Na+ glucose symporter
* Na+ down its electrochemical gradient provides energy to mvoe glucose against its concentration gradient
* ransom oscolations between conformations, reversible but conformational changes to move the solute only occur after
* both sites are occupied: cooperative binding of Na+ and glucose
* both sites empty: both Na+ and glucose dissociate
* ransom oscolations between conformations, reversible but conformational changes to move the solute only occur after
* both sites are occupied: cooperative binding of Na+ and glucose
* both sites empty: both Na+ and glucose dissociate
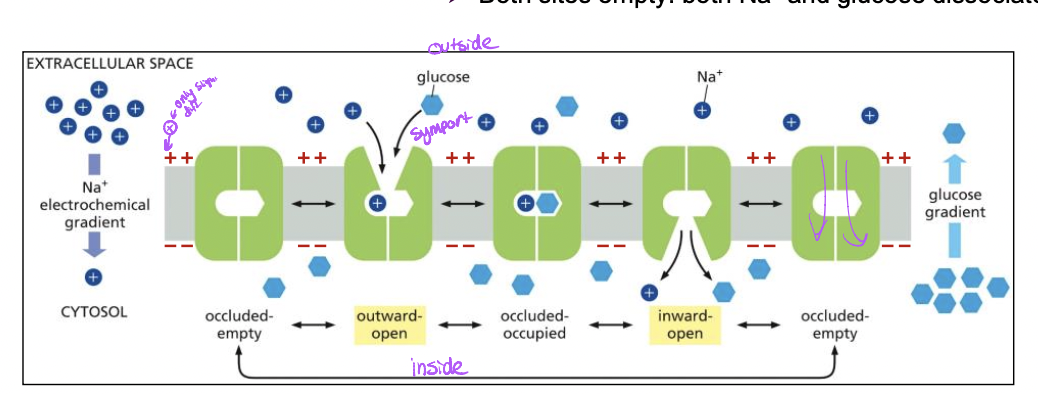
61
New cards
Antiport example: Na+ and H+ exchanger
* cytosolic pH needs to be regulated for optimal enzyme function
* but excess H+ occurs in the cytosol
* from acid forming reactions
* leaks out of lysosome
* transporters maintain cytosolic pH
* Na+ down concentration gradient provides energy to move H+ against concentration gradient
* but excess H+ occurs in the cytosol
* from acid forming reactions
* leaks out of lysosome
* transporters maintain cytosolic pH
* Na+ down concentration gradient provides energy to move H+ against concentration gradient
62
New cards
how is the Na+ electrochemical gradient maintained with sym and anti-porter moving it down concentration gradient?
Na+ k+ pump (active transport using alternate energy)
63
New cards
ATP-driven pumps
* active transport
* use energy from atp hydrolysis to transport solues
* 3 types: P-type pump, ABC transporter, V-type pump
* use energy from atp hydrolysis to transport solues
* 3 types: P-type pump, ABC transporter, V-type pump
64
New cards
P-type pumps
* require ATP
* phosphorylate during pumping cycle
* Pee themselves during pump cycle
* flippases
* transport phospholipids
* phosphorylate during pumping cycle
* Pee themselves during pump cycle
* flippases
* transport phospholipids
65
New cards
Examples of P-type pumps
* animal plasma membrane Na+ K+ pump
* move Na+ and K+ against their electrochemical gradient
* 3 Na+ out 2 K+ in
* Na+ gradient is used to transport nutrients into cell and maintain pH, cell volume
* move Na+ and K+ against their electrochemical gradient
* 3 Na+ out 2 K+ in
* Na+ gradient is used to transport nutrients into cell and maintain pH, cell volume
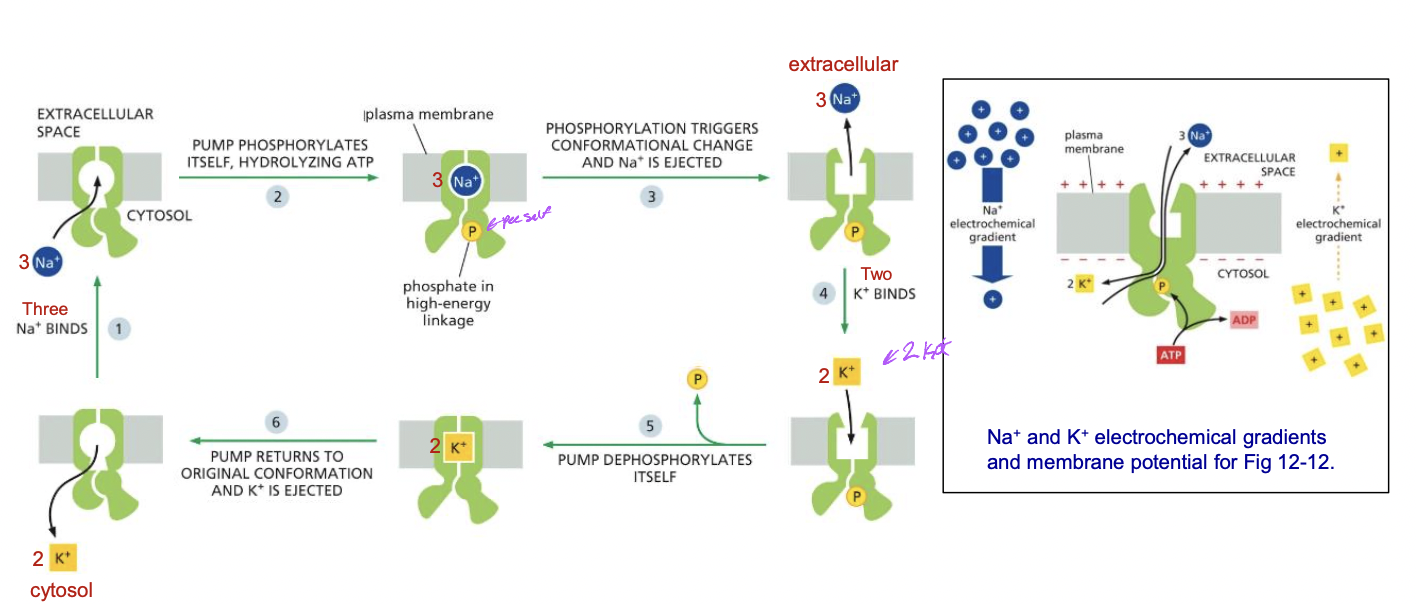
66
New cards
P-type pumps and electrochemical gradients
* function in generating and maintaining electrochemical gradients
* H+ pump in plant cell
* H+ pump in plant cell
67
New cards
H+ P-type pump
* generates H+ electrochemical gradient
* used for H+ driven symport/antiport
* membrane potential
* used for H+ driven symport/antiport
* membrane potential
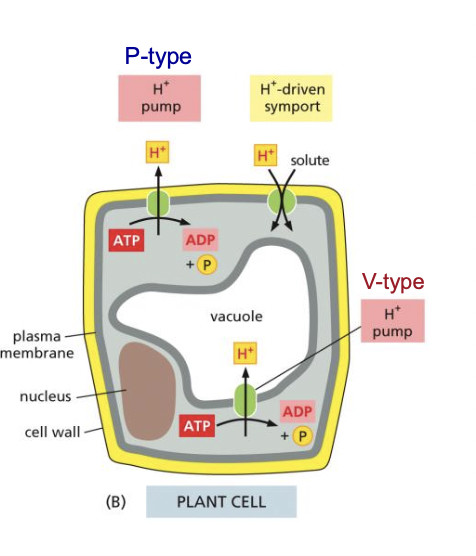
68
New cards
ABC transporter
* uses 2 ATP to pump small molecules across cell membrane
* move toxins out of body
* move toxins out of body
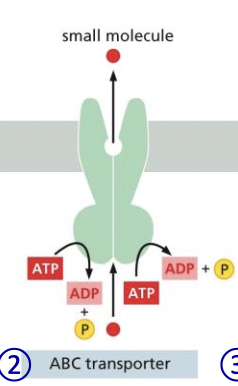
69
New cards
V-type PROTON pump
* uses ATP to pump H+ into organelles to acidify the lumen
* in lysosome, plant vacuol
* no peeing, does a spin
* in lysosome, plant vacuol
* no peeing, does a spin
70
New cards
F-type ATP synthase
* structurally related to V-type proton pump, but opposite mode of action
* H+ in, P+ ADP that become ATP instead of using ATP
* Uses H+ gradient to drive the synthesis of ATP
* in mitochondria, chloroplasts, bacteria
* H+ in, P+ ADP that become ATP instead of using ATP
* Uses H+ gradient to drive the synthesis of ATP
* in mitochondria, chloroplasts, bacteria
71
New cards
V-type vs F-type
V: uses atp to pump H+ against gradient
F: uses H+ gradient to generate ATP
\
F: uses H+ gradient to generate ATP
\
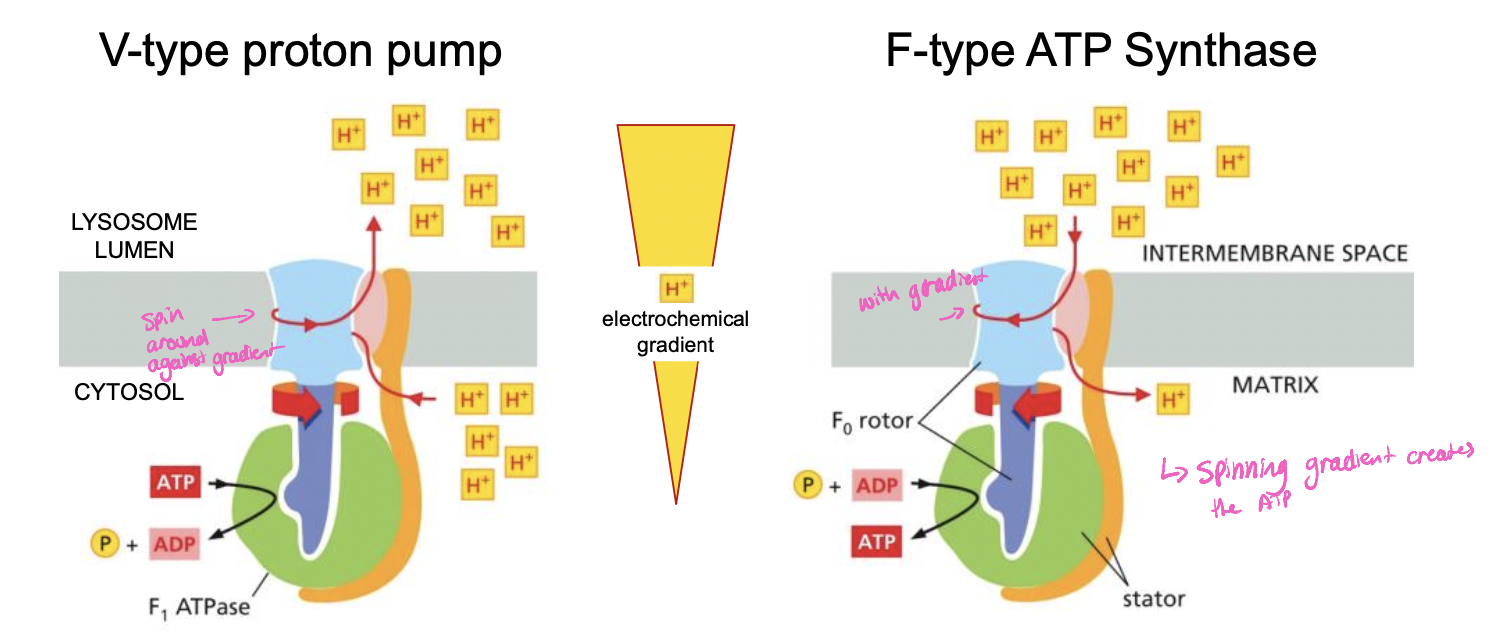
72
New cards
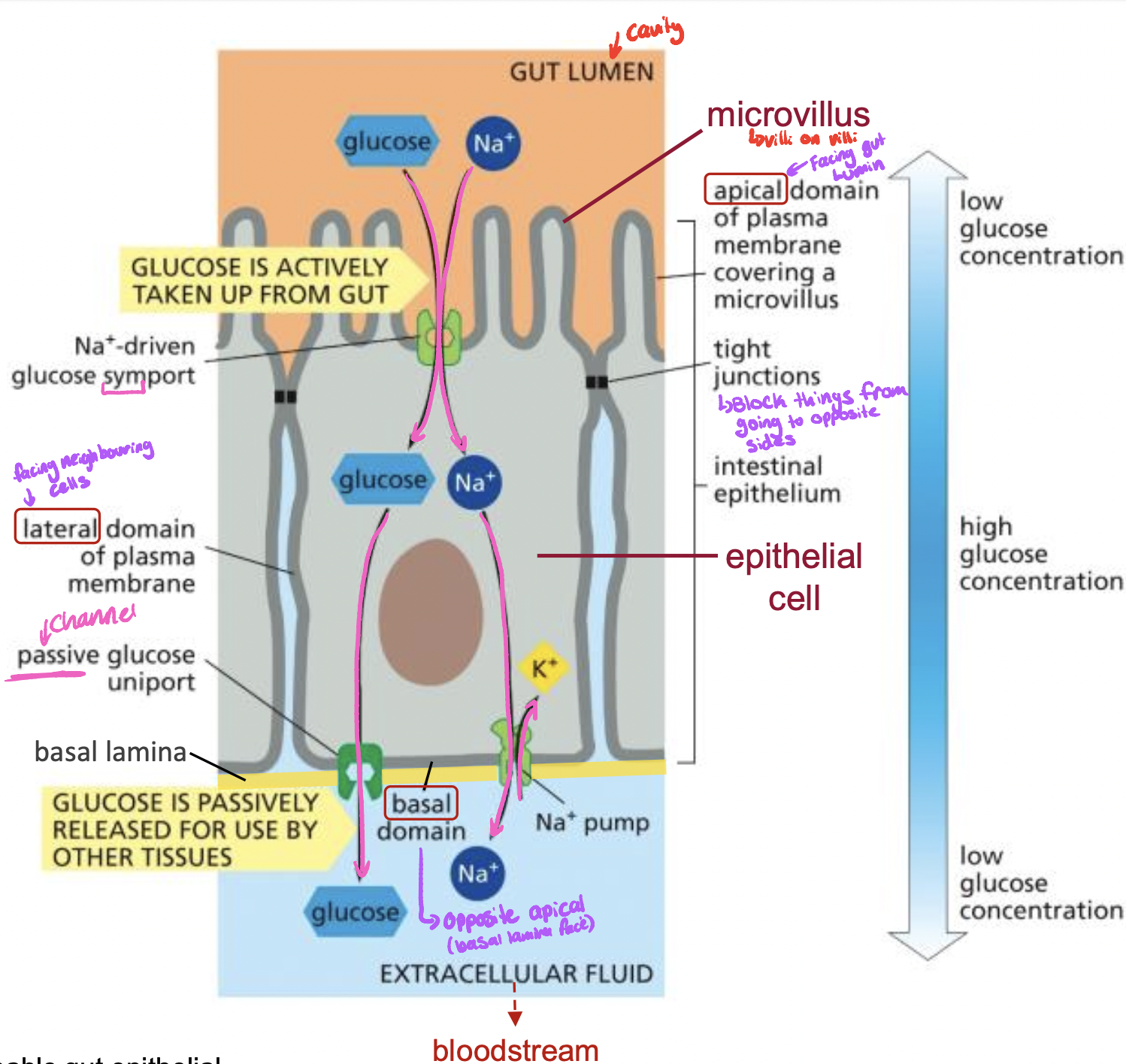
Transporters work together to transfer Glucose from the intestine to the blood stream
1. goes through Na+ & glucose symporter
2. a) glusoce goes through a passive glucose uniport into the extracellular fluid
b) Na+ is now to high in middle, so Na+ K+ pump takes out Na+ and puts in K+ into the epithelial cell
73
New cards
Glucose path when transporting in to blood stream
* gut lumen (apical domain)
* epithelial cell (lateral domain)
* extracellular fluid (basal domain)
* epithelial cell (lateral domain)
* extracellular fluid (basal domain)
74
New cards
Tight junctions
* restruct transport proteins
* block things from going from basal side to apical side, proteins will stay on which ever side they are on
* example: glucose transport: symporter on apical side, na+ k+ pump and glucose uniport on basolateral membrane
* block things from going from basal side to apical side, proteins will stay on which ever side they are on
* example: glucose transport: symporter on apical side, na+ k+ pump and glucose uniport on basolateral membrane
75
New cards
Membrane potential
* difference in electrical charge on two different sides of membrane
* used by gradient driven pumps to carry out active transport (symport and antiport)
* important for electrical signaling
* used by gradient driven pumps to carry out active transport (symport and antiport)
* important for electrical signaling
76
New cards
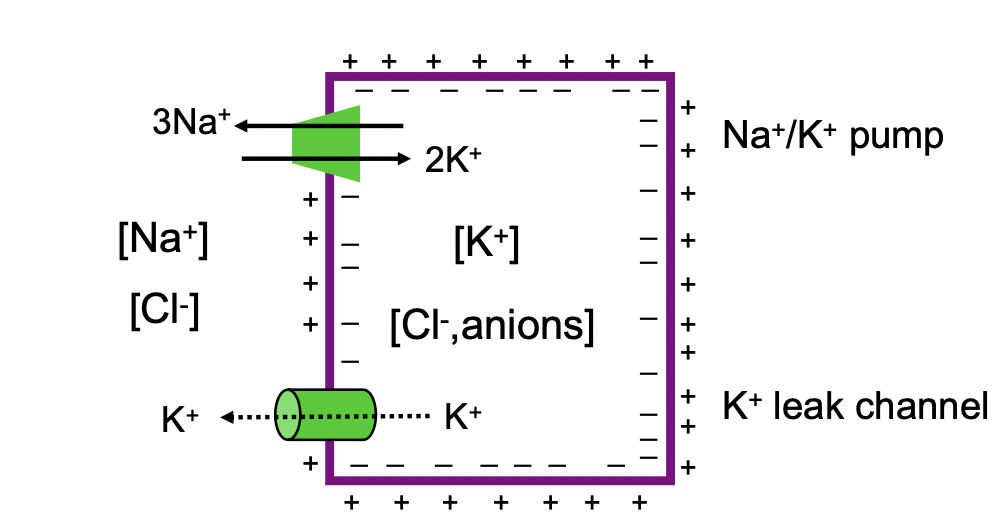
Generation of membrane potential in animal cells overview
1. K+ leak channel
1. major role in membrane potential
2. outward flow of K+
2. Na+ K+ pump
1. maintains Na+ gradient with low Cytosolic Na+, K+ concentration with high cytosolic K+
2. electrogenic 3 Na+ ions pumped out of cell, 2 K+ ions pumped into the cell
1. net is 1 ion pumped out (more out than in)
77
New cards
Membrane potential generation results in
* more + outside (Na+ and K+ since some K+ is leaving through leak channels even when some is being pumped in)
* more - on inside (Cl- and fixed anions)
* this is what forms the membrane potential
* more - on inside (Cl- and fixed anions)
* this is what forms the membrane potential
78
New cards
EQM= resting membrane potential in animal cells is
\-20 mV to -200 mV
* from the perspective of outside so inside is more -
* from the perspective of outside so inside is more -
79
New cards
Generation of membrane potential (plant cells)
* plasma membrane p-type pump (H+)
* generates H+ electrochemical gradient
* membrane potential
* used by gradient driven pumps to carry out active transport
* electrical signalling
* regulate pH
* generates H+ electrochemical gradient
* membrane potential
* used by gradient driven pumps to carry out active transport
* electrical signalling
* regulate pH
80
New cards
Plant cell membrane potential range
\-120 to 160 mV (smaller range than animal)
81
New cards
Intracellular compartments
* membrane enclosed organelles are over 50% of cell volume
* includes:
* endosome (endocytosis)
* lysosome (digestive enzyme)
* cytosol
* peroxisome (oxidative reactions)
* free riboomes (not compartment, protein synthesis)
* nucleus (DNA synthesis, RNA)
* Endoplasmic reticulum
* golgi apparatus (modifies proteins and lipids)
* mitochondrion (ATP synthesis)
* includes:
* endosome (endocytosis)
* lysosome (digestive enzyme)
* cytosol
* peroxisome (oxidative reactions)
* free riboomes (not compartment, protein synthesis)
* nucleus (DNA synthesis, RNA)
* Endoplasmic reticulum
* golgi apparatus (modifies proteins and lipids)
* mitochondrion (ATP synthesis)
82
New cards
Volumes of different organelles in cells for liver,
cytosol is half the volume
* protein synthesis and degradation
* many metabilic pathways
* cytoskeleton
* has lots of smooth ER because it is involved in detoxification
* protein synthesis and degradation
* many metabilic pathways
* cytoskeleton
* has lots of smooth ER because it is involved in detoxification
83
New cards
Oranelles definition
* discrete structure or subcompartment of a euk. cell that is specialized to carry out a specific function
* most are membrane enclosed but they dont have to be
* examples
* nucleus
* ER
* golgi
* nucleolus
* centrosome
* used to be defined by if they could be seen on a microscope so some arent membran enclosed but still stick together enough to be seen like one
* most are membrane enclosed but they dont have to be
* examples
* nucleus
* ER
* golgi
* nucleolus
* centrosome
* used to be defined by if they could be seen on a microscope so some arent membran enclosed but still stick together enough to be seen like one
84
New cards
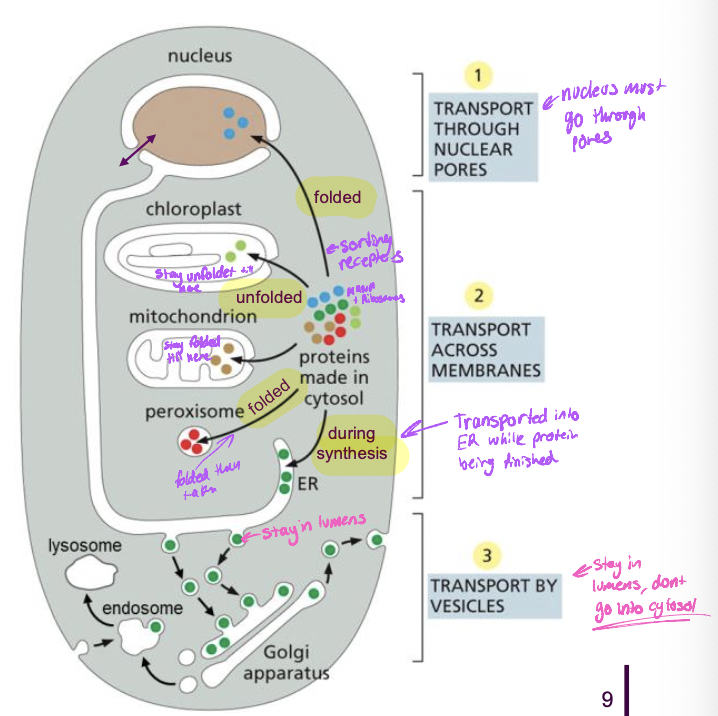
Protein sorting overview
* proteins are nuclear encoded (DNA creates RNA to create proteins)
* mRNA arrives in cytoplasm to translate
* cytosolic proteins stay in cytoplasm so they have no sorting signal
* proteins that go to other organelles have signal sequences
* not something added specially but its encoded within the protein
* mRNA arrives in cytoplasm to translate
* cytosolic proteins stay in cytoplasm so they have no sorting signal
* proteins that go to other organelles have signal sequences
* not something added specially but its encoded within the protein
85
New cards
How are proteins sorted overview
* sorted by signal sequence
* stretch of AA sequence within protein to direct protein to right compartment
* signal specifies
* specific destination in the cell
* specific signal sequences direct proteins to nucleus, mitochondria, ER peroxisomes
* sequences are recognized by
* sorting receptors that take proteins to their destinations
* proteins that take other proteins to where the signal goes
* stretch of AA sequence within protein to direct protein to right compartment
* signal specifies
* specific destination in the cell
* specific signal sequences direct proteins to nucleus, mitochondria, ER peroxisomes
* sequences are recognized by
* sorting receptors that take proteins to their destinations
* proteins that take other proteins to where the signal goes
86
New cards
which locations require proteins folded vs which dont
* folded: Nucleus, Peroxisome
* unfolded: mitochondria and chloroplasts
* during synthesis (while being translated) goes into ER/lumen system
* unfolded: mitochondria and chloroplasts
* during synthesis (while being translated) goes into ER/lumen system
87
New cards
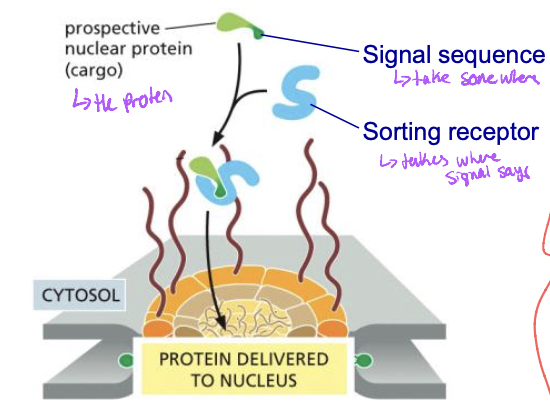
Post translational sorting
* proteins are nuclear-encoded
* fully synthesized in cytosol before sorting
* folded nucleus and peroxisomes
* unfolded mitochondria and chloroplasts
* fully synthesized in cytosol before sorting
* folded nucleus and peroxisomes
* unfolded mitochondria and chloroplasts
88
New cards
Co-translational sorting (while translating)
* proteins nuclear encoded
* have ER signal sequence
* associated with ER during protein synthesis in the cytosol
* moving into ER while being translated
* ER signal sequence is hydrophobic
* go to ER which is entry point for endomembrane system
* golgi, ER, endosomes, lysosomes
* have ER signal sequence
* associated with ER during protein synthesis in the cytosol
* moving into ER while being translated
* ER signal sequence is hydrophobic
* go to ER which is entry point for endomembrane system
* golgi, ER, endosomes, lysosomes
89
New cards
Transport through nuclear pores (post translational and folded)
* transports through nuclear pores
* theres signals to move into or out of nucleus
* theres signals to move into or out of nucleus
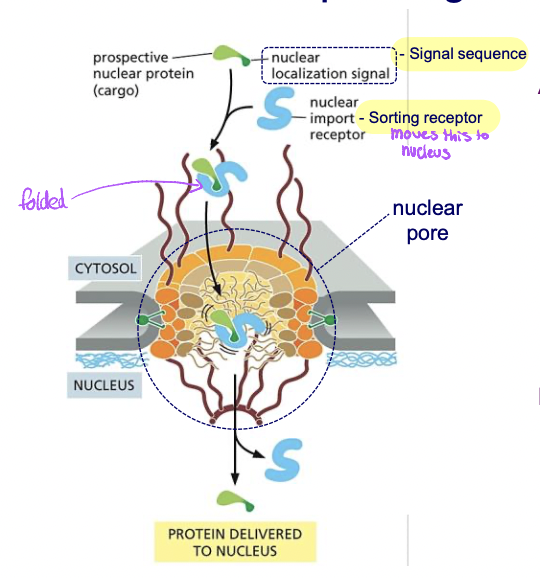
90
New cards
Peroxisomes sorting
* contain enzymes for oxidative reactions
* detoxify toxins, break down fatty acid molecules
* enzymes imported into the peroxisome through a transmembrane protein complex
* folded and post translational when transported
* detoxify toxins, break down fatty acid molecules
* enzymes imported into the peroxisome through a transmembrane protein complex
* folded and post translational when transported
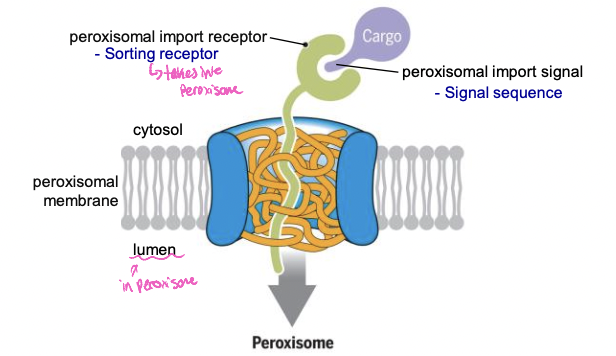
91
New cards
Sorting to mitochondria and chloroplasts
* Post translational and unfolded
* have own genomes and ribosomes
* most proteins for these are nuclear encoded still
* translated in cytosol and targeted by signal sequence for import
* proteins are unfolded for import by hsp70 chaperone proteins
* helo other proteins fold and stay folded or unfoled
* have own genomes and ribosomes
* most proteins for these are nuclear encoded still
* translated in cytosol and targeted by signal sequence for import
* proteins are unfolded for import by hsp70 chaperone proteins
* helo other proteins fold and stay folded or unfoled
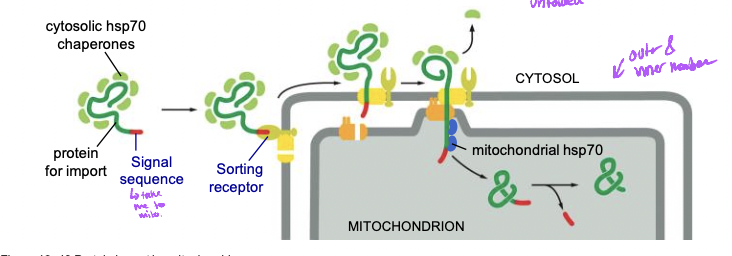
92
New cards
93
New cards
ER
* Rough= synthesis of soluble transmembrane proteins for the endomembrane
* smooth= phospholipid synthesis
* smooth= phospholipid synthesis
94
New cards
Sorting to the ER
* mRNA arrives in cytoplasm
* translation starts on ribosomes in the cytosol
* translating protein with ER signal sequence makes it stop producing protein and ribosome moves it to the ER
* then it restarts when in the ER
* proteins entering the ER:
* soluble proteins (go all the way in)
* transmembrane proteins stick in membrane
* translation starts on ribosomes in the cytosol
* translating protein with ER signal sequence makes it stop producing protein and ribosome moves it to the ER
* then it restarts when in the ER
* proteins entering the ER:
* soluble proteins (go all the way in)
* transmembrane proteins stick in membrane
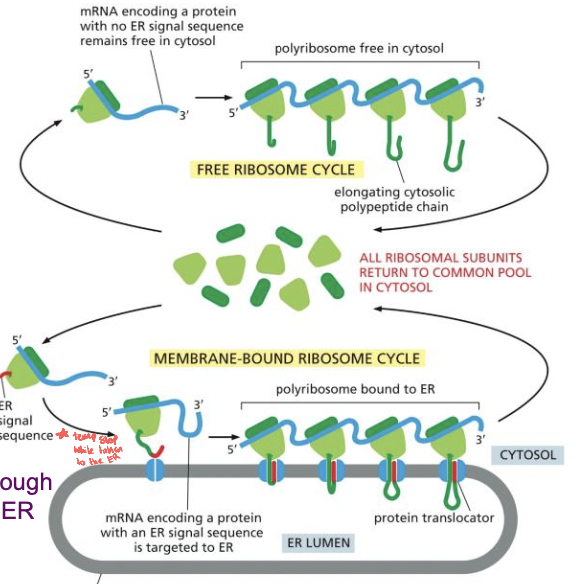
95
New cards
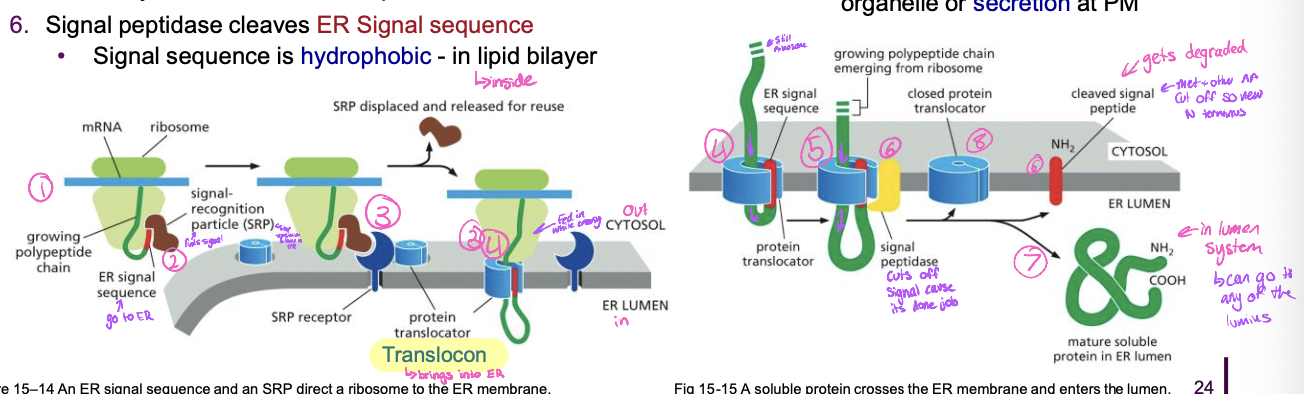
Co-translational translocation: **soluble protein**
* goes completely inside lumen
1. translation begins at N-terminus, ER signal sequence emerges
1. recognized bt SRP and this stops translation
2. SRP-ribosome complex → SRP receptor → translocon
1. translocon opens
3. protein synthesis resumes with protein transfer into the ER lumen
4. signal peptidase cleaves ER signal sequence
1. signal sequence in hydrophobic so it stays in the lipid bilayer
5. protein released into ER lumen
6. translocon closes
1. translation begins at N-terminus, ER signal sequence emerges
1. recognized bt SRP and this stops translation
2. SRP-ribosome complex → SRP receptor → translocon
1. translocon opens
3. protein synthesis resumes with protein transfer into the ER lumen
4. signal peptidase cleaves ER signal sequence
1. signal sequence in hydrophobic so it stays in the lipid bilayer
5. protein released into ER lumen
6. translocon closes
96
New cards
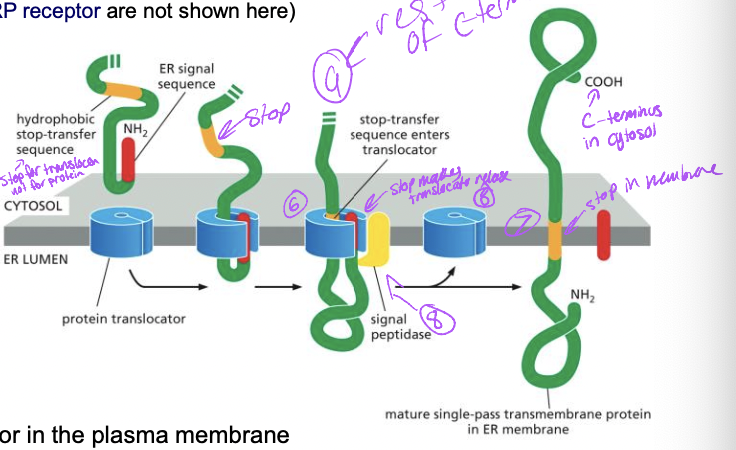
Co-translational translocation: **Transmembrane Protein**
* N-terminal ER signal sequece
1. translation begins at N-terminus, ER signal sequence emerges
1. recognized bt SRP and this stops translation
2. SRP-ribosome complex → SRP receptor → translocon
1. translocon opens
3. protein synthesis resumes with protein transfer into the ER lumen
4. stop transfer sequence enters translocon
1. internal hydrophobic segment
1. membrane spanning a-helix
5. protein transfer stops adn transmembrane domain is released into lipid bilayer
6. signal peptidase cleaves n-terminal ER signal sequence adn translocon closes
7. rest of c-terminus is synthesized
1. translation begins at N-terminus, ER signal sequence emerges
1. recognized bt SRP and this stops translation
2. SRP-ribosome complex → SRP receptor → translocon
1. translocon opens
3. protein synthesis resumes with protein transfer into the ER lumen
4. stop transfer sequence enters translocon
1. internal hydrophobic segment
1. membrane spanning a-helix
5. protein transfer stops adn transmembrane domain is released into lipid bilayer
6. signal peptidase cleaves n-terminal ER signal sequence adn translocon closes
7. rest of c-terminus is synthesized
97
New cards
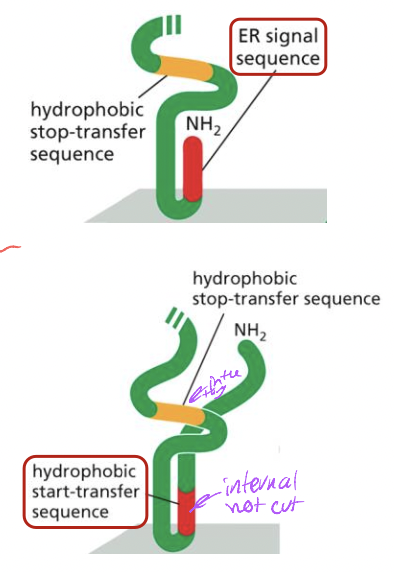
2 types of ER signal sequences
* N-terminal ER signal sequence
* at n-terminus of protein
* removed by signal peptidase
* Internal ER signal sequence
* start transfer sequence
* not removed-remains part fo protien
* membrane spanning a-helix
* at n-terminus of protein
* removed by signal peptidase
* Internal ER signal sequence
* start transfer sequence
* not removed-remains part fo protien
* membrane spanning a-helix
98
New cards
Transmembrane protein with internal signal sequence
1. translation begins, interal start-transfer sequence
2. SRP-ribosome complex → SRP receptor → translocon
1. translocon opens
3. protein synthesis resumes with protein transfer into the ER lumen
4. stop transfer enters translocon
5. protein transfer stops
6. start transfer sequence adn stop transfer sequence are released into bilayer
7. translocon closes
8. protein synthesis complete
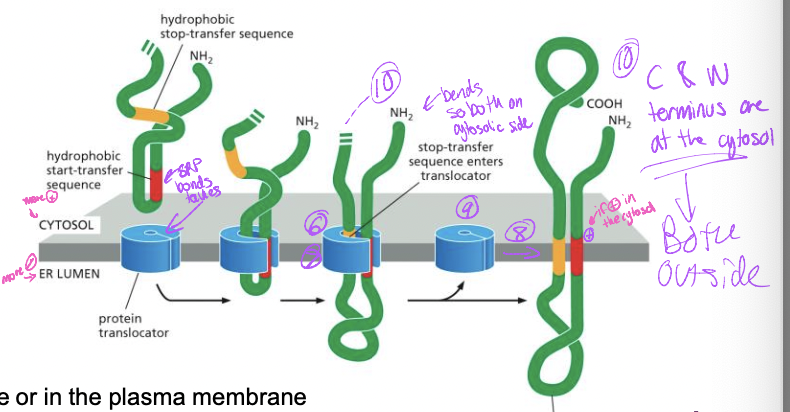
99
New cards
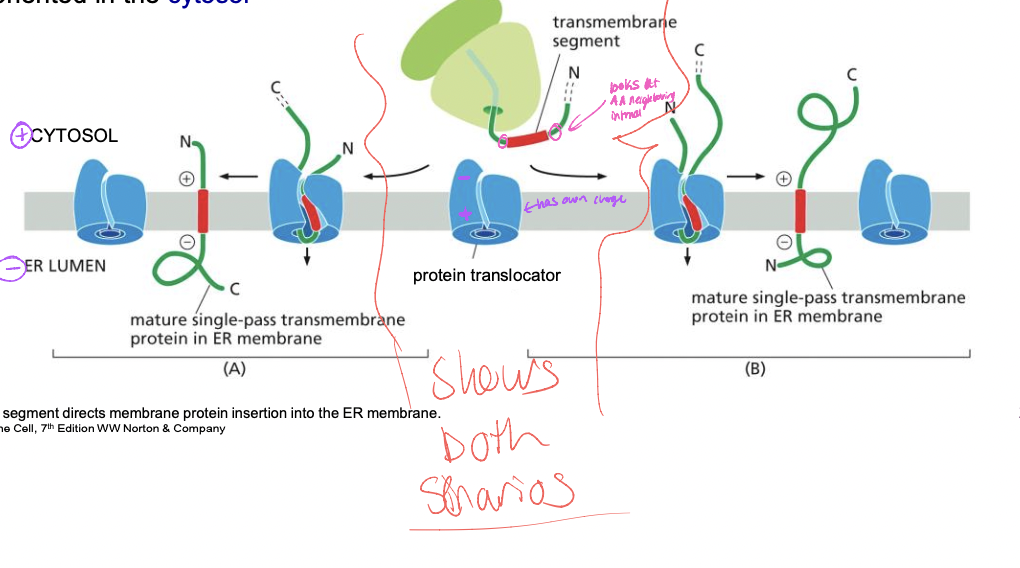
Orientation of transmembrane segment depends on
* flanking positively charged amino acids
* the N or C terminus side can be negative
* cytosol is +, lumen is -
* the positive needs to be on + - on -
* the N or C terminus side can be negative
* cytosol is +, lumen is -
* the positive needs to be on + - on -
100
New cards
Intercellular compartments are dynamic
* golgi, ER, endosomes, lysosomes are in endomembrane system
* exchange lipids, proteins
* exchange lipids, proteins