DNA to RNA to Protein
1/86
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
87 Terms
Nucleotide structure
Responsible for transmission of genetic information, preservation of genetic information, roles of nucleosides and nucleotides, energy storage, energy transmission, signaling, act as antioxidants
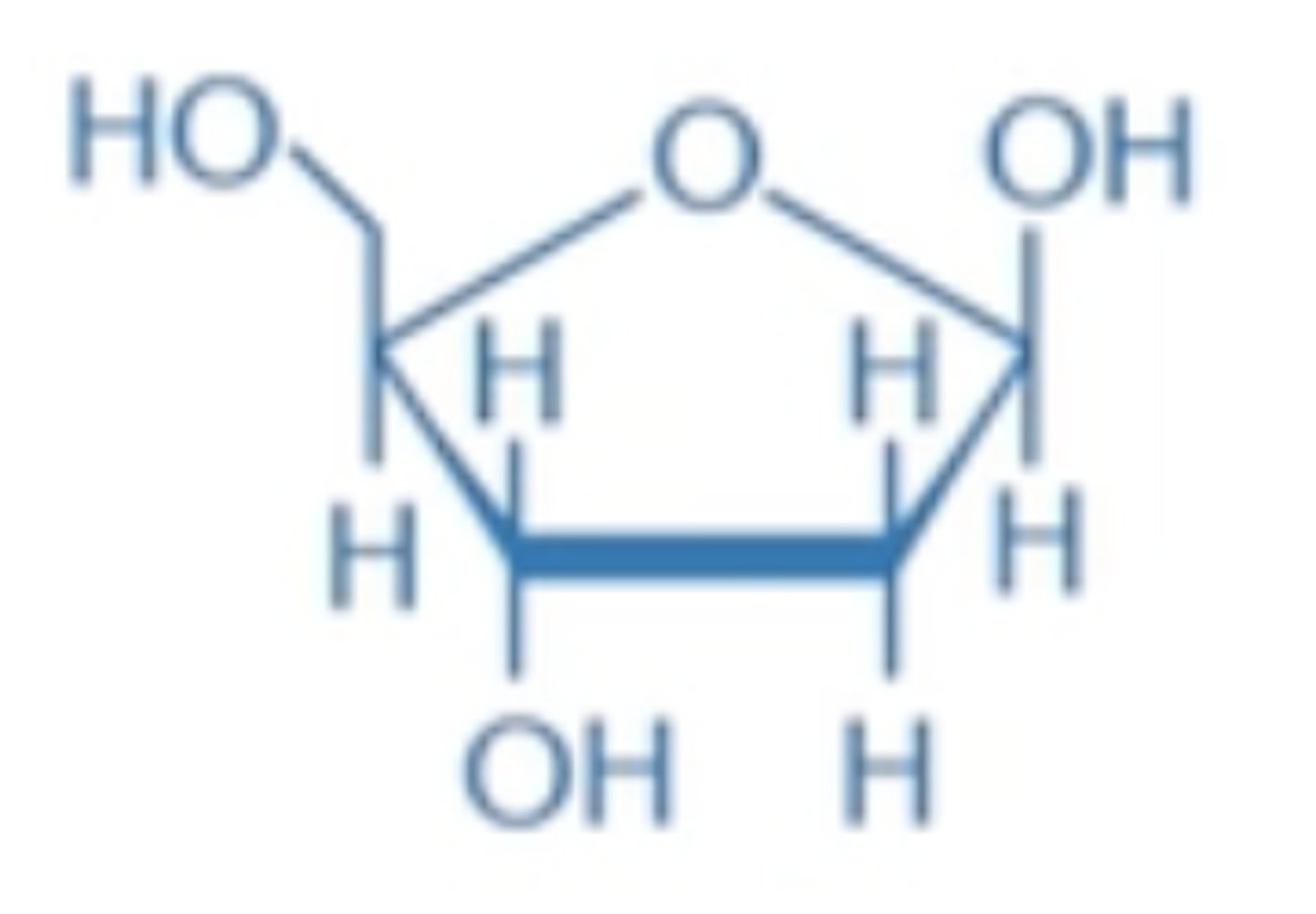
ribose
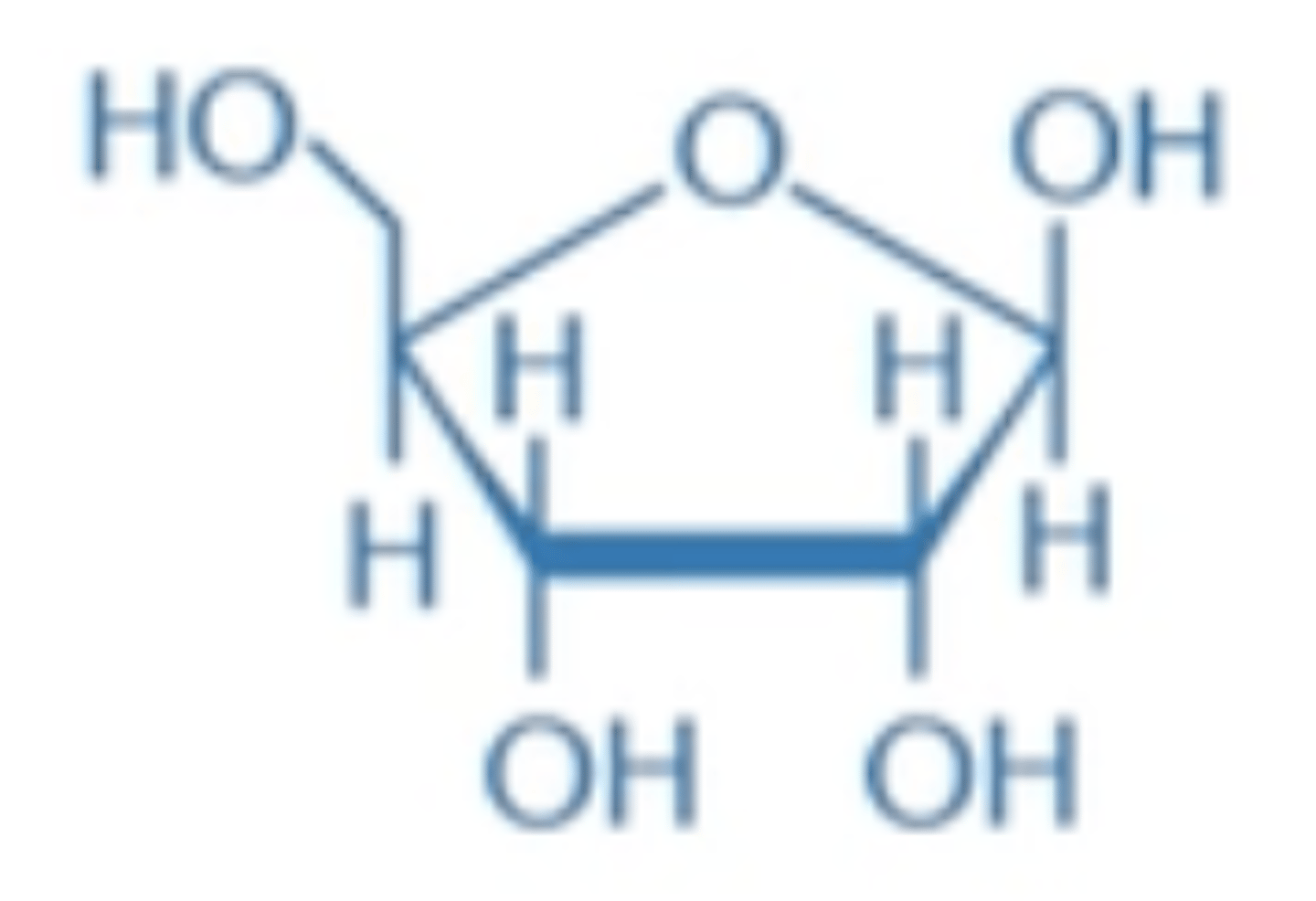
deoxyribose
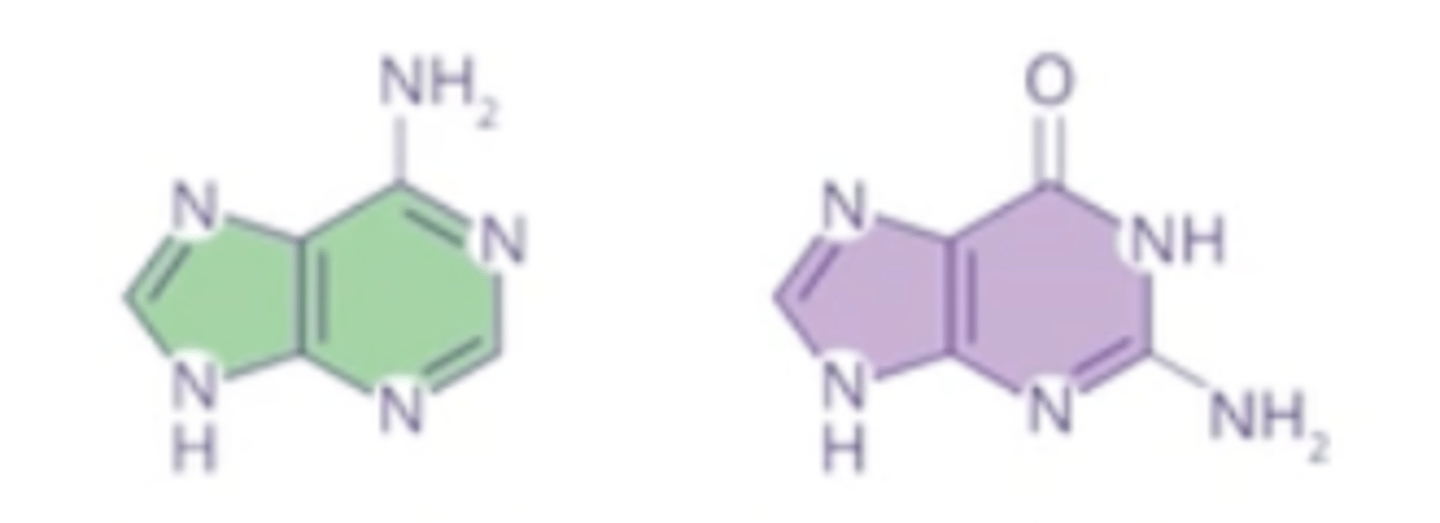
purines
A and G
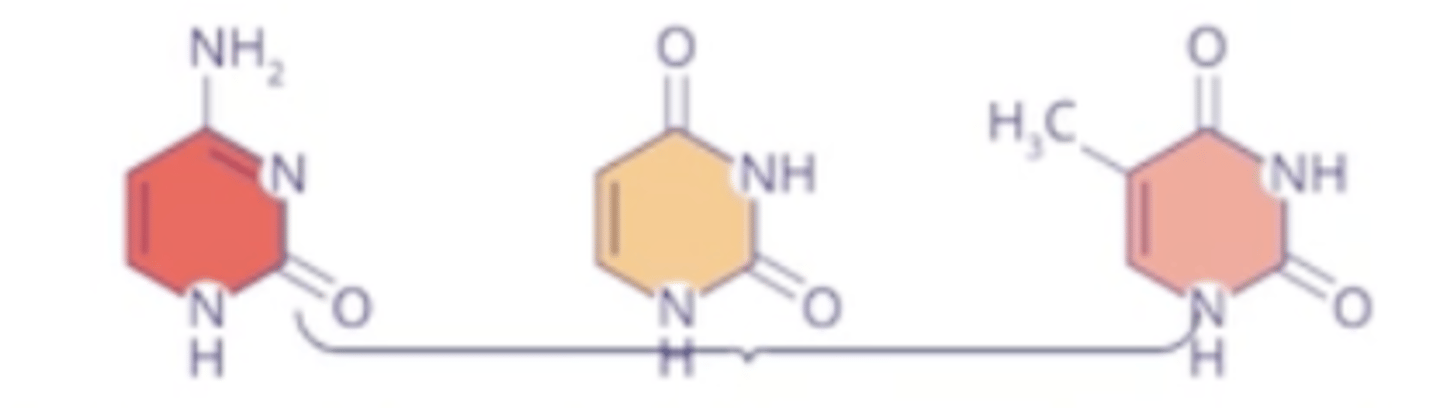
pyrimidines
C, T, and U
nitrogenous bases
Attach to the carbon 1'
Phosphate groups
One, two, or three attached to the carbon 5'
Nucleoside
Sugar and a base, but no phosphate group
Ribonucleic acids
- Nucleotides with ribose sugars
- Many can be attached to each other to make RNA
Deoxyribonucleic acids
- Made from nucleotides with deoxyribose
- Form DNA
Sugar-phosphate Backbone
The covalent bonds that connect nucleotides join the phosphate of one nucleotide to the sugar of another
The double helix
- Formed by phospho-diester bonds and form by the loss of two phosphates with each attachment
- Nitrogenous bases are hydrophobic aromatic rings that want to be away from water
- Charged phosphates are hydrophilic and very happy to be near water
Double stranded property of DNA
- Formed by intermolecular interactions = hydrogen bonds between nitrogenous bases
- Base pairing: purine always pairs with the pyrimidine
Antiparallel
- Two strands run in opposite directions
- As a result, the enzymes will operate in opposite directions
DNA denaturation
The process of separating two DNA strands into single strands
Helicase
- Separates DNA strands
- Breaks hydrogen bonds between nitrogenous bases
DNA denaturation by temperature
- Will be complete at 95 ˚C
- G-C hydrogen bonds break at higher
- A-T break hydrogen bonds at lower
Reannealing
- The process of two single DNA strands that have been separated by helicase or heat coming back together to form the original double stranded DNA
- Favored by hydrogen bonding and hydrophobic regions that want to stay away from water
Hybridization
- When a complementary (sometimes shorter or tagged) strand of DNA is annealed (paired) to another of interest
- If the sequences are not complementary, this will not occur
Central Dogma
Explains the flow of information in all biological systems
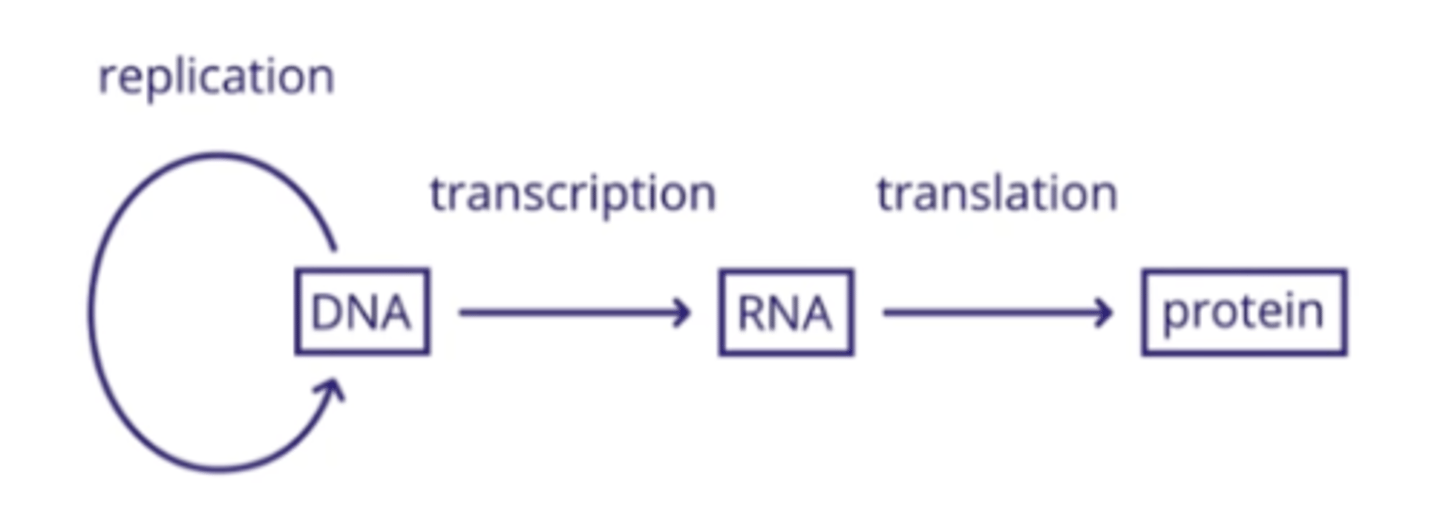
Anticodons
- Found on tRNA
- During the process of translation, each codon is recognized by a complementary one of these
Codons
- 64 possible
- Each is specific and unambiguous, corresponding to precisely one amino acid
- But the genetic code is degenerated: has built in redundancy so that more than one of these can correspond to the same amino acid
- How we have only 20 amino acids
Wobble
- Due to degeneracy of genetic code
- When multiple codons code for the same amino acid, most often those codons those codons share the same first two bases
- The third nucleotide of the codon is the variable one
- Due to the tRN anticodon literally wobbling in matching the final nucleotide to the mRNA codon
- An evolutionary development that protects us against mutations
- If a mutation occurs in the wobble position, or a slightly wrong tRNA is brought in, that change is likely to be a silent mutation (no effect on the polypeptide sequence)
Point mutation
A mutation affecting only on nucleotide in a gene sequence
Missense mutation
- A mutation where one amino acid is substituted for another
- Alter the primary amino acid sequence of the protein
- We call them expressed mutations
Nonsense mutations
- Are even more serious than missense mutations
- They change a codon from an amino acid to code for a premature stop codon instead
- Also known as truncation mutations
Silent Mutation
does not affect the protein
Frameshift Mutation
involves the deletion or insertion of a base which results in a change to the gene’s reading frame
Deleterious mutation
genetic variation that is known to be associated with an increases risk of disease
AUG
- Start codon on the mRNA
- Codes for methionine
- It signals where to start translation of mRNA to protein
Stop codons
UAA
UGA
UAG
Initiation (transcription)
- The start of transcription
- involves the:
promoter region of the gene
Transcription factors
TATA box (euk)
The Pribnow box (pro)
Helicase
RNA pol
Promoter region of the gene
RNA pol binds onto the DNA many base paris in advance of the actual start site of transcription
Transcription factors
proteins that may bind to the promoter region to help signal that RNA pol should bind there as well
TATA box
- Most euk and pro genes have a sequence of bases that is rich in T and A nucleotides that RNA pol recognizes and binds onto
- Is located 25 to 35 base pairs upstream of the actual start site
The Pribnow box
- In prokaryotes
- About -10 base pairs from the start site
- Not a TATA box
- rich in Ts and As and also the site of RNA pol binding
Helicase
- Unwinds the DNA, breaks the H-bonds between nucleotides
- A subunit of RNA pol itself
RNA pol
- Reads the base of the gene and attaches a complementary RNA nucleotide onto the DNA
- Keeps reading the DNA and matching the correct RNA base pair, while simultaneously catalyzing a new phosphodiester backbone bond between the growing RNA strand and the next RNA nucleotide
elongation (transcription)
involves: RNA pol, Sense strand/coding strand, Antisense strand/template strand
Sense strand/coding strand
The strand of DNA that looks exactly like the synthesized strand of RNA
Antisense strand/template strand
The strand of DNA that is the opposite of the synthesized strand of RNA that was read by RNA pol to create complementary RNA base pair match
Termination in prokaryotes (transcription)
Rho independent or Rho dependent
Rho
A protein that can bind on the synthesized strand of RNA
Rho-dependent
- Rho binds to the synthesized strand of RNA, which introduces steric strain which tugs n the RNA and pulls it away from the RNA pol
- The RNA is now ready to be immediately translated and the RNA pol finishes the job by sealing together the DNA strands and detaching to go find another gene to transcribe
Rho-independent
- G-C rich sequence in DNA is then synthesized as a G-C rich sequence of mRNA
- They create a hairpin loop (fold) that causes steric strain, tugging RNA away from the DNA
termination in eukaryotes (transcription)
includes 5' modification, 3' poly A tail, and splicing
5' modification
- The addition of a methylguanosine cap (guanine nucleotide attached backwards)
- Prevents the degradation of the mRNA transcript
- Prevents it from fitting in the active site of the exonuclease that would have degraded the mRNA transcript
3' poly A tail
- Long sequence of adenine residues is added to end of the mRNA
- Prevents degradation
- The exonuclease would have to break away each adenine one by one, which would take more time before it reaches the signaling the part of the mRNA
- Longer = more iterations of translation of the mRNA, signal stays around longer
- Shorter = message lasts around less time
splicing
Excising introns: introns removed before the mRNA exits the nucleus
rRNA
- Structural RNA that composes the large and small subunits of the ribosomes themselves
- Do not code
Structure of ribosomes
- The small subunit binds first
- The large subunit contains the A, P, and E sites
Initiation (translation)
The small subunit binds to the mRNA before the start codon
initiation in Prokaryotes
The small subunit binds to the shine-dalgarno sequence
initiation in Eukaryotes
The small subunit binds to the 5' cap from the post-transcriptional modification
translation
- doesn't start until AUG is recognized by the small subunit
- Then the large subunit initiates the binding at the A, P, and E sites
Elongation (translation)
An amino acid bound to the tRNA binds to the A site and is moved over to the P site via peptidyl transferase
Termination (translation)
- Determined by stop codons
- Release factor comes and binds to the A site to release the ribosome from the mRNA and the tRNAs
Chaperone proteins
Condense the polypeptide into a 3D shape so that it becomes a recognizable protein
Chromatin structure
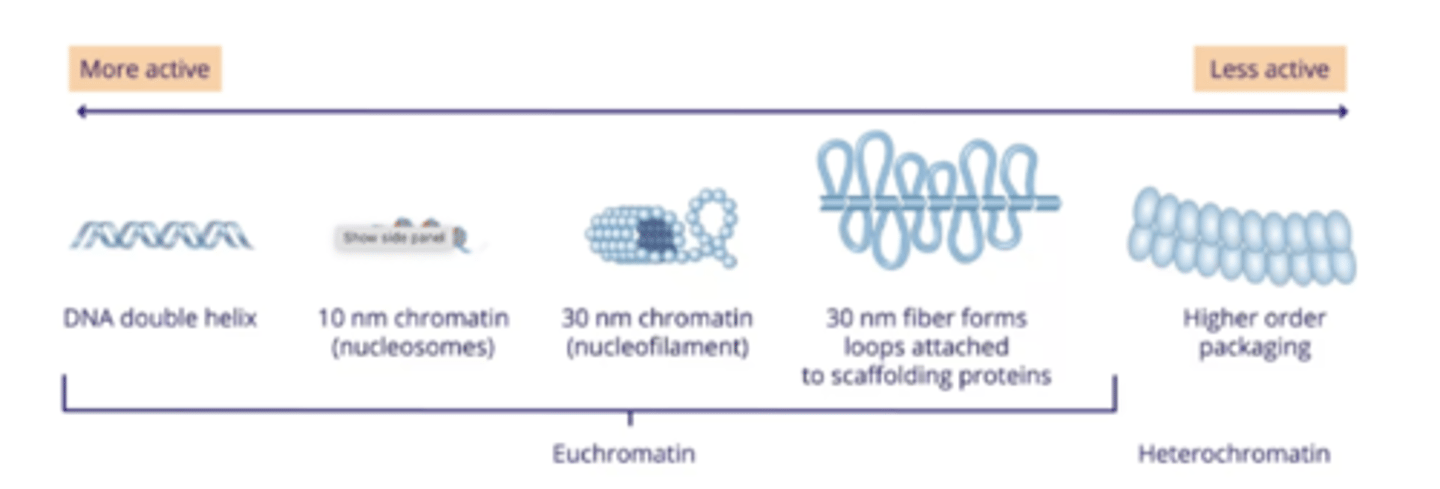
Histones
- Protein that DNA molecules are wound around
- Around 200 bp of DNA are wounds around one complex
Nucleosome
The complex of a histone protein wrapped by 200 bp of DNA
Euchromatin
- When the nucleosome complexes look like beads on a string
- Allows the DNA molecule itself to be easily accessed
- Crucial to allow RNA pol to bind to DNA and initiate transcription
Heterochromatin
Further compacted euchromatin
DNA methylation
- Way to alter the structure of chromatin
- Cytosine and adenine bases in DNA can be methylated
- Hinders the ability of RNA pol to transcribe and is also associated with more compacted DNA
Histone acetylation, methylation, and phosphorylation
- Decrease their affinity for DNA and makes the DNA molecule more accessible
- Conversely, removing these groups makes the chromatin more compact so it's less available for transcription
Histone displacement
- Slide apart
- Allow the DNA sequence between them to be accessed more easily
Transcription factors
Bind to response elements (specific DNA sequences) which are located in the promoter region of every gene
General transcription factors
- Serve crucial roles in transcription machinery
- Recruit RNA pol and unwind DNA helix
Selective transcription factors
- Bind to specific response elements that are found in only some genes
- The response elements they bind to are often found hundreds of bases away from the promoters of the gene (enhancers)
give the cell an excellent way to regulate which genes are expressed
- In the presence of specific signals, they will bind to their response elements in the enhancer region and either promote or inhibit the transcription of those genes
Post-transcriptional regulation
The nucleus can regulate which mRNA transcripts and how many are exported from the nucleus
Untranslated regions or UTRs
- On both the 5' and 3' end of the mRNA transcript
- Generally not translated into protein
- Serve critical functions in regulating translation
- 5' UTR: ribosome relies on it to bind to to begin translation
- 3' UTR: often forms secondary structures that help stabilize the transcript or are recognized by specific enzyme that degrade the transcript
Oncogenes
- Promote cell division
- Overexpressed in cancer cells
- More cell division → tumor
- Normally are suppressed in nondividing cells, preventing tumor formation
- Many kinds of cancers are the result of the overexpression of these genes
Tumor suppressor genes
When incorrectly suppressed, cells either divide more frequently (increase likelihood of tumor formation) or accumulate DNA damage (making more susceptible to tumor-promoting mutations)
Gatekeeper
- Directly supports growth
- Cell cycle regulator genes
- Checkpoint control genes
- Apoptosis related genes
Caretaker
- Maintain overall genetic stability
- DNA repair proteins
PCR
- Use a set of primers (complementary DNA sequences that we design) to locate sequences of DNA on the target site
- DNA strands go through cycles of denaturing, hybridization, and replication
Solution used for PCR
Contains DNA fragments, dNTP (ATCG), enzymes, and primers
Denaturing step of PCR
- Uses incredibly high temperatures to heat up solution and separate double stranded DNA sequences into two parental single-stranded sequences
- To about 94-96 ˚C to break down hydrogen bonds
Hybridization step of PCR
- Solution is cooled
- DNA strands will anneal to the primers to form a short DNA hybrid
Replication step
- DNA pol comes in to extend the hybrid DNA
- Reaction mixture will be heated to at least 70˚C in order for Taq pol to function properly
- Sequences then form dual stranded hybrid that can then be broken apart and the cycle can be repeated with a new template
Taq polymerase
- Used in PCR to make sure polymerase doesn't denature at very high temperatures
- From bacteria that live in volcano vents on the ocean floor
Gel electrophoresis
- Figure out how many base pairs long a segment of DNA is
- Works by moving fragments of DNA through an agarose gel plate
- Smaller pieces of DNA can move faster than larger ones
- DNA fragments all move forward because of the negatively charged phosphodiester backbone
- Can compare the fragments' movement to a DNA ladder (a separate solution of carrying fragments of DNA that we already know the exact sizes of
- Only allows us to determine the size of the fragment
Southern blotting
- Use probes to identify the target DNA after it has been run on a gel
- Useful for identifying if the target gene is in the sample
- DNA is blotted with a sheet containing a probe designed to hybridize with our desired DNA, after it's run through gel electrophoresis
- Nitrocellulose paper is used to block the DNA segment out of the gel, and onto the membrane
- A solution containing a fluorescently labeled hybridization probe for our target sequence is then placed on the membrane itself
- Since the primer only binds to a specific sequence that we design, the presence of a band after imaging the blot means that we've identified our gene of interest
Northern blotting
used to identify RNA
Western blotting
used to identify proteins
Subcloning
- The process of inserting genetic material into a plasmid, to make recombinant DNA, and is a form of cloning
- The problem is that there are many different plasmids that we can insert DNA sequences into
- Knock-in: pAcc-B plasmid
Restriction enzymes
Are site-specific, so enzymes will only latch onto a particular sequence of DNA on our plasmid