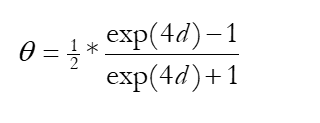BIN300 W1- genetic distances
1/11
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
12 Terms
genetic distances
number of cross-overs between loci
recombinations vs cross-overs
Think of recombination as the entire process of recipe mixing for new dishes. A crossover is like physically swapping ingredients (e.g., exchanging spices between two recipes) — a specific part of the process.

genetic distance d
average number crossover per chromsome segment
morgan
genetic distance where on average 1 crossover occurs
recombination fraction Ɵ
fraction of times of uneven number of crossovers
recombination → are 2 loci inherited together or not?
Max Ɵ = 0.5
physical distances between loci
number of kbase pairs (nucleotides
recombination distance Ɵ
probability of uneven number of cross-overs (max. Ɵ = 1/2)
genetic distance in cMorgan
1 cM
1 cM is on average 1 crossover/100 meiosis
mapping function in relation to genetic distance
Ɵ = f(d)
Ɵ=d (Morgan mapping function)
generally non-linear function
describes relationship between d and Ɵ
Morgan and Haldane mapping function graph
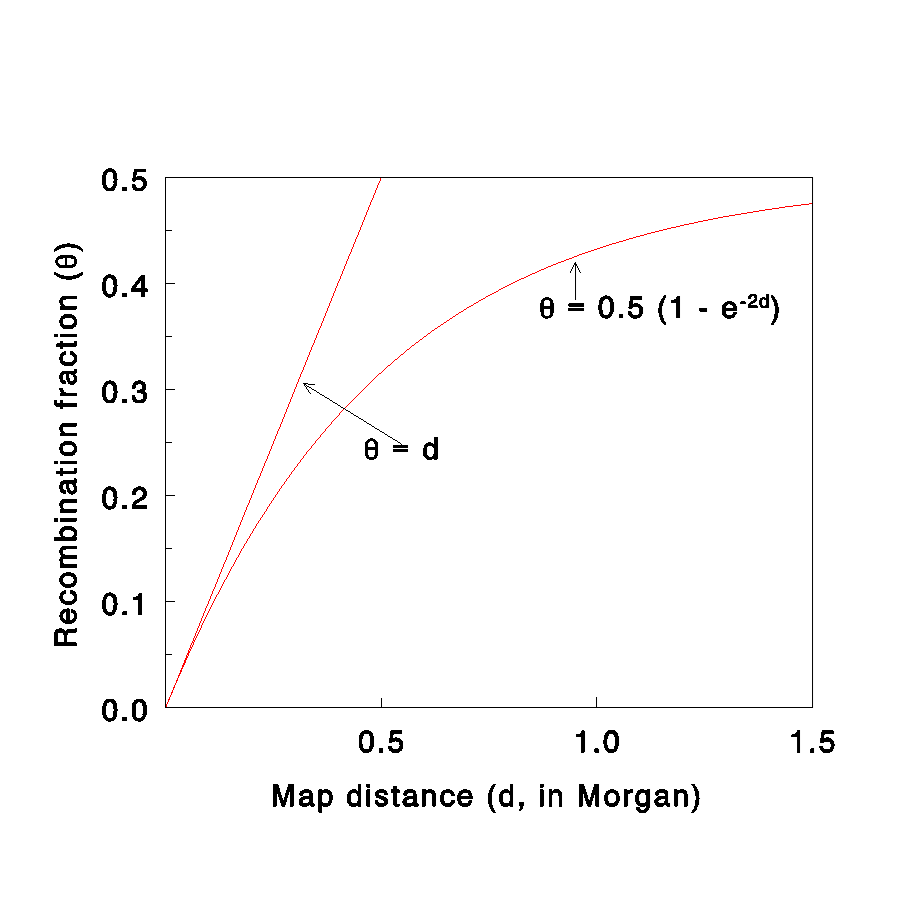
haldane mapping function
assumes independent cross over probs
no interference
Ɵ=1/2(1-exp(-2d))
d=1/2ln(1-2Ɵ)
addition rule for 3 loci A_B_C
dAC = dAB + dBC
ƟAC=ƟAB+ƟBC-2ƟABƟBC
Kosambi mapping function
assumes interference
crossover at locus X affects prob of recombination at locus X+d
interference decreases with distance