1. Regulation of gene expression : transcription in prokaryotic cells
1/54
Earn XP
Description and Tags
Molecular Biology for prokaryotic cells
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
55 Terms
1.1 General structure of an operon
Promoter, +1 = Transcription Starting Site (TSS), Shine Dalgardo (SD) sequence and a Terminator
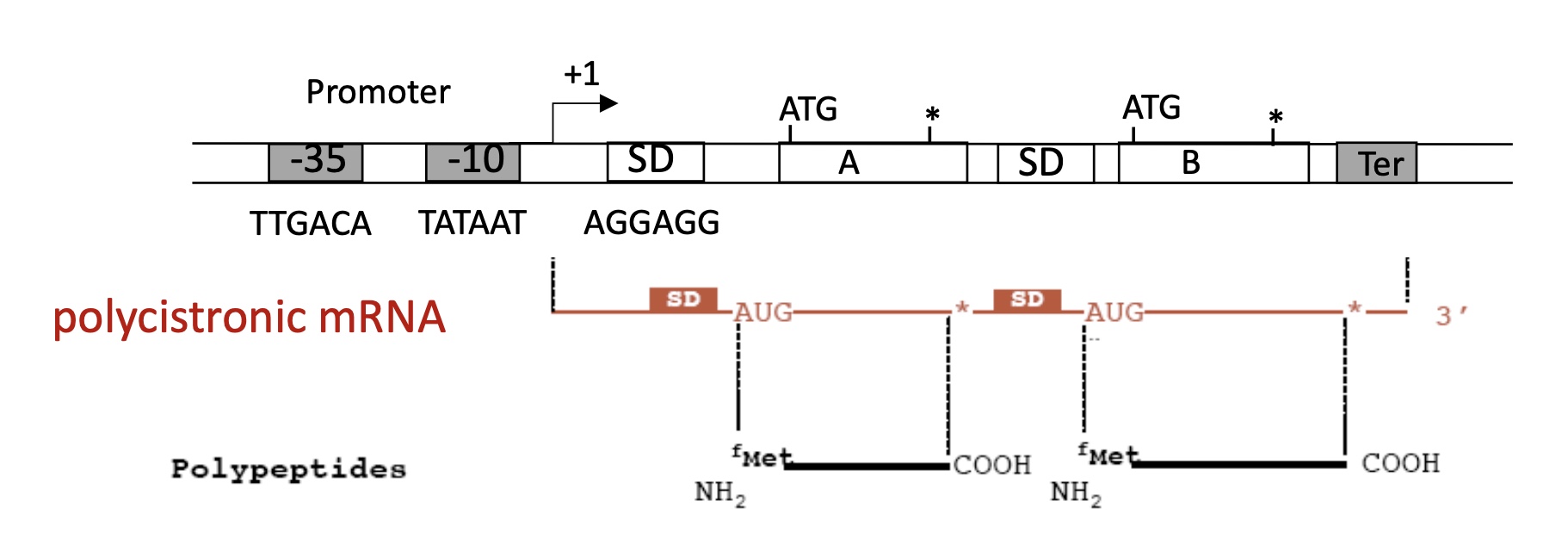
1.1 General structure of a Promoter
2 consensus sequences :
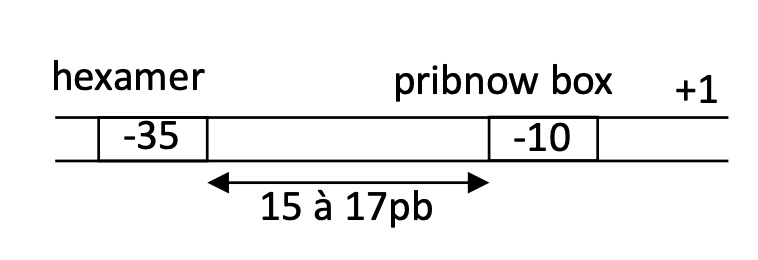
Hexamer - 35
Pribnow box -10
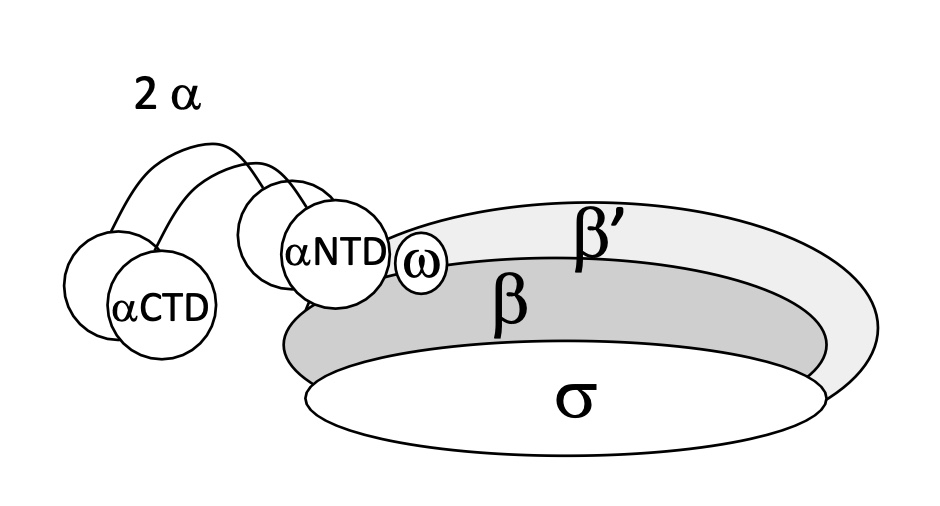
1.1 ARN pol structure
5 sub units :
X2 α
β
β’
ω
= Core enzyme
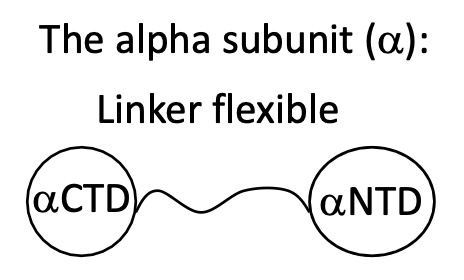
1.1 Role and structure of α
2 extremities αCTD & αNTD
αNTD —> associates to β
αCTD —> interacts w/ DNA 7 transcription regulatory proteins
1.1 Role of β & β’ sub-units
contain active site to remain attached to DNA
non specific binding to DNA strand
1.1 Function of ω sub-unit
Facilitates assembly of RNA polymerase
1.1 Why does ARN binds w/ sigma σ sub-unit ? What is the complex called ?
To initiate the transcription as σ sub-unit is responsible for recognising the consensus sequences on the promoter
Core enzyme + σ = Holoenzyme (6 sub-units)
1.1 Specificity and function of sigma σ sub-unit
contains many sub-regions that’s have a high specificity for the promoter and their consensus sequences
prevent transcription starting from anywhere
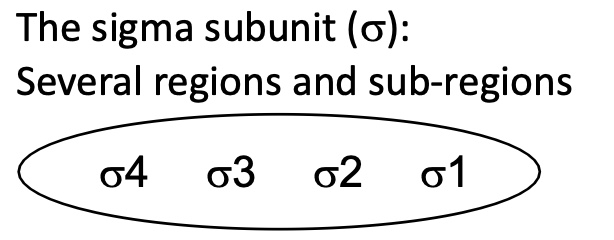
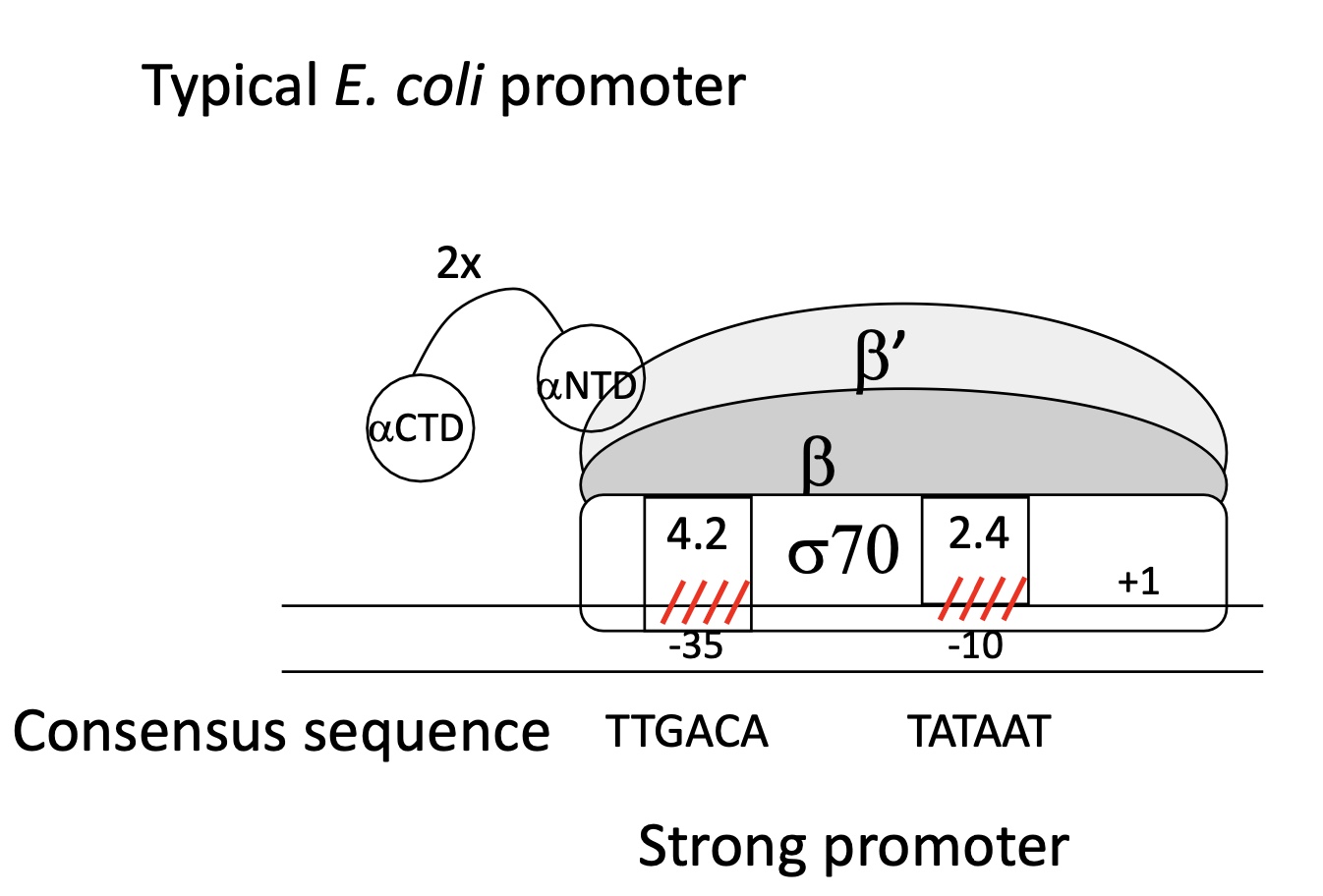
1.1 How can a promoter be a strong promoter
Hexamer & Prinbow box have a sequence identical/very similar to consensus sequence
Strong binding between domains of σ sub-unit with the sequences
Strong association of RNA pol w/ promoter
initiation of transcription
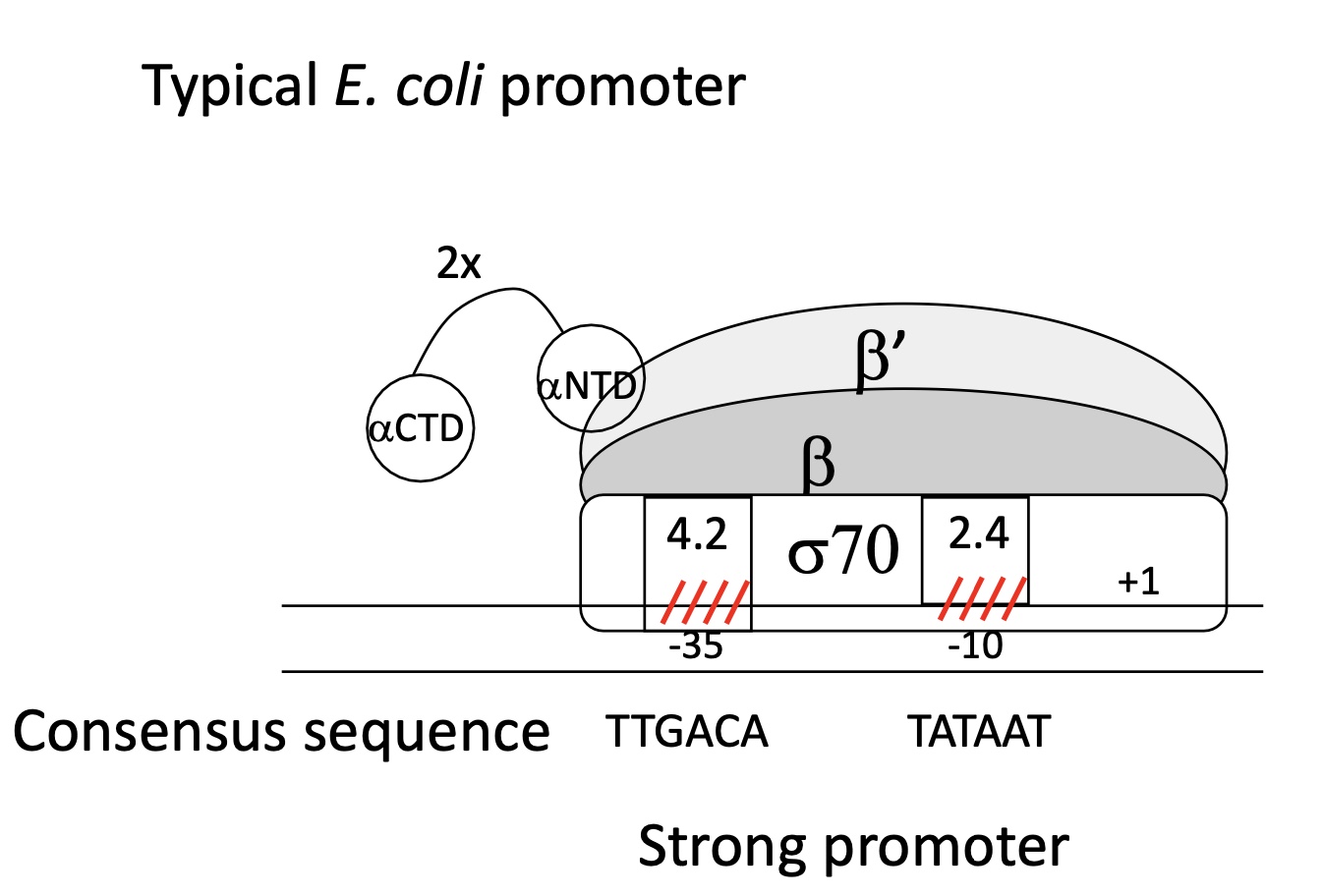
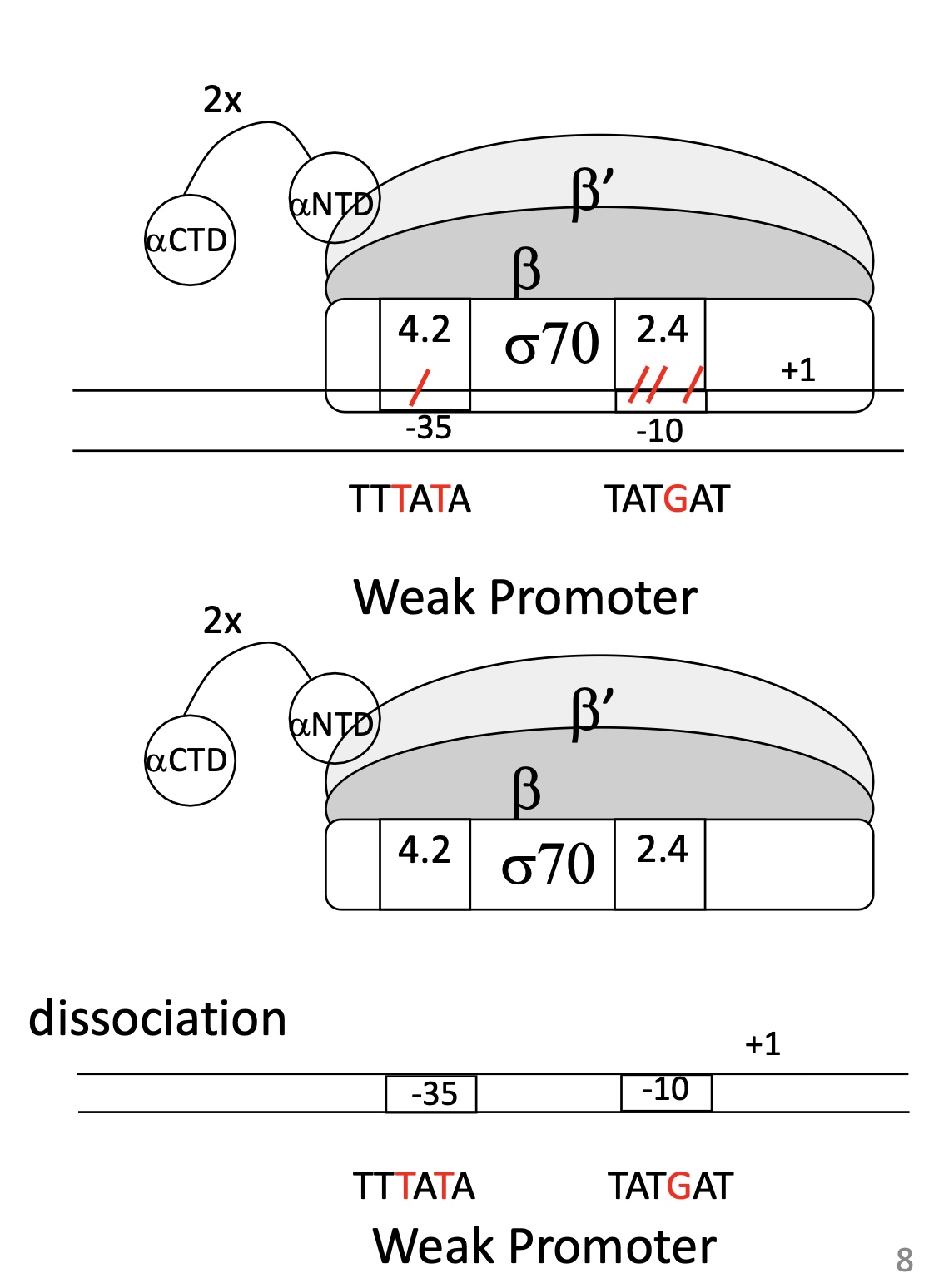
1.1 What makes a weak promoter
Hexamer & Prinbow box sequences are different from consensus sequences
Weak binding between domains of σ sub-unit
Weak association of RNA pol w/ promoter
Dissociation of ARN pol from promoter —> no transcription
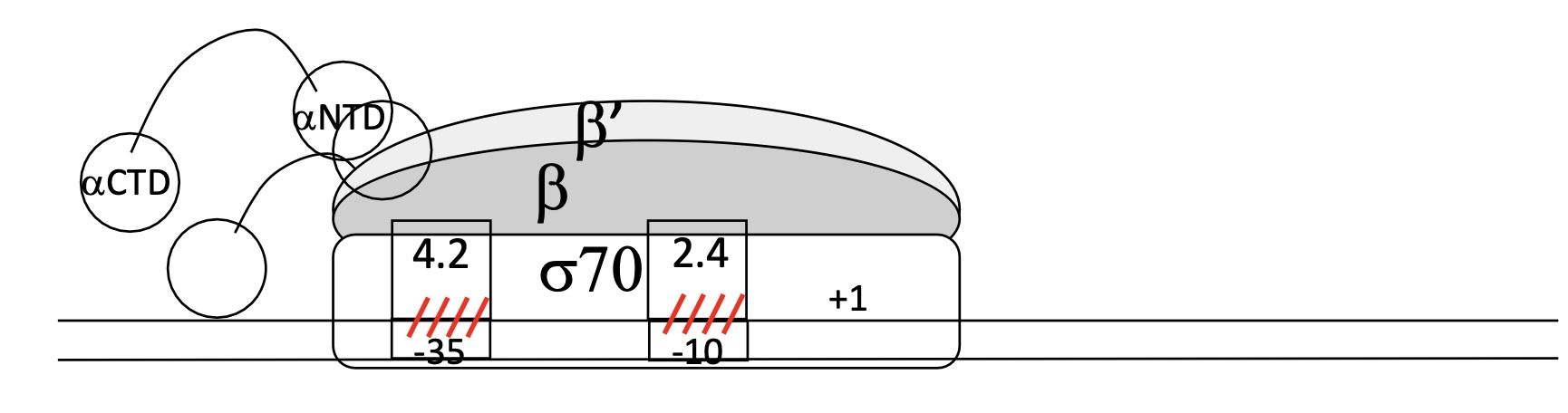
1.1 E.g. of σ sub-unit, and the domains that bind to Hexamer & Prinbow box on the promoter in E.coli
σ70 = σ sub-unit
2 domains ":
4.2 binds to Hexamer
2.4 binds to Prinbow box
1.1 What are the 3 steps of INITIATION of transcription ?
Fixation of ARN pol (holoenzyme) on the promoter = closed complex (double strand of DNA remains closed)
Double strands of DNA are separated = open complex
Promoter escape (holoenzyme —> core enzyme)
1.1 What is the closed complex in the first step of INITIATION of transcription
RNA pol/Holoenzyme is associated to the promoter of the DNA
DNA double strand remains closed
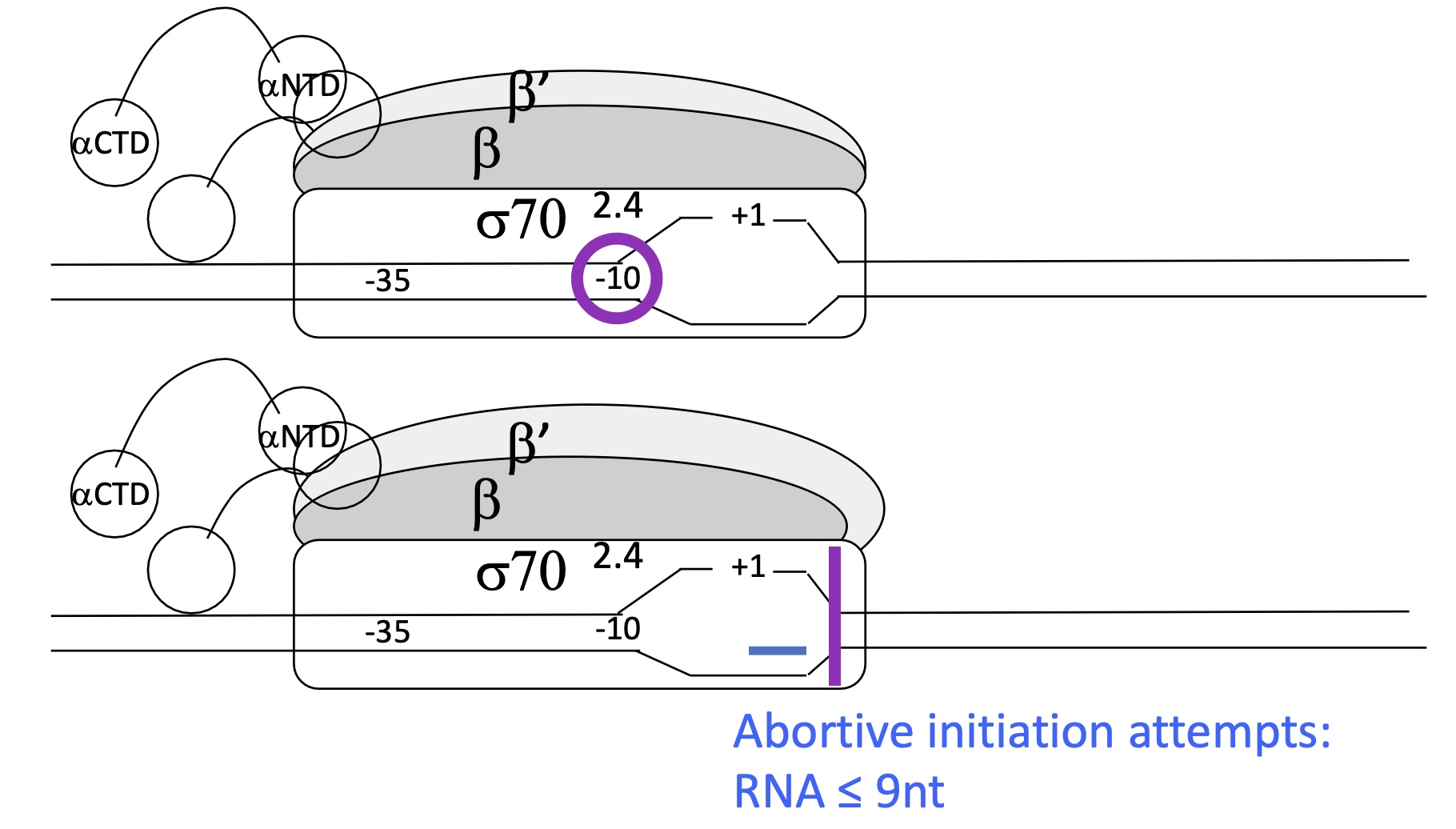
1.1 Explain what happens in the 2nd step of INITIATION (open complex) of transcription
DNA is opened by 13bp at the Prinbow box ( as only 2 H bonds between T-A compared 3 in C-G) by the Holoenzyme
Transcription blocked by RNA Pol/Holoenzyme at an RNA size of ≤ 9 nucleotides as
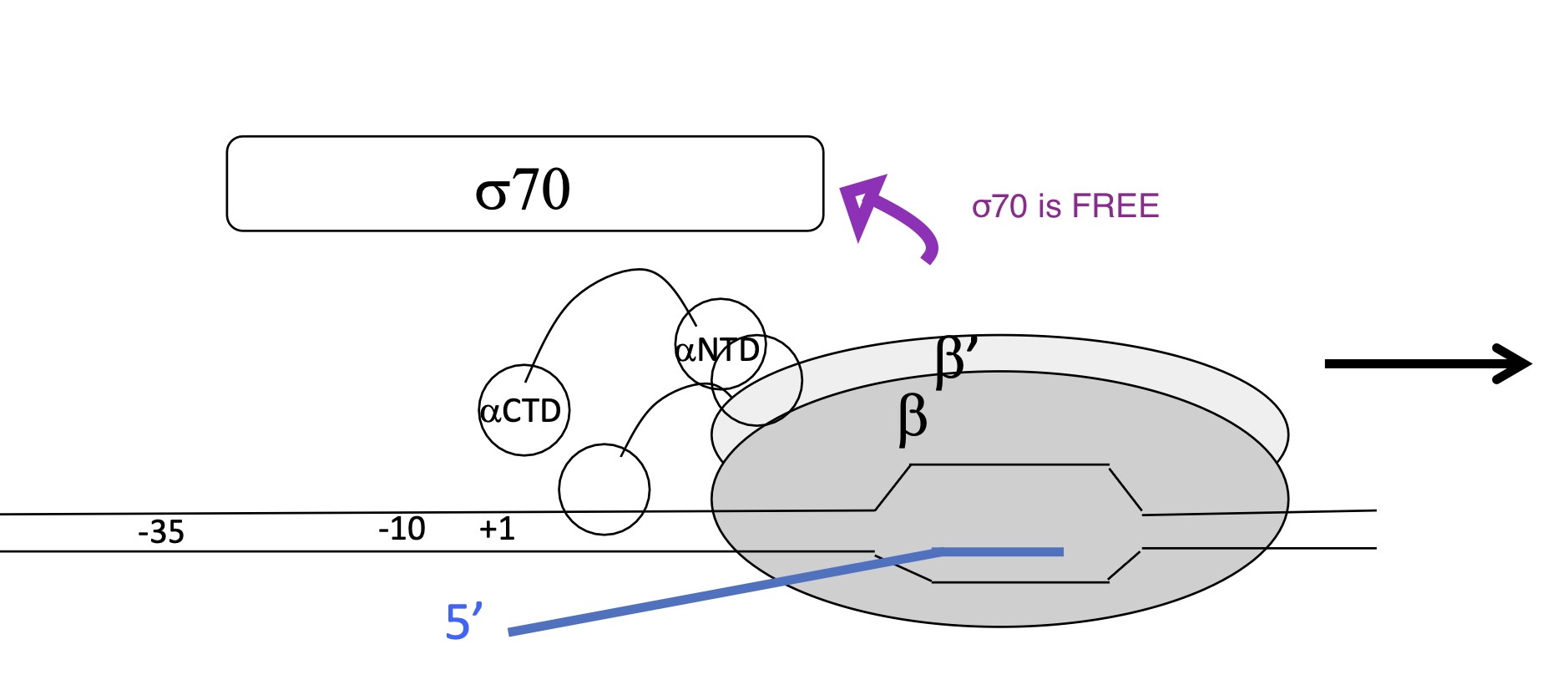
1.1 Explain promoter escape (3rd step in INITIATION of transcription)
After 10bp have been transcribes the σ70 sub-unit dissociated from core enzyme
RNA pol/Core enzyme is no longer fixated to the promoter
RNA pol/Core enzyme moves down DNA and Strats elongation w/ complementary base pairing
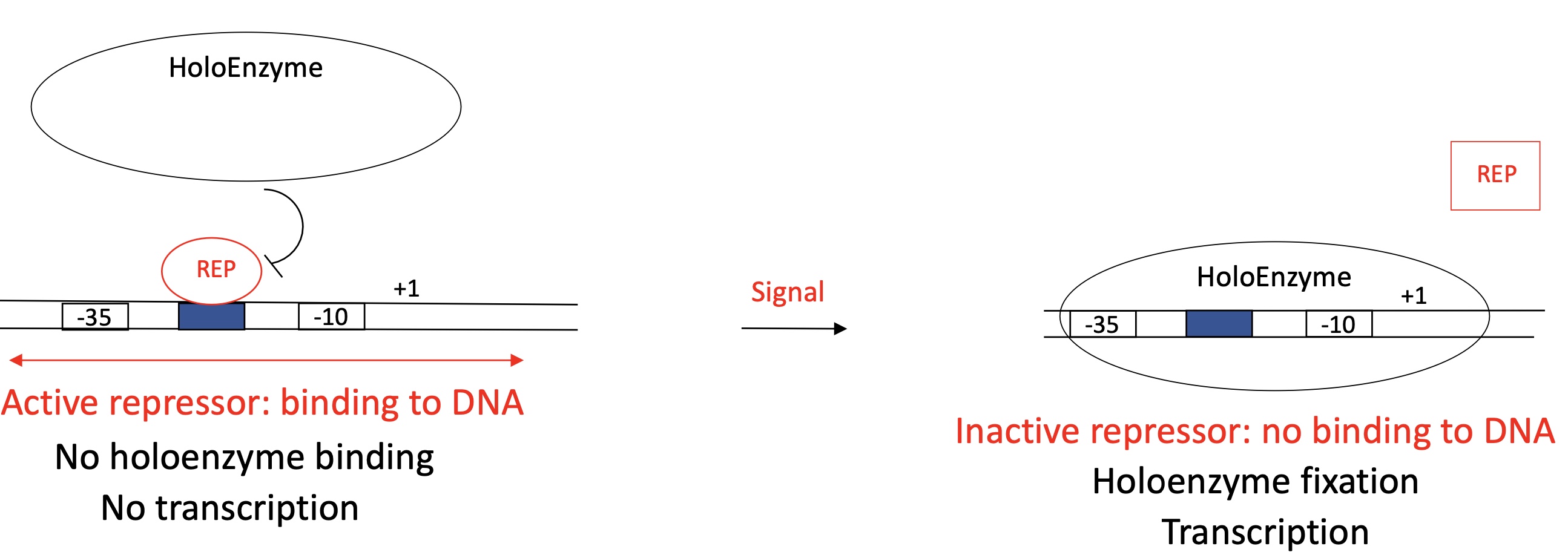
1.2 Repressor general model to control the promoter ; location in between -35 & -10 and function
Repressor binding site in between -35 et -10
Active repressor :
Repressor protein binds to REP binding site = active repressor
Inhibition of binding of RNA pol/holoenzyme
No transcription
Inactive repressor :
REP protein doesn’t bind to REP binding site due to a signal
Holoenzyme can bind to the promoter
Initiate transcription
1.2 Repressor general model to control the promoter ; location after -10 and function
Rep binding site further down after -10
Active repressor :
REP protein binds to REP binding site
Holoenzyme can bind to the promoter but is blocked by Rep protein
Holoenzyme can’t move forward
No transcription
Inactive repressor :
Rep doesn’t bind to REP binding site due to signal
No blockage from REP protein
Holoenzyme not blocked and move forward
Transcription occurs
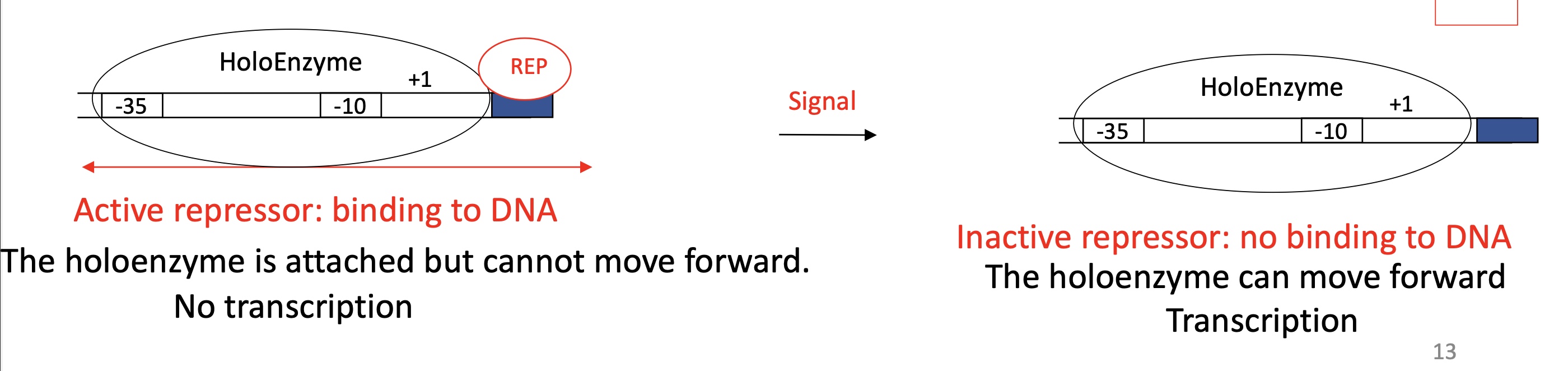
1.2 General activator model to control the promoter ; Fixation of weak promoter
Activator binding site before -35
Inactive activator
ACT protein doesn’t bind to ACT binding site
Holoenzyme binds but quickly dissociates from DNA
No transcription
Active activator
ACT binds to ACT binding site due to a signal
Stabilises intégration of holoenzyme w/ the promoter
Transcription takes place
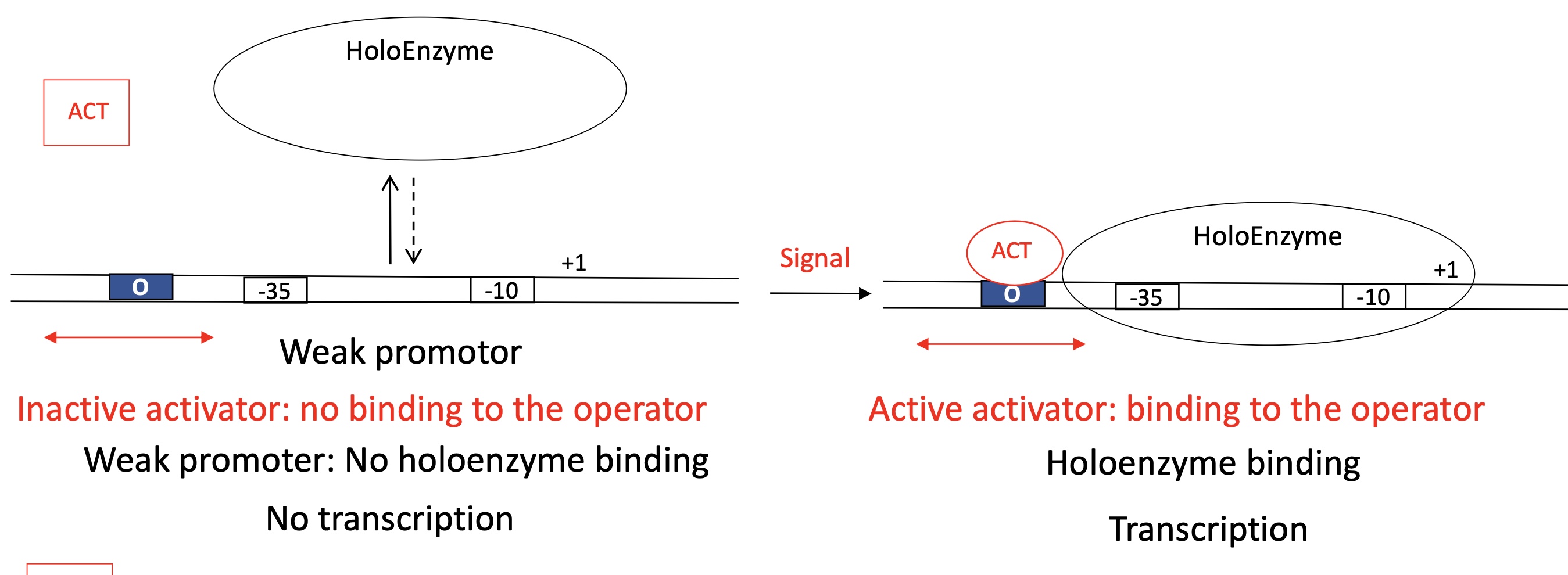
1.2 General activator model to control the promoter ; Transition from close —> open complex for a weak promoter
ACT binding site before -35
Inactive Activator
ACT doesn’t bind to ACT binding site
Holoenzyme is bound to promoter
Unable to open double stranded DNA —> remains in closed complexe
No transcription
Active Activator
ACT binds to ACT binding site
Allows Holoenzyme to enter the open complex
DNA is opened
Transcription takes place
1.2 Class of Activators
Class I : fixation of holoenzyme
Class II : closed —> open complex
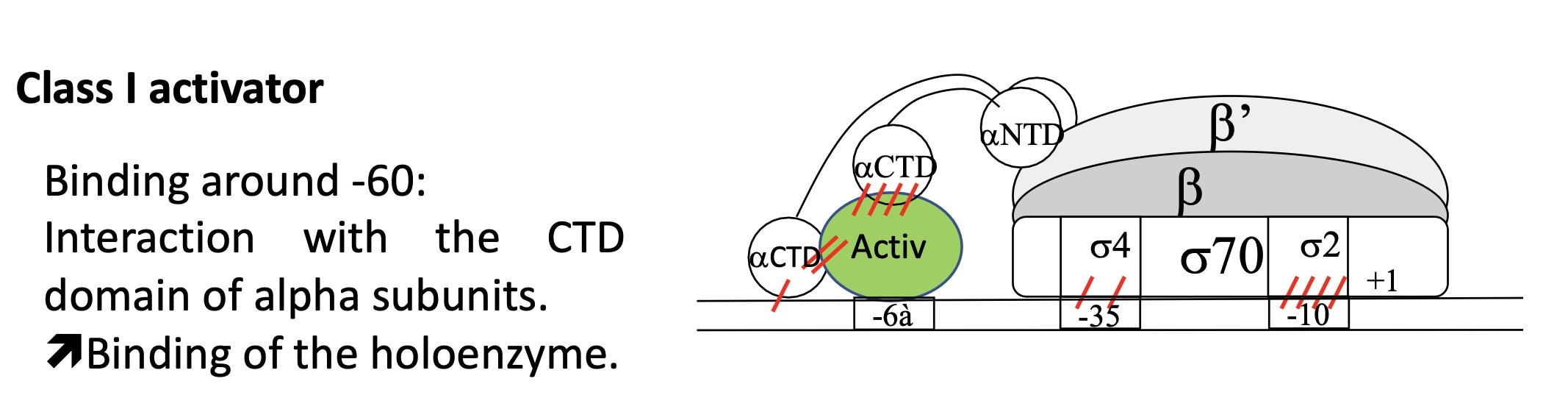
1.2 E.g of Class I Activators
fixation around -60
interaction of αCTD with ACT protein to stabilise interacation of holoenzyme w/ promoter
1.2 E.g of Class II Activator close to holoenzyme
= The majority of ACT
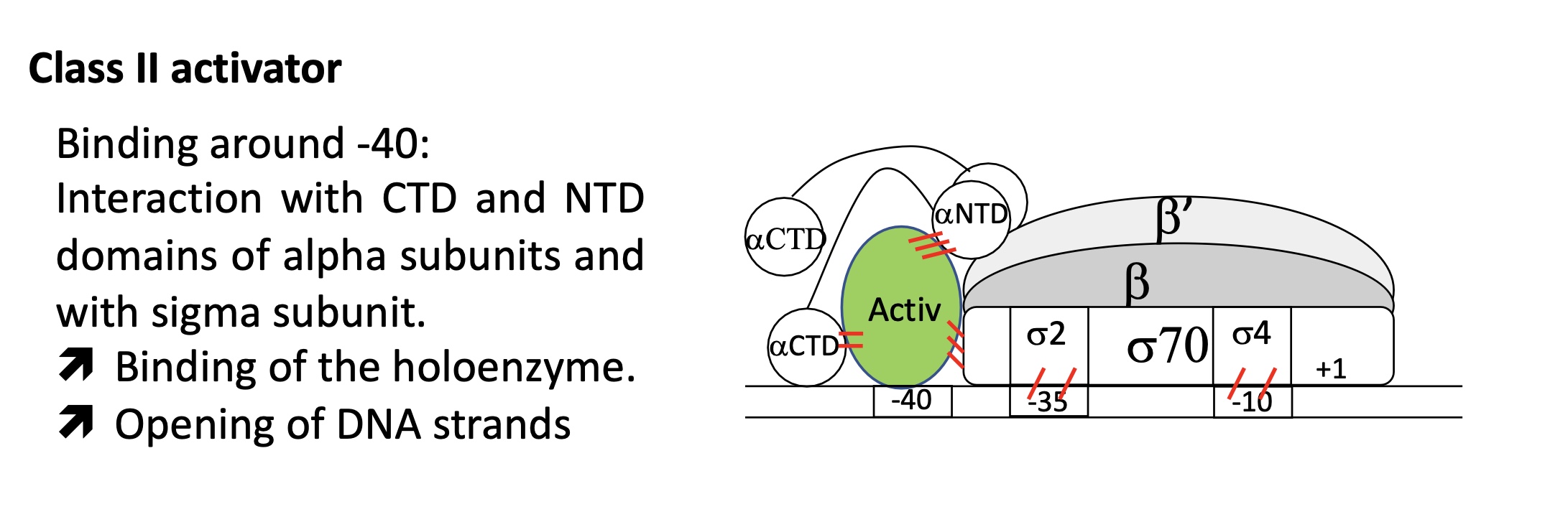
fixation around -40
ACT interacts w/ αCTD, αNTD & σ sub units
stabilises interaction of holoenzyme w promoter
allows transition from closed to open complex
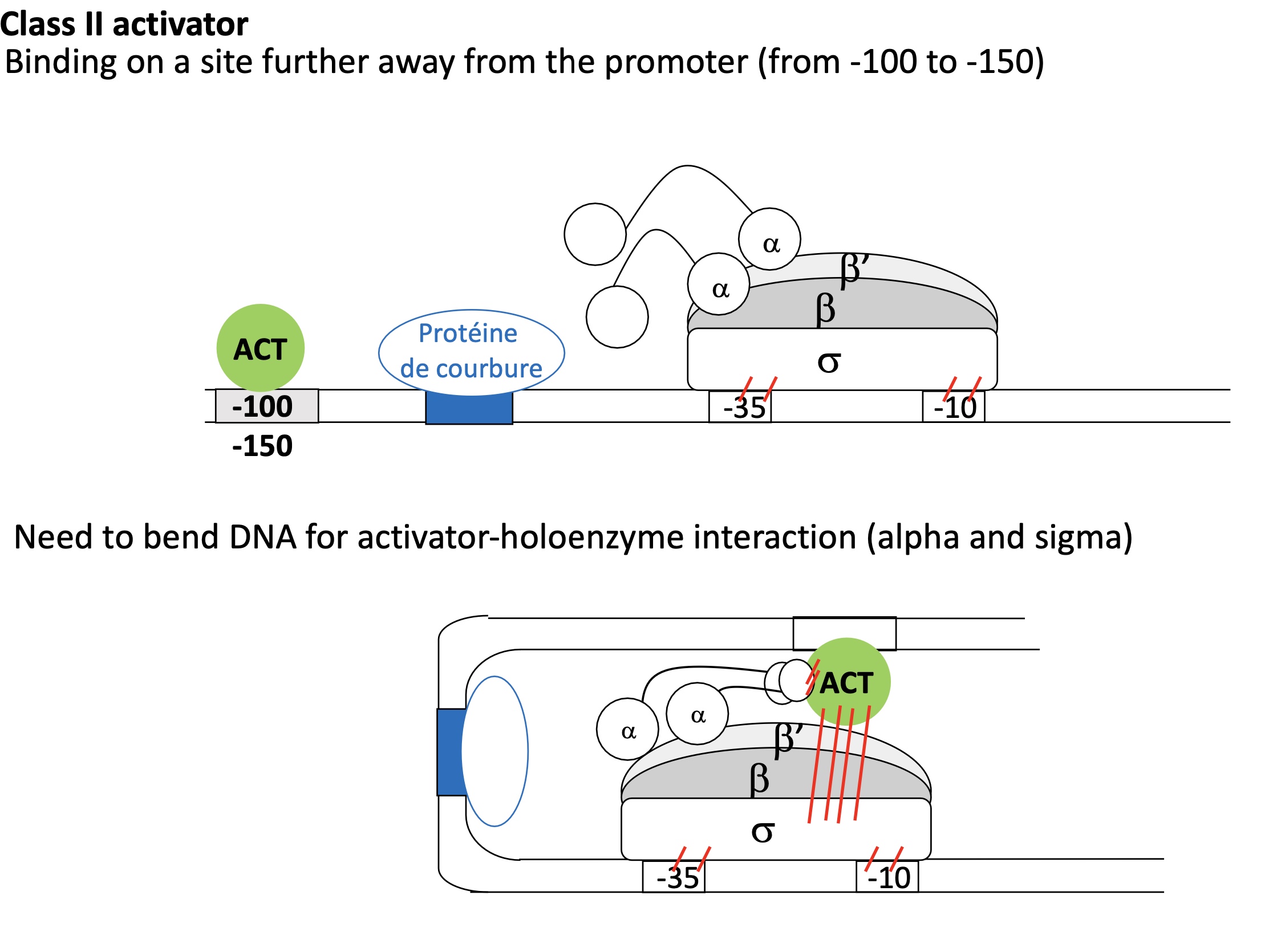
1.2 E.g of Class II Activator far from holoenzyme
ACT binding site is distant from Holoenzyme
Bending protein blinds to its binding site before the ACT binding site —> DNA bends
ACT protein bound to its binding site can now interact w/ holenzme
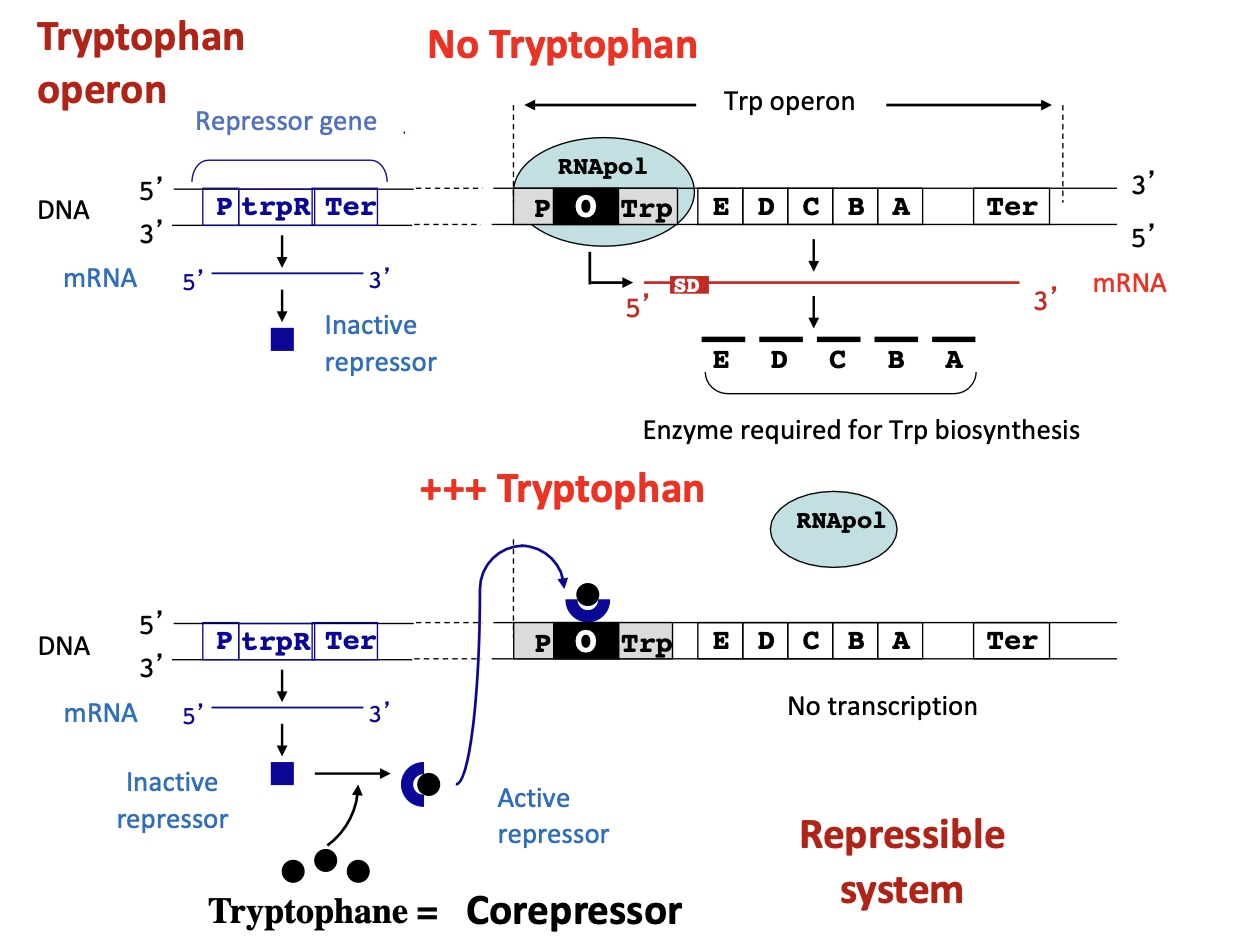
1.3.1 Operon tryptophan
Inactive repressor Trp R : absence of Trp
transcription of REP
no binding of REP w/ Trp
no binding to operator —> no transcription of enzymes needed for biosynthesis of Trp
Active repressor TrpR : presence of Trp
Trp binds to REP
complex binds to operator so holoenzyme can’t bind to the promoter
no transcription
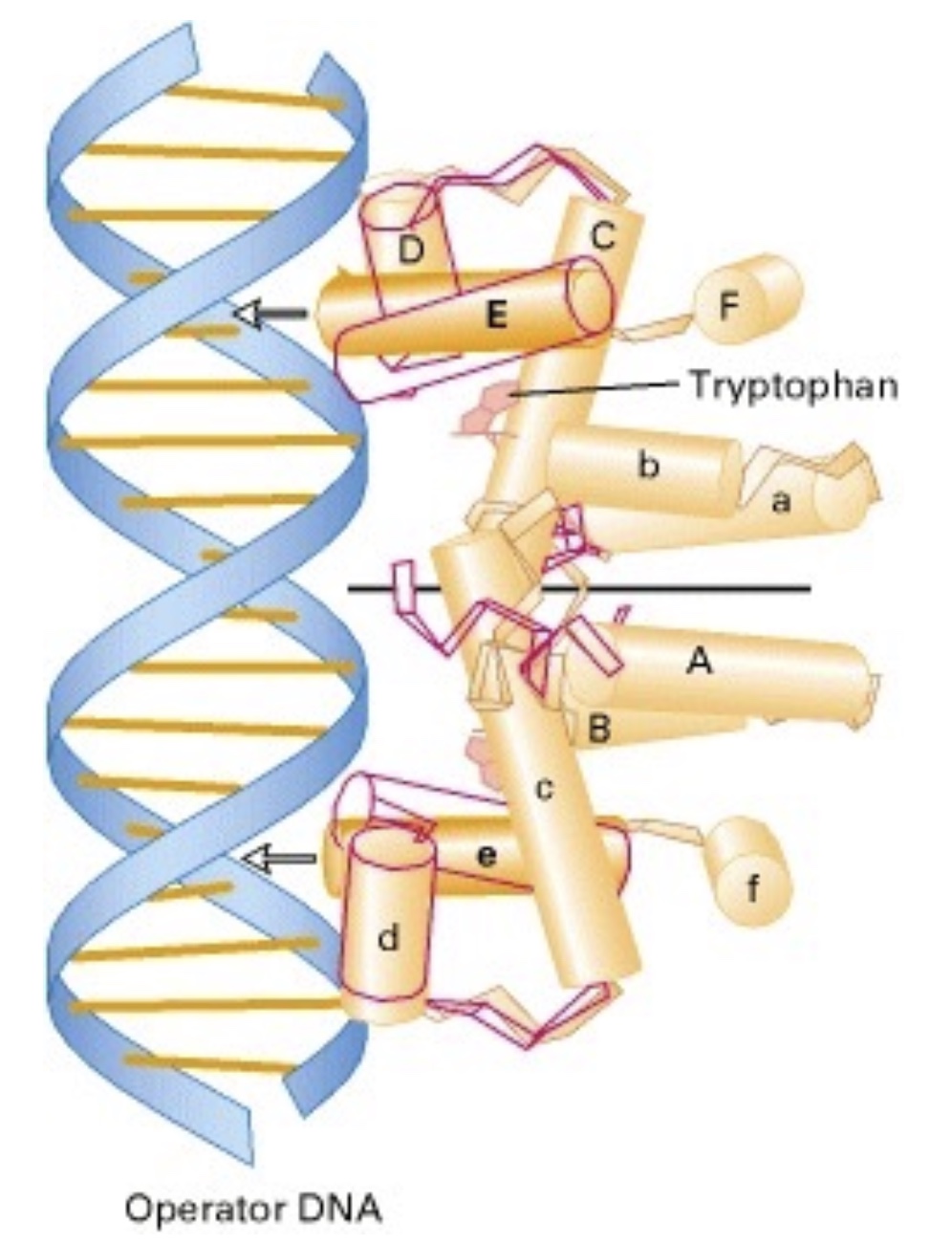
1.3.1 Trp R structure
dimeric
di-α helix
bound w/ Trp —> TrpR can bind w/ major groove of DNA ( conformational change allows this)
1.3.1 Can TrpR repress other genes ?
YES
Trp (70 times)
aroH (2 times) ; genes involved in synthesis of aromatic AA
TrpR (3 times) ; self regulation
1.3.1 How is there a differences in repression efficiency between the same repressor and different genes ?
TrpR has a different affinity for the operators of each gene (less similarity —> weaker fixation —> less repression)
The position of the operator on each gene : stronger when operator is around -10 and weaker around -35)
Depends on the strength of the promoter : if strong promoter —> strong competition between TrpR & holoenzyme —> less repression [and vise versa]
![<p>TrpR has a different affinity for the operators of each gene (less similarity —> weaker fixation —> less repression)</p><p>The position of the operator on each gene : stronger when operator is around -10 and weaker around -35)</p><p>Depends on the strength of the promoter : if strong promoter —> strong competition between TrpR & holoenzyme —> less repression [and vise versa]</p>](https://knowt-user-attachments.s3.amazonaws.com/1c5666d2-d0b6-4d2d-b3c7-b565e8b6dba4.jpg)
1.3.2 Principal of repression by blocking the escaping of the holoenzyme (step 3 of initiation of transcription) e.g protein p4 in bacteriophage
bacteriophage injects genome in bacteria
viral cycle : expression of “early” genes first but no expression of the “late” genes
expression of “late” genes later but no “early” genes
1.3.2 What is a p4 protein and where is the p4 binding sites located on the DNA?
p4 = “early” gene protein
binding site located inform of promoter of the “early” and “late” genes
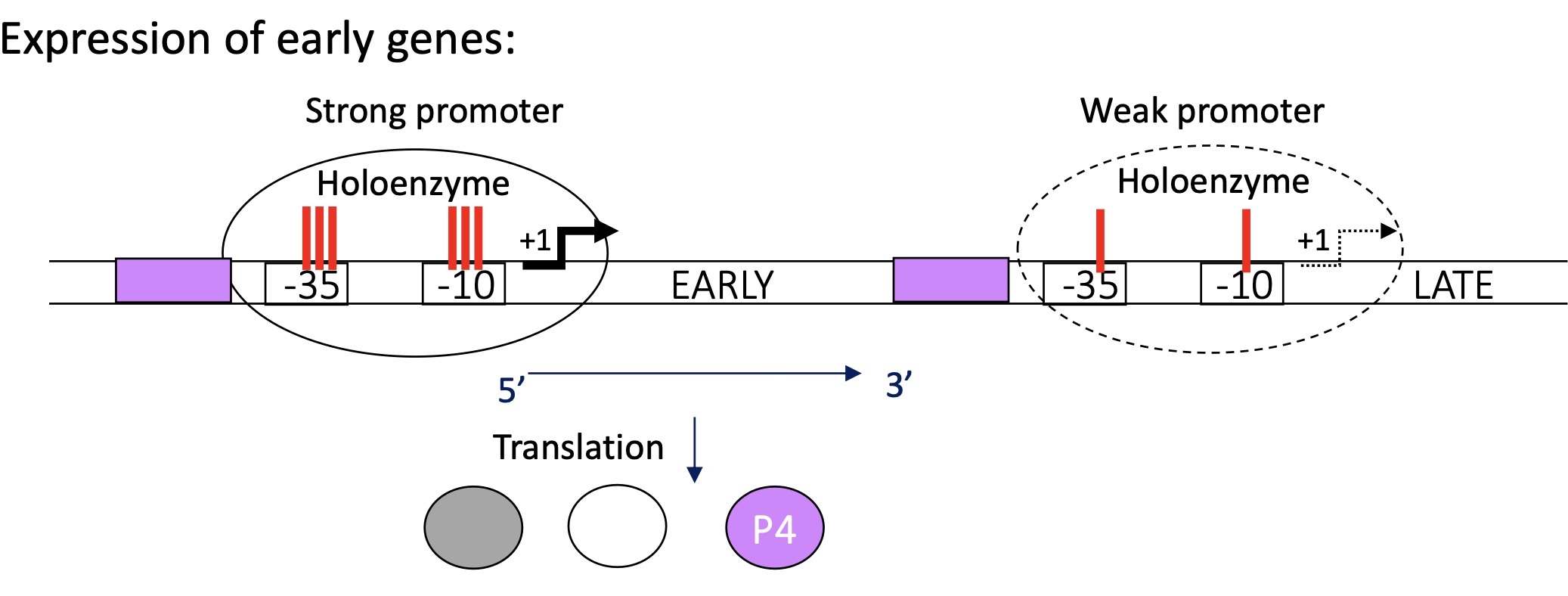
1.3.2 Expression of “early” genes in bacteriophage using p4
At the beginning :
promoter for “early” genes = strong
strong binding between holoenzyme & promoter of “early” genes e.g. p4
strong expression of p4
promoter for “late” genes = weak
no transcription of “late” genes at the moment
1.3.2 Expression of “late” genes in bacteriophage using p4
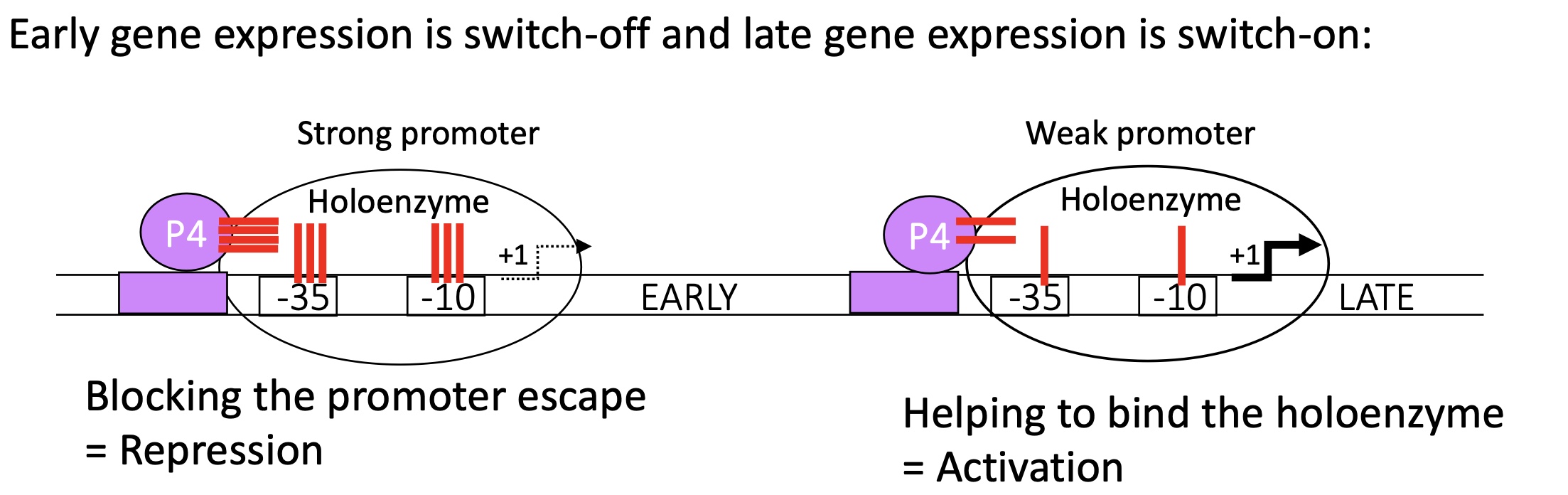
After expression of “early genes” :
promoter for “early” genes = strong
p4 binds to its binding site infront of its promoter for “early” genes
blocks the holoenzyme from escaping as the binding of the holoenzyme w/the promoteur is very strong
no transcription of “early” genes
promoter for “late” genes = weak
p4 binds to its binding site infant of the promoter for “late”genes
stabilises the fixation of there holoenzyme
transcription of “late” genes
1.3.2 what is p4 to “early” and “late” genes
to “early” genes p4= repressor
to “late” genes p4 = activator
1.3.3. lactose operon ; how many operators (binding sites for lacI) does it contain and genes does it code for
2 operators ; O1 & O2
3 sequences :
lac z —> beta-galactosidase
lac y —> perméase
lac A —> transacetylase
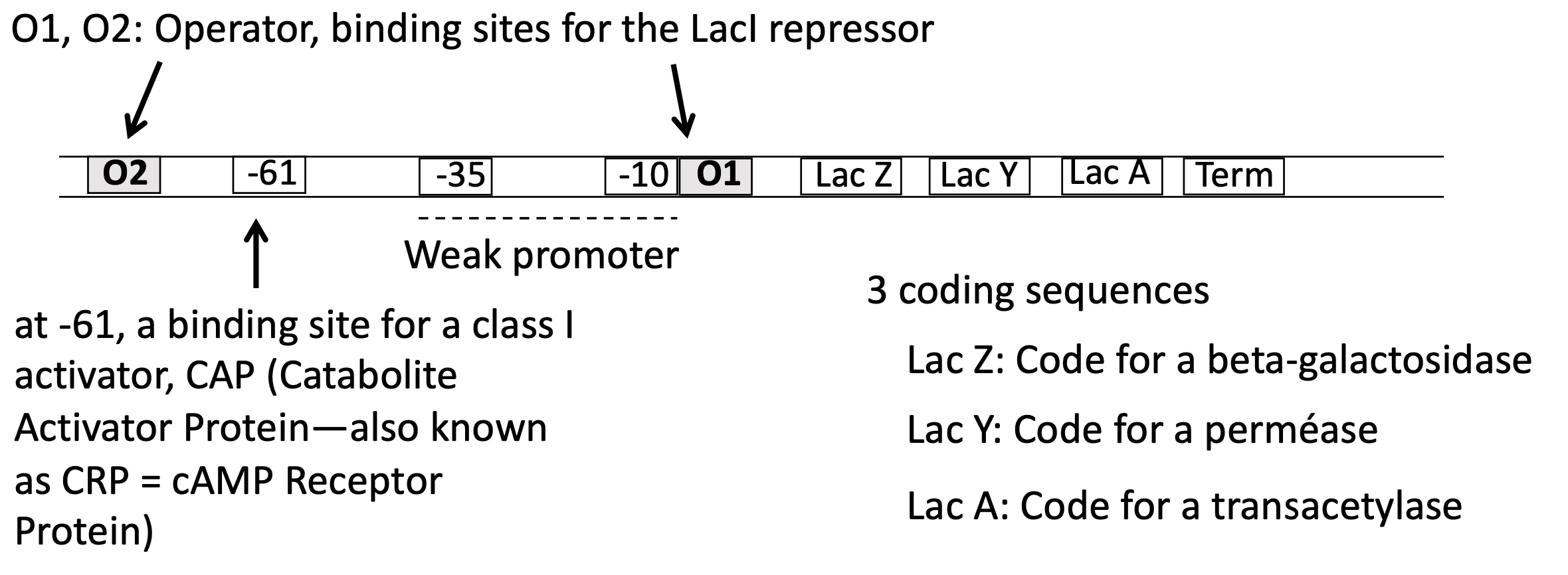
1.3.3 is the promoter for lactose operon weak or strong ?
weak
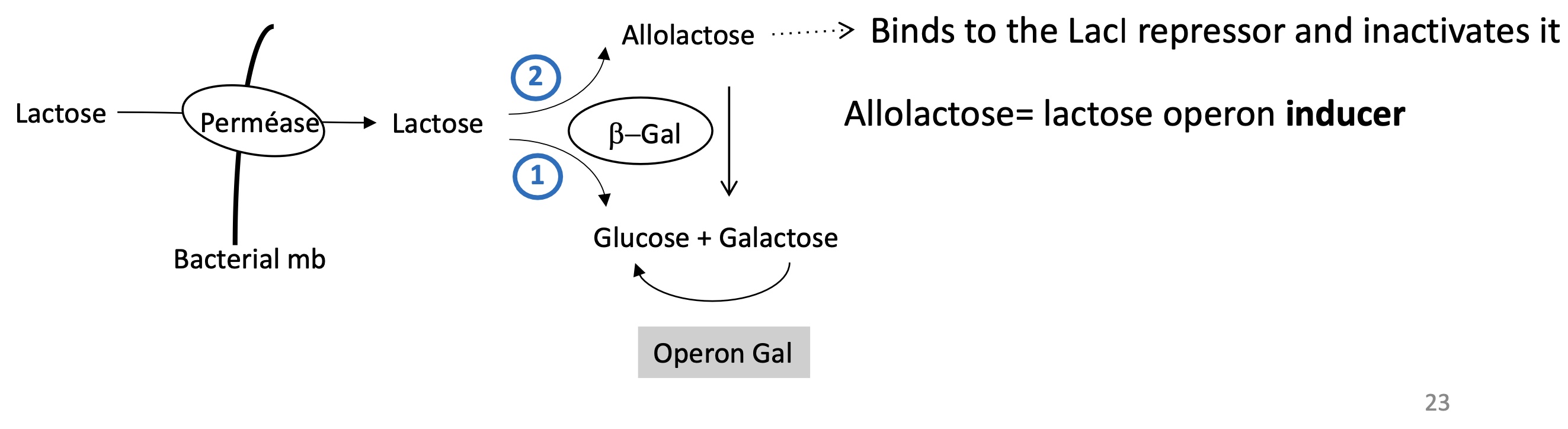
1.3.3. allolactose role in regulating initiation & LacI
Lactose enters the bacteria via permease
lactose is transformed into allolactose and galactose + glucose by β-Gal
allolactose = ligand that binds to LacI REP —> inactivating LacI
allolactose = inductor of lactose operon
1.3.3 by what is the lactose operon regulated by ?
LacI will repress the transcription in the absence of lactose
CAP will activate the transcription when little glucose is available
1.3.3 what is the CAP protein
Class I activator ( stabilises interaction of holoenzyme w/ promoter )
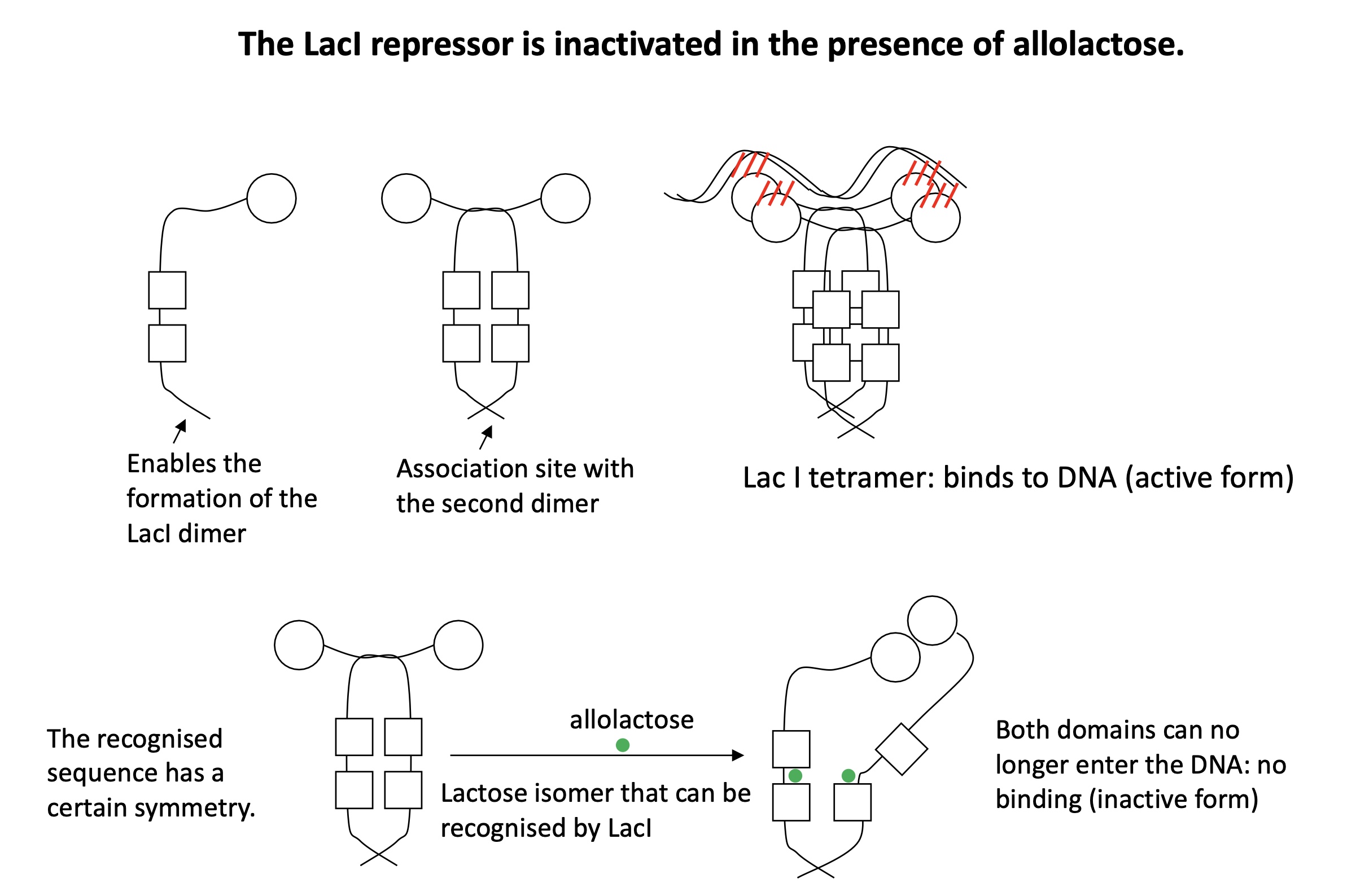
1.3.4 how does the structure of LacI REP change in presence of allolactose
LacI forms a homodimer = active form and can bind to DNA to form a tetramer
When allolactose present it binds to to the homodimer of LacI = change of conformation inactivates the REP & can no longer bind to DNA
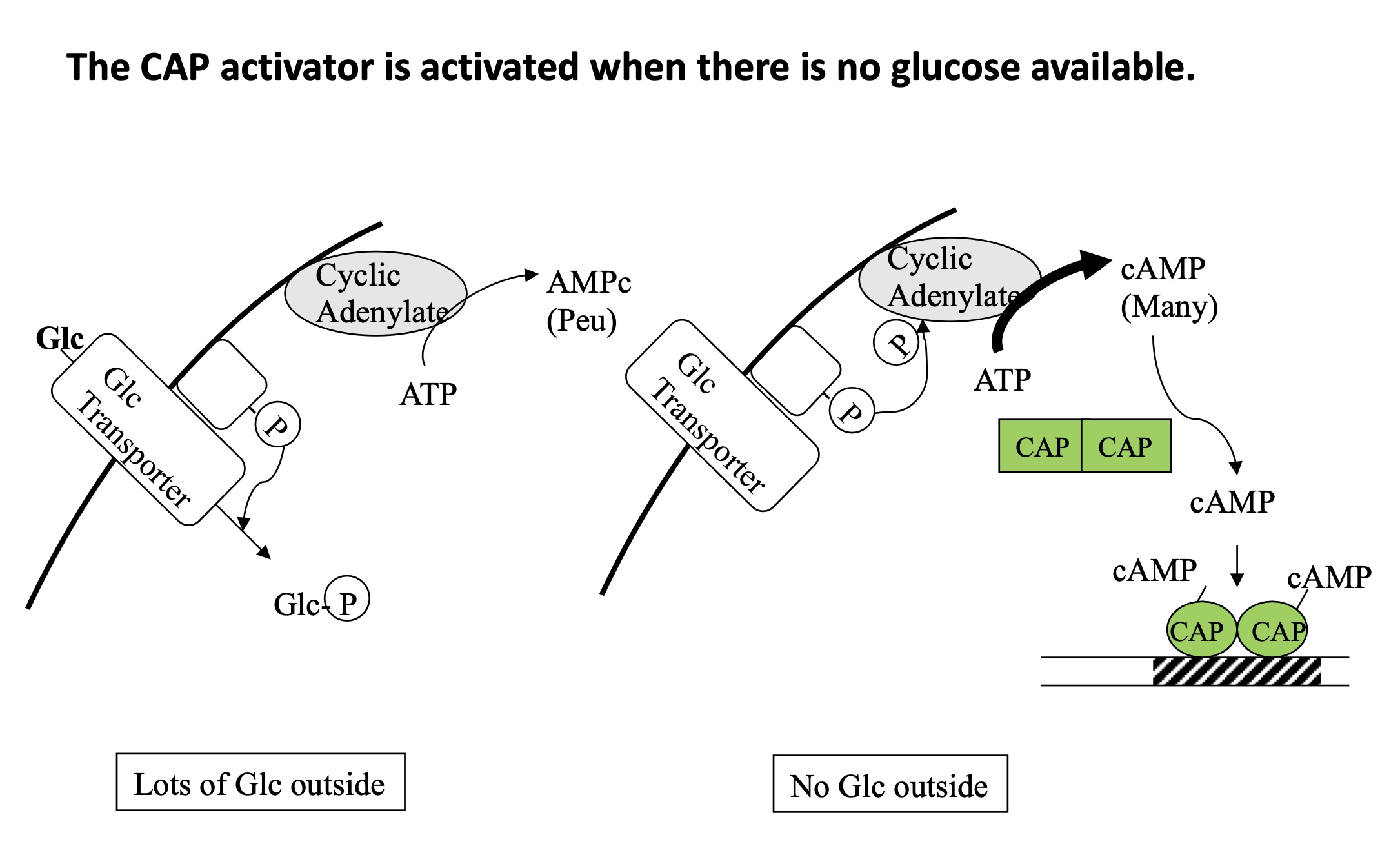
1.3.4 How is CAP regulated ?
Lots of Glc outside of the bacteria —> inactive CAP
Glc in imported by a transporter
Glc is phosphorylated so cannot exit the bacteria
cyclic adenylate transforms ATP to cAMP (very little)
No Glc outside of the bacteria —> active CAP
no transcription of Lactose operon
cyclic adenylate is phosphorylated and transforms lots of ATP into cAMP
cAMP binds to CAP dimer —> active form
CAP binds to activator domain of the promoter
transcription of lactose operon
Glc already present —> no need for lactose
No Glc present —> lactose needed to be metabolised into Glc
1.3.4 Is lactose operon transcribed in the absence of lactose
NO
LacI = active and can bind to operator site 1 —> blocking holoenzyme from binding to promoter
LacI binds to operator site 2 —> increasing repression as there’s complete blockage
—> no transcription
1.3.4 Is lactose operon transcribed in presence of lactose & glucose
YES but very little
lactose —> allolactose which binds to LacI —> inactivating REP LacI —> holoenzyme can bind to WEAK promoter -35 & -10
CAP inactived by Glc —> so weak interaction between holoenzyme and promoter
—> little to no transcription
1.3.4 Is lactose operon transcribed in presence of lactose & absence of glucose
YES
lactose —> allolactose which binds to LacI REP —> no inhibition of transcription
cyclic adenylate phosphorylated —> strong production of cAMP —> cAMP binds to CAP dimer activating it —> CAP binds to activator binding site stabilising holoenzyme-promoter interaction
—> good transcription
1.3.4 so what is an example of a class one activator couples to a repressor ?
lactose operon
activated by CAP = Class I activator
inhibited by LacI = REP
1.3.5 what 2 regulators are an example of a class II activator (closed —> open complex)
luxR regulates the expression of lux operon
FadR regulates the expression of fabA and fabL genes
1.3.5 explain how LuxR regulates lux operon
free bacteria in water : lux operon is off —> no bioluminescence
bacteria colonising squid : luc operon on —> transcription of enzymes for luciferase —> bioluminescence
1.3.5 what is the mecanism of luxR when the bacteria is free in the water
bacteria produces small amount of AHL (=Acyl homoserine lactose a ligand for luxR)
AHL diffuses out of the bacteria and dilutes in the water
little AHL present in bacteria —> inactive LuxR —> no transcription of enzymes for luciferase
—> no bioluminescence
1.3.5 what is the mecanism of luxR when the bacteria has colonised a squid
bacteria is in a closed compartment/specialised organ
AHL produced by bacteria remains in the bacteria so high [AHL] in bacteria
AHL binds to luxR —> dimer formed
luxR binds to activator site -42 allowing stabilisation of holoenzyme and for it to go from closed to open complex
—> bioluminescence
1.3.5 what are the genes fabA and fadL involved in and what regulates them ?
fabA = Fatty Acid Biosynthesis : Fatty Acid (FA) synthesis for membrane lipids
this is expressed when there are no exogenous/exterior FA’s
fadL = Fatty Acid Degradation : FA Brocken down to be used for membrane lipids or into acetyl-CoA
this is expressed when there are exogenous FA’s
Both ones are regulated by FadR protein
1.3.5 how is FadR activated
active form = unbound from FA
inactive form = bound to FA
1.3.5 what strength are the primers of fabA and fadL
weak : fabA
strong : fadL
1.3.5 what happens to the expression of fabA and fadL when there is presence of FA
exogenous FA present
fabA :
FA binds to fadR —> inactive form
cannot bind to fadR binding site in-front of the weak fabA promoter
no fixation of holoenzyme
—> no transcription —> no FA biosynthesis
fadL :
FA binds ro fadR —> inactive form
doesn’t bind to fadR binding site in-front of the strong fadL promoter
holoenzyme binds to promoter
—> transcription —> FA degradation
1.3.5 what happens to the expression of fabA and fadL in absence of FA
fabA :
no binding of FA to fadR —> active form
FadR binds to FadR binding site infant of the weak promoter of fabA
stabilises holoenzyme-promoter interaction & promotes transition from closed to open complex
—> transcription —> FA biosynthesis
fadL :
FadR = active as no FA bound to it
FadR binds to 2 operators located between -35 & -10 of the strong promoter of fadL
steric hindrance ; prevents holoenzyme from binding to -35 & -10
—> no transcription —> no FA degradation
1.3.6 what is an examples of the use of 2 activators working together?
respiration : nitrate & fumarate reductase operon
1.3.6 how does anaerobic respiration work ?
when O2 is absent E.coli uses other e- acceptors like :
nitrate NO3- —> produces less ATP than O2
fumarate —> produces less ATP than NO3-
use of nitrate and fumarate requires enzymes : nitrate and fumarate reductase