Biochemistry - DNA & Biotechnology
1/88
Earn XP
Description and Tags
Chapter 6
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
89 Terms
2 forms of nucleic acids in eukaryotic cells
DNA & RNA (polymers with distinct roles = create molecules integral to life in organisms)
DNA
- macromolecule essential to understand how nucleotides/nucleosides are constructed
- polydeoxyribonucleotide composed of many monodeoxyribonucleotides linked together
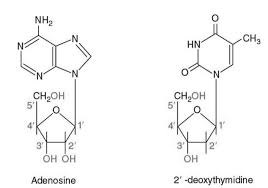
Nucleosides
- composed of 5-ring sugar (pentose) bonded to nitrogenous base
- formed by covalently linking base to C-1’ of sugar (sugar carbon)
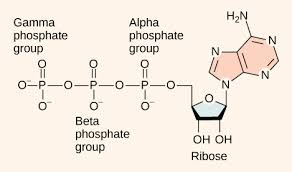
Nucleotides
- formed when 1 or more phosphate groups are attached to C-5’ of nucleoside
- named according to number of phosphates present (example: ADP & ATP)
- building blocks of DNA
Nucleic acid
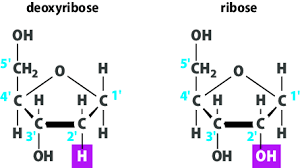
classified according to pentose they contian:
ribose pentose: RNA
deoxyribose pentose: DNA
Adenine
Nucleoside: adenosine (deoxyadenosine)
Nucleotides: AMP (dAMP), ADP (dADP), ATP (dATP)
Guanine
Nucleoside: guanosine (deoxyguanosine)
Nucleotides: GMP (dGMP), GDP (dGDP), GTP (dGDP)
Cytosine
Nucleoside: cytidine (deoxycytidine)
Nucleotides: CMP (dCMP), CDP (dCDP), CTP (dCTP)
Uracil
Nucleoside: uridine (deoxyuridine)
Nucleotides: UMP (dUMP), UDP (dUDP), UTP (dUTP)
Thymine
Nucleoside: (deoxythymidine)
Nucleotides: (dTMP), (dTDP), (dTTP)
Sugar-phosphate backbone
- composed of alternating sugar & phosphate groups
- read 5’ to 3’
- phosphate group links 3’ carbon of sugar to 5’ phosphate group of next incoming sugar
- phosphates carry negative charge = DNA & RNA have overall negative charge
Polarity in backbone
5’ end: -OH or phosphate group bond to C-5’ of sugar
3’ end: free -OH on C-3’ of sugar
DNA strand written ways
1. 5’-ATG-3’ or ATG
2. backward: 3’-GTA-5’
3. showing phosphate positions: pApTpG
4. “d” for deoxyribose: dAdTdG
Purines in nucleic acids
Adenosine
Guanine
(both in DNA & RNA)
Pyrimidines in nucleic acids
Cytosine
Uracil
Thymine
(cytosine in both DNA & RNA)
(uracil in RNA)
(thymine in DNA)
Biological aromatic heterocycles examples
purine and pyrimidine
Aromatic stable ring system rules
1. cyclic compound
2. planar compound
3. conjugated compound (alternating single and double bonds/lone pairs = at least one unhybridized p-orbital for each atom in ring)
4. 4n+2 pi electrons in compound (Huckel’s Rule)
Benzene aromatic compound
- all carbon atoms are sp2-hybridized
- equally overlapping orbitals
= delocalized electrons for 2 pi electron clouds (1 above & 1 below ring plane); characteristic of aromatic molecules = unreactive
Heterocycles
ring structures that contain at least 2 different elements in ring
example: purine & pyrimidine contain nitrogen in aromatic rings
Watson-Crick model
3D structure of DNA; double helical nature & specific base-pairing
key features of watson-crick model
1. 2 antiparallel strands of DNA; one strand with polarity 5’ to 3’ down page, other has polarity 5’ to 3’ up page
2. sugar-phosphate backbone on outside of helix, nitrogenous base on inside of helix
3. complementary base-pairing: A & T (2 H bonds), G & C (3 H bonds) = stability
4. Chargaff’s Rules: amount of A = amount of T and amount of G = amount of C [therefore, purine amount = pyrimidine amount]
![<p>1. 2 antiparallel strands of DNA; one strand with polarity 5’ to 3’ down page, other has polarity 5’ to 3’ up page<br>2. sugar-phosphate backbone on outside of helix, nitrogenous base on inside of helix<br>3. complementary base-pairing: A & T (2 H bonds), G & C (3 H bonds) = stability<br>4. Chargaff’s Rules: amount of A = amount of T and amount of G = amount of C [therefore, purine amount = pyrimidine amount]</p>](https://knowt-user-attachments.s3.amazonaws.com/6b5a39a6-b1a7-42a9-bdc4-ce890a2b7ed2.png)
B-DNA
- double helix of most DNA
- makes a turn every 3.4 nm
- contains about 10 bases within that span
- major and minor grooves identified between interlocking strands (often site of protein binding)
Z-DNA
- zig-zag appearance
- left-handed helix
- makes a turn every 4.6 nm
- contains 12 bases within each turn
- high GC-content or high salt concentration contributes to formation
- unstable and difficult to research = no biological activity attributed
Denaturing of DNA
- disrupt hydrogen bonding and base-pairing = melting of double helix into two separate single strands (no covalent bonds break during this process)
- caused by heat, alkaline pH, chemicals (formaldehyde/urea)
Reannealed
denatured DNA brought back together, if denaturing condition is slowly removed
Histones
- DNA that makes up chromosome wound up around group of small basic proteins
- form chromatin
- 5 histone proteins in eukaryotic cells
- 2 copies of each histone protein: H2A, H2B, H3, H4 form histone core
- 200 base pairs of DNA wrapped around protein complex = nucleosome
- H1 seals off DNA as it enters & leaves nucleosome = stability
- example of nucleoprotein (proteins associating with DNA) ~others are acid-soluble & simlate transcription
Heterochromatin
- small percentage of chromatin compacted during interphase
- appears dark under light microscopy & transcriptionally silent
- consists of DNA with highly repetitive sequences
Euchromatin
- dispersed chromatin
- light under light microscop
- contains genetically active DNA
Telomere
- repeating unit (TTAGGG) at end of DNA
- some of sequence lost in each round of replication, replaced by telomerase enzyme
- progressive shortening of telomeres = aging
- high GC-content = strong strand attractions at end of chromosomes = prevent unraveling
Telomerase
enzyme highly expressed in rapidly dividing cells
Centromere
- region of DNA in center of chromosomes
- site of constriction (form noticeable indentations)
- during cell division, 2 sister chromatids remain connected at centromere until microtubules separate chromatids during anaphase
DNA replication
- highly regulated to ensure close to 100% perfect copy of genome
- necessary for reproduction of species & any dividing cell

human genome
has ~3 billion base pairs packed into multiple chromosomes
Replisome (replication complex)
- set of specialized proteins that assists DNA polymerases
origins of replication
DNA starts to unwind; begins process of replication
replication forks
generation of new DNA proceeds in opposite directions (= forks on both sides of origin)
Bacterial chromosome
- closed, double-stranded circular DNA molecule with single origin of replication
= 2 replication forks move away from each other around circle
= production of 2 identical circular molecules of DNA
Eukaryotic v Prokaryotic replication
Eukaryotic:
- copy more bases
- slower process
- efficiency: each uekaryotic chromosome contains 1 linear molecule of double-stranded DNA with multiple origins of replication
Prokaryotic:
- copy less bases
- faster process
- efficiency: single origin of replication

As replication forks move toward each other…
sister chromatids are created & remain connected at centromere
Helicase
enzyme responsible for unwinding DNA; generates 2 single-stranded template strands ahead of polymerase (strands held apart by single-stranded DNA-binding proteins) and causes positive supercoiling that strains the DNA helix
Single-stranded DNA-binding proteins
binds to unraveled strand (prevents DNA strands reassociation & degradation by nucleases) to hold strands apart
Supercoiling (positive)
wrapping of DNA on itself as helical structure is pushed toward telomeres during replication; alleviated by DNA topoisomerases
DNA topoisomerases
work ahead of helicase, nick one or both strands = relaxation of torsional double-stranded DNA molecules = introduce negative supercoils
parental strands
serve as templates for generation of new daughter strands
= semiconservative process
Semiconservative
one parental strand is retained in each of 2 resulting identical double-stranded DNA molecules
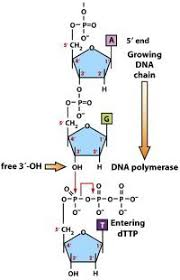
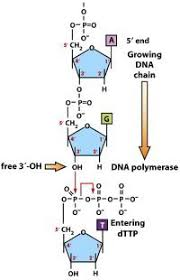
DNA polymerase
- read DNA template (parental strand) in 3’ to 5’ direction & synthesize new daughter (complementary) strand in 5’ to 3’ direction
= new double helix with antiparallel orientation
leading strand
copied in continous fashion; same direction as the advancing replication fork; read in 3’ to 5’ direction & complement synthesized in 5’ to 3’ direction
lagging strand
copied in fragments; opposite direction of replication fork; read in 5’ to 3’ & synthesized by DNA polymerase by producing Okazaki fragments
Okazaki fragments
as replication fork moves forward, additional space clears for DNA polymerase to fill in…once okazaki fragment is complete, it turns around to find another gap to fill
(lagging strand)
DNA Replication Steps
1. lay down RNA primer
2. primase synthesizes short primer in 5’ to 3’ direction to start replication on each strand
3. leading strand with one (or more) primer & lagging strand with multiple for each okazaki fragment
4. DNA polymerase III (prokaryotes) or DNA polymerase a, d, e (eukaryotes) synthesize daughter strands of DNA in 5’ to 3’ direction
5. incoming nucleotides: 5’ deoxyribonucleotide triphosphates: dATP, dCTP, dGTP, dTTP
6. new phosphodiester bond made; free pyrophosphate (PPi) released
7. DNA polymerase I (prokaryotes) or RNase H (eukaryotes) removes RNA to maintain integrity of genome
8. DNA polymerase I (prokaryotes) or DNA polymerase d (eukaryotes) adds DNA nucleotides where RNA primers were [fills in gaps]
9. DNA ligase seals end of DNA molecules together = 1 continuous strand of DNA
![<p>1. lay down <u>RNA primer</u><br>2. <u>primase</u> synthesizes short primer in 5’ to 3’ direction to start replication on each strand<br>3. <u>leading strand</u> with one (or more) primer & <u>lagging strand</u> with multiple for each <u>okazaki fragment</u><br>4. <u>DNA polymerase III</u> (prokaryotes) or <u>DNA polymerase </u><em><u>a, d, e</u></em> (eukaryotes) synthesize <u>daughter strands</u> of DNA in 5’ to 3’ direction<br>5. incoming <u>nucleotides</u>: 5’ deoxyribonucleotide triphosphates: dATP, dCTP, dGTP, dTTP<br>6. new <u>phosphodiester bond</u> made; free <u>pyrophosphate</u> (PP<sub>i</sub>) released<br>7. <u>DNA polymerase I</u> (prokaryotes) or <u>RNase H</u> (eukaryotes) removes RNA to maintain integrity of genome<br>8. <u>DNA polymerase I</u> (prokaryotes) or <u>DNA polymerase d</u> (eukaryotes) adds DNA nucleotides where RNA primers were [fills in gaps]<br>9. <u>DNA ligase</u> seals end of DNA molecules together = 1 continuous strand of DNA</p>](https://knowt-user-attachments.s3.amazonaws.com/dda130e6-1dcf-4b7e-91c9-2ca94938ed4c.png)
5 ‘classic‘ DNA polymerases in eukaryotic cells
a
B
y
d
e
further research revealed: z through m
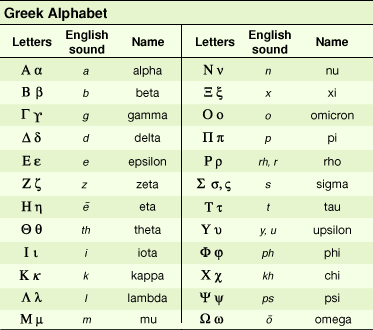
DNA polymerases a, d, e
synthsize leading & lagging strands
~d also fills gaps when RNA primers are removed
DNA polymerase y
replicates mitochondrial DNA
DNA polymerase B and e
important in process of DNA repair
DNA polymerases d and e
assisted by PCNA protein: assembles into trimer = forms sliding clamp (helps strengthen interaction between DNA polymerases and template strand)
DNA damage & repair
- damage: breaking of backbone, structural/spontaenous alteration of bases, incorporation of incorrect base during replication
- DNA is susceptible to damage: if not corrected, will be copied & passed on
- Cell has multiple processes to catch & correct genetic errors = maintains integrity & stability of genome from cell to cell and generation to generation
Cancer cells
- proliferate excessively: able to divide without stimulation from other cells ~no longer subject to normal controls on cell proliferation
- can migrate by local invasion or metastasis (migration to distant tissues by bloodstream or lymphatic system)
- over time, accumulate mutations
Oncogenes
- mutated genes that cause cancer (before mutation, refered to as proto-oncogenes)
- encode cell cycle-related proteins
- abnormal alleles encode proteins (more active than normal proteins) = rapid cell cycle advancement
- mutation in only one copy is sufficient for tumor growth; considered dominant
Src - sarcoma
- src = first gene discovered in proto-oncogene category
- sarcoma = category of connective tissue cancers
tumor suppressor genes (antioncogenes)
- encode proteins that inhibit cell cycle or participate in DNA repair
- function to stop tumor progression
mutation of antioncogenes
- promotes cancer through loss of tumor suppression activity
- inactivation of both alleles = loss of function [even one copy of normal protein = inhibits tumor formation]
Proofreading
complementary strands incorrectly pair bases
= unstable H bonds between strands
- unstability detected as DNA passes through polymerase
- incorrect base is excised and replaced with correct base
Methylation
- template strand existed in cell longer = more heavily methylated
- plays role in transcriptional activity of DNA
- efficient process that corrects most errors in sequence during replication
[DNA ligase: closes gaps between okazaki fragments but lacks proofreading ability = increased likelihood of mutations in lagging strand than leading strand]
Mismatch repair
- cells with machinery in G2 phase of cell cycle
- enzymes encoded by genes MSH2 and MLH1: detect & remove missed errors from S phase
- enzymes are homologous of MutS and MutL in prokaryotes ~serve similar funciton
Nucleotide & Base excision repair
Repair mechanism: recognize damage/lesion, remove damage, use complementary strand as template to fill gap
Cell machinery: recognizes two DNA damage types in G1 and G2, fixes them through nucleotide/base excision repair
Nucleotide Excision Repair (NER) mechanism
cut-and-patch process:
- specific proteins scan DNA molecule
- recognize lesion due to bulge in strand
- excision endonuclease nicks phosphodiester backbone of damaged strand on both sides of thymine dimer
- removes defective oligonucelotide
- DNA polymerase fills gap by synthesizing DNA in 5’ to 3’ direction (using undamaged strand as template)
- nick sealed by DNA ligase
thymine dimer:
- UV light = formation of dimers between thymine residues in DNA
- thymine dimers interfere with DNA replication & normal gene expression
- distorts shape of double helix
- thymine dimer eliminated from DNA by NER

Base Excision Repair (BER)
- detection system for small, non-helix disorting mutations in other bases
BER:
- affected base is recognized
- removed by glycosylase enzyme
- leaves apurinic/apyrimidinic (AP) site (abasic site)
- recognized by AP endonucleaase
- removes damaged sequence from DNA
- DNA polymerase & DNA ligase fill gap and seal strand
Alterations to bases occur with other cellular insults
- thermal energy absorbed by DNA = cytosine deamination (loss of amino group from cytosine)
= conversion of cytosine to uracil
- uracil cant’t be found in DNA = detected as error
Recombinant DNA technology
- allows DNA fragment from any source to be multiplied by gene cloning or polymerase chain reaction (PCR)
- provides means of analyzing/alter genes/proteins
- provides reagents necessary for genetic testing (carrier detection & prenatal diagnosis of genetic diseases)
- useful for gene therapy
- can provide a source for a specific protein in almost unlimited quantities
DNA cloning
- technique that can produce large amounts of desired sequence (homogenous DNA) for other applications
- DNA to be cloned: present in small quantity & part of heterogenous mixture containing other DNA sequences
- requires investigator to ligate DNA of interest to piece of nucleic acid = ‘vectore’ or ‘recombinant vector’
- bacteria grown in colonies, colony with recombinant vector is isolated
[isolation accomplished by ensuring recombinant vector includes gene for antibiotic resistance; antibiotics kill off all colonies that don’t contain recombinant vector = resulting colony grown in large quantities]
- bacteria made to express gene of interest or lysed to reisolate replicated recombinant vectors (processed by restriction enzymes to release cloned DNA from vector)
vector
- baterial or viral plasmid that can be transferred to a host bacterium after inserting DNA of interest
Restriction enzymes (restriction endonucleases)
- enzymes that recognize specific double-stranded palindromic DNA sequences
[palindromic: 5’ to 3’ sequence of one strand is identical to 5’ to 3’ sequence of antiparallel strand]
- isolated from bacteria (natural source; protected via restriction & modification system that protects bacteria from DNA virus infection)
- can cut through backbone of double helix of specific identified sequence
- many commercially available to labs = process DNA in specific ways
- some produce offset cuts
= sticky ends on fragments
= advantageous in facilitating recombination of restriction fragment with vector DNA
(vector of choice can be cut with same restriction enzyme = fragments inserted directly into vector)
![<p>- enzymes that recognize specific double-stranded palindromic DNA sequences<br>[palindromic: 5’ to 3’ sequence of one strand is identical to 5’ to 3’ sequence of antiparallel strand]<br>- isolated from bacteria (natural source; protected via restriction & modification system that protects bacteria from DNA virus infection)<br>- can cut through backbone of double helix of specific identified sequence<br>- many commercially available to labs = process DNA in specific ways<br>- some produce offset cuts<br>= sticky ends on fragments<br>= advantageous in facilitating recombination of restriction fragment with vector DNA<br>(vector of choice can be cut with same restriction enzyme = fragments inserted directly into vector)</p>](https://knowt-user-attachments.s3.amazonaws.com/208dd574-5a5a-44ae-8318-a42332b13b1a.png)