Cell Biology EXAM 2
1/175
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
176 Terms
Vesicular Transport
membrane enclosed vesicles transport proteins from one topologically equivalent space to each other
cargo might be in the lumen or membrane of the vesicle
transport proteins face outside of the vesicle/cell membrane
Exocytosis
when vesicles release their contents to the extracellular space
Endocytosis
when vesicles intake their contents from the extracellular space
How does “stuff” get from the ER to the golgi?
the ER sends stuff within vesicles made of itself to the golgi
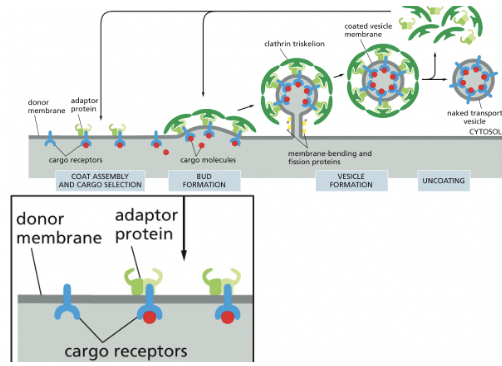
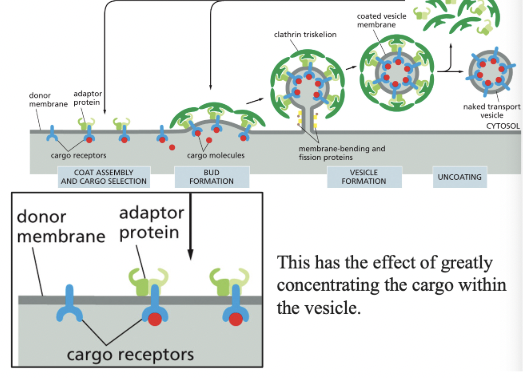
Coat Proteins
select different cargo (vesicles) & shape the transport vesicles that mediate the various steps in the secretory & endocytic pathways
changes where vesicles go (makes sure it doesn’t deliver things incorrectly)
Clathrin
Coat protein that signals endocytosis from outside of cell
assembly of this drives vesicle formation (3 heavy chains & 3 light chains)
Adaptor Proteins
select cargo into Clathrin-coated vesicles
Phosphatidylinositol Phosphates (PIPs)
mark organelles & membrane domains, helping to regulate where vesicles form
some groups on phospholipid are phosphorylated (what’s phosphorylated is different for each kind)
different organelles have different distributions of PIPs
different PIPs are recognized by different proteins
Membrane-Bending Proteins
help deform the membrane during vesicle formation
ex) BAR-domain dimer
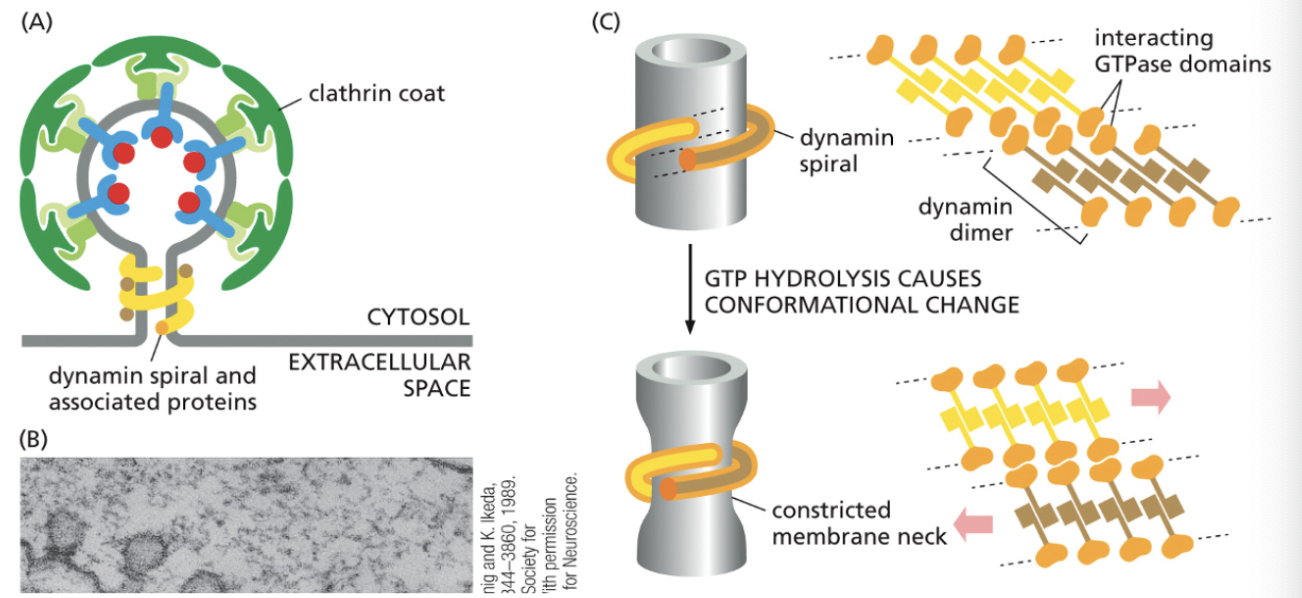
What type of proteins regulate the pinching off/uncoating of coated vesicles?
CYTOPLASMIC PROTEINS
Dynamin spiral & associated proteins (connected via interacting GTPase domains)
GTP hydrolysis causes conformational change
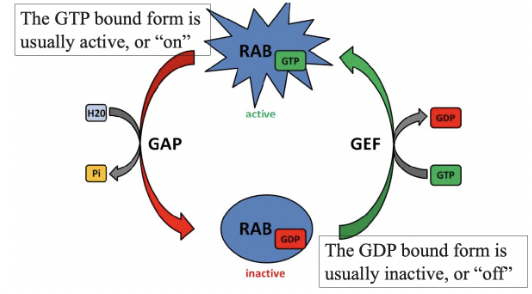
Rab Proteins
small GTPases that guide transport vesicles to their target membranes
each target organelle will have its own version of Rab proteins
alternate between GTP & GDP bound forms
their GTPase activity is low on their own
GTPase Activating Protein (GAP)
cause a small GTPase to hydrolyze GTP to GDP, so the GTPase is in its GDP bound form
GDP Dissociation Inhibitor (GDI)
keeps Rab proteins in their GDP bound form
blocks the action of GEF, keeping Rab in its GDP-bound form
Rab hydrolyzes GTP during this process
Guanine Exchange Factor (GEF)
causes a small GTPase to release GDP & bind to GTP, so GTPase is in its GTP bound form
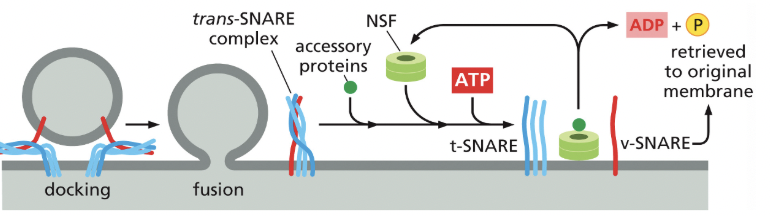
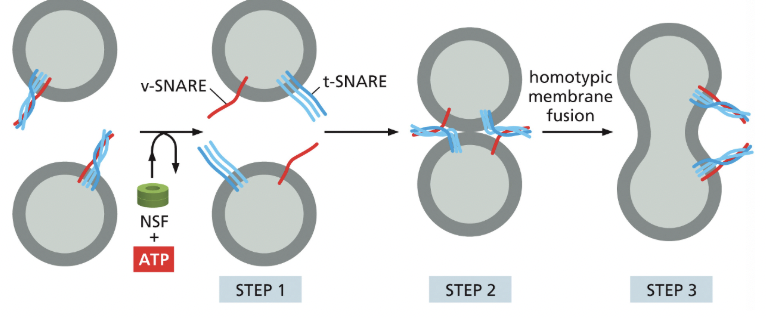
SNAREs
integral membrane proteins that mediate membrane fusion
v-SNAREs are on the vesicle
t-SNAREs are on the target membrane
when they interact, they tightly coil together, pulling the membranes close together, squeezing out any water, allowing the membranes to directly touch; facilitating fusion of the membranes
interacting SNAREs need to be pried apart before they can function again
NSF
an ATPase that uses energy from ATP to pull apart the tightly coiled SNAREs, so they can be recycled
What does the Golgi Apparatus do?
transport from the ER (endoplasmic reticulum) goes through the golgi
accepts proteins from the ER
modifies the proteins
sorts them & send them to their destinations like cell exterior, cell membrane, endosome, lysosome, back to ER, etc.
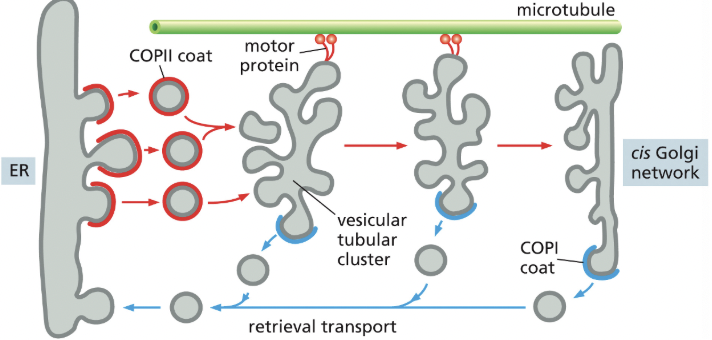
What do proteins leave the ER in?
in COPII-coated transport vesicles
only proteins that are properly folded & assembled can leave the ER
Tubular Clusters
formed by vesicles from ER that fuse with one another on their way to the golgi
mediate transport from the ER to the golgi apparatus
Retrieval Pathway
COP1 coated vesicles bud off of tubular clusters & head back to the ER, returning ER specific components
COP1 proteins come from the golgi network
KDEL
amino acid sequence that marks proteins as belonging in the ER→ part of the retrieval pathway
removing KDEL from an ER protein results in the protein being secreted
adding KDEL to a protein normally secreted results in it accumulating in the ER
the KDEL receptor proteins let go of the KDEL sequence when it gets to the ER, prob because of change in pH
the receptor can then be sent back to the golgi
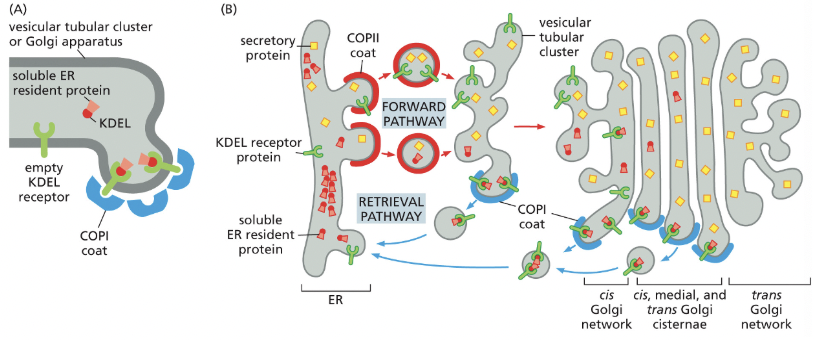
cis-Golgi Network
collection of fused vesicular tubular clusters from the ER→ the different compartments of the Golgi contain different sets of resident enzymes (all membrane bound) to perform different functions
the cis face of the Golgi is on the ER side
in fibroblasts, golgi faces direction the cell is crawling
cis golgi network function = SORTING (phosphorylation of oligosaccharides on lysosomal proteins)
What are the steps of oligosaccharide chain process in the Golgi Apparatus?
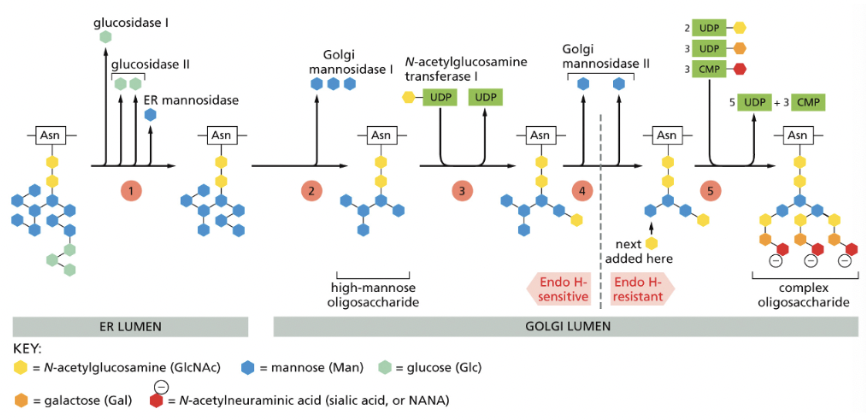
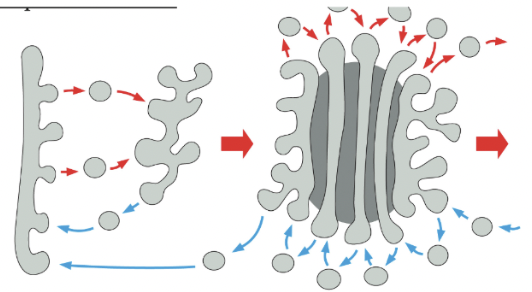
Vesicle Transport Mechanism (transport through Golgi)
Golgi cisternae are static compartments, which contain a characteristic complement of resident enzymes
Passing of molecules from cis to trans through the Golgi is accomplished by forward-moving vesicles, which bud from one cisterna & fused with the next in a cis-to-trans direction
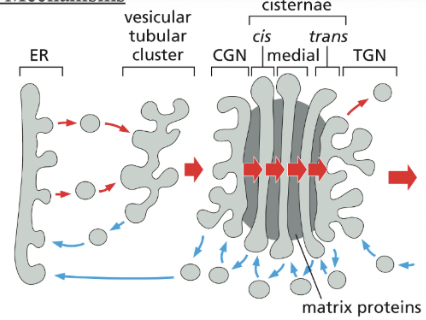
Cisternal Maturation Mechanism (transport through Golgi)
In the cisternal maturation mechanism, each Golgi cisterna maturates as it migrates outward through the stack
At each stage, the Golgi resident proteins that are carried forward in a maturing cisterna are moved backwards (blue arrows) to an earlier compartment in COPI-coated vesicles
When a newly formed cisterna moves to a medial position, “leftover” cis golgi enzymes would be extracted/transported retrogradedly to a new cis cisterna behind
likewise the medial enzymes would be received by retrograde transport from the cisternae just ahead
a cis cisterna would mature to a medial & trans cisterna as it moves outward
Golgi Reassembly and Stacking Proteins (GRASPs)
tether adjacent cisternae to each other within the Golgi
golgi matrix proteins that help organize the stack
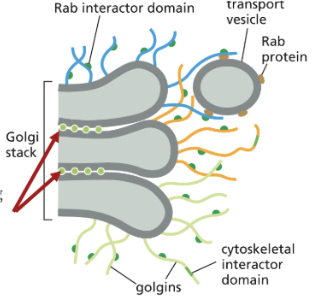
Filamentous Golgins
anchored to golgi membranes that capture transport vesicles by binding to Rab proteins on the vesicle surface
golgi matrix protein that helps organize the stack
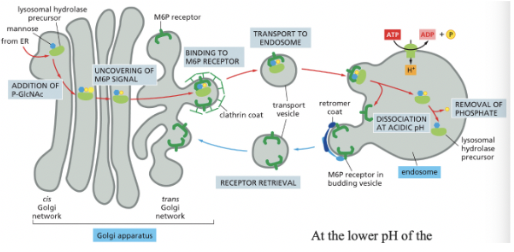
Signal-Mediated Diversion to Lysosomes (via Endosomes)
GlcNAc Phosphotransferase: recognizes a cluster of neighboring amino acids (signal patch) on the surface of lysosomal hydrolases
happening in the Golgi & serves to convert the terminal mannose to M6P (signal for sorting lysosomal hydrolases)
A mannose 6-phosphate receptor sorts lysosomal hydrolases in the trans golgi network
**lysosome = membrane-bound organelle containing hydrolytic enzymes & digests worn out cellular components/extracellular materials taken in by endocytosis
M6P modification binds to an adapter protein→ the M6P modification causes proteins to be destined for the lysosome
at the lower pH of the endosome, the hydrolases dissociate from the M6P receptors
What happens when defects occur in the GlcNAc Phosphotransferase
Causes a lysosomal storage disease in humans
if hydrolases aren’t marked for sorting to the lysosome, then undigested materials accumulate in the endosome, forming large inclusions in the cell
Most severe cases = all organ systems are affects & life expectancy is <10 years
Signal-Mediated Diversion to Secretory Vesicles (for Regulated Secretion)
Secretory vesicles bud from the trans golgi network
when they originally bud from the trans golgi network, it has clathrin (this is lost when the secretory vesicle becomes mature)
clathrin-coated vesicles retrieve excess membrane & lumenal content present in immature secretory vesicles → concentrates the cargo
Secretory vesicles wait near the plasma membrane until signaled to release their contents (ex. insulin is triggered by Ca²+)
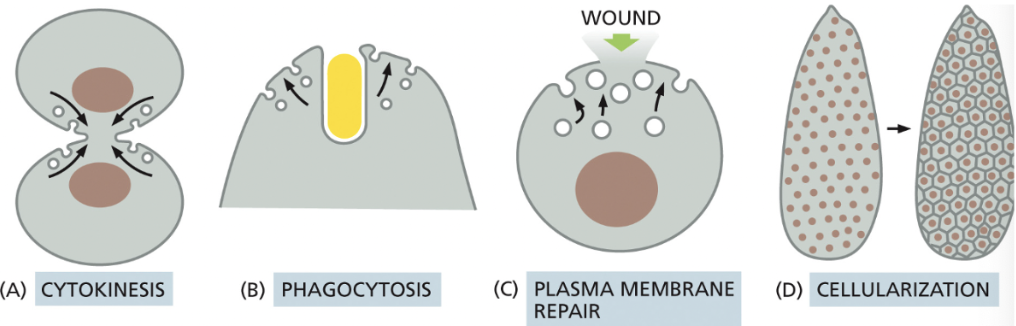
What are some regulated exocytosis events that enlarge the plasma membrane? (4)
**Need to do this when a cell isn’t growing; need amt of membrane being removed from plasma membrane to = amt of membrane being added via vesicle fusion
Cytokinesis→ at the end of this, the combined surface area of the two daughter cells is greater than the mother cell, thus, the membrane must be added
Phagocytosis of large particles→ requires addition of membrane to the surface, since a large amt was taken in
If a cell is wounded (creating a hole in the plasma membrane), a large amount of membrane may be delivered to the surface to facilitate repair
Cellularization→ during early Drosophila development, there are many rounds of nuclear division without cytokinesis, resulting in syncytium: thousands of nucleus surrounded by one plasma membrane
this is followed by cellularization, when every nucleus gets its own plasma membrane
What happens on the trans side of the Golgi network?
polarized cells direct proteins from the trans Golgi network to the appropriate domain of the plasma membrane
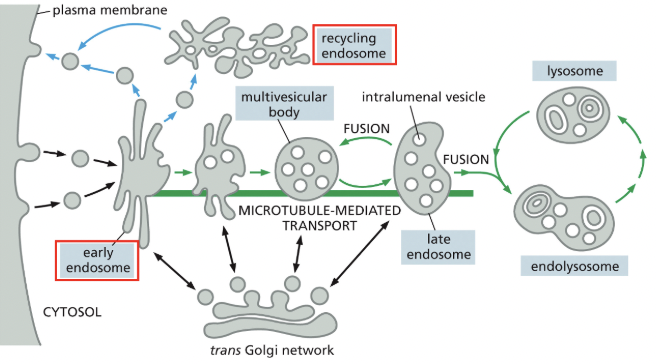
Direct sorting in the trans Golgi network vs. Indirect sorting via early endosomes
Transport into the cell from the plasma membrane = endocytosis
near the cell periphery, endocytic vesicles fuse with an early endosome, which is the primary sorting station
**glucose causes black arrows & insulin causes blue arrows
vesicles bud off the early endosome to recycle components back to the plasma membrane; this can happen directly or indirectly via the recycling endosome (which can store those components until needed)
as the early endosome matures, any membrane proteins bound for degredation become internalized in intraluminal vesicles, resulting in the multivesicular body/late endosome
the late endosome eventually fuses with the lysosome, whose hydrolytic enzymes act to digest contents
Caveolae (in fibroblast membrane)
often stable invaginations that can serve as a reservoir of plama membrane for cells exposed to mechanical forces that stretch the membrane
Receptor-Mediated Endocytosis
cells use this to import selected extracellular macromolecules
ex) cells import cholesterol in LDL particles by this mechanism
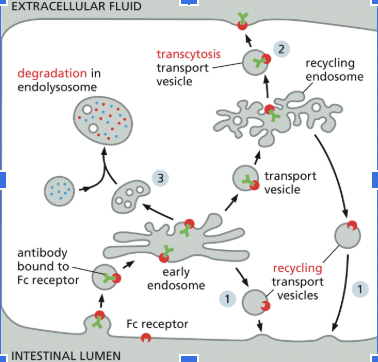
Possible Fates for Transmembrane Receptor Proteins that have been Endocytosed (3)
Back to where it came from
Transcytosis: delivered to another part of the plasma membrane
Degradation in the endolysome
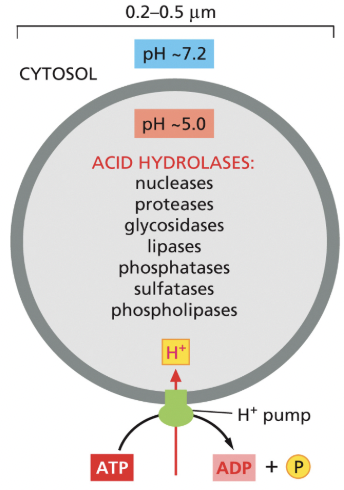
Lysosomes
principal sites of intracellular digestion
most of the lysosome membrane proteins are highly glycosylated, which helps to protect them from the lysosomal proteases in the lumen
proton pumps maintain an acidic pH inside the lysosome
Late Endosome
has hydrolase & completely intact intralumenal vesicle
becomes the endolysosome
Endolysosome
When the intralumenal vesicles start having “cracks” in their membranes
can become itself or become the lysosome
Lysosome
when the endolysosome digests its contents
could develop intralumenal vesicles and become an endolysosome after
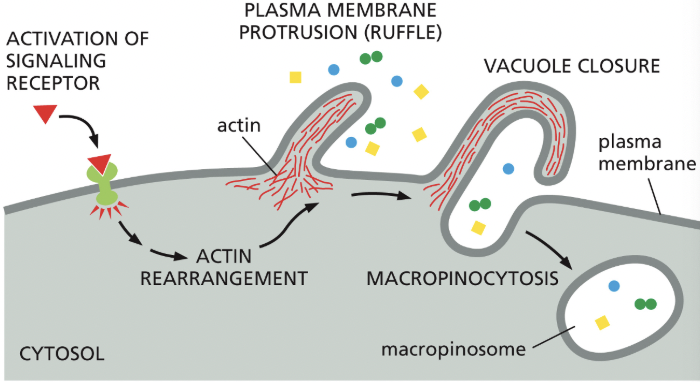
Macropinocytosis
cells can acquire nutrients from the extra fluid through this
non-specific, “cell drinking”
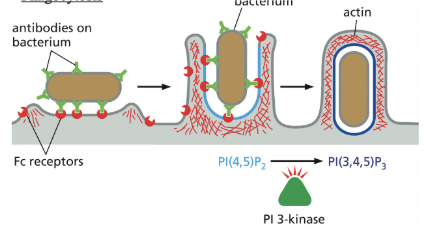
Phagocytosis
cargo recognition by cell-surface receptors
specific process for engulfing large particles are bacteria or dead cells
Pseudopod extension & phagosome formation are driven by actin polymerization & reorganization, which respond to the accumulation of specific phosphoinositides in the membrane of the forming phagosome
PI(4,5)P2 stimulates actin polymerization = promotes pseudopod formation
then, PI(3,4,5)P3 depolymerizes actin filaments at the base
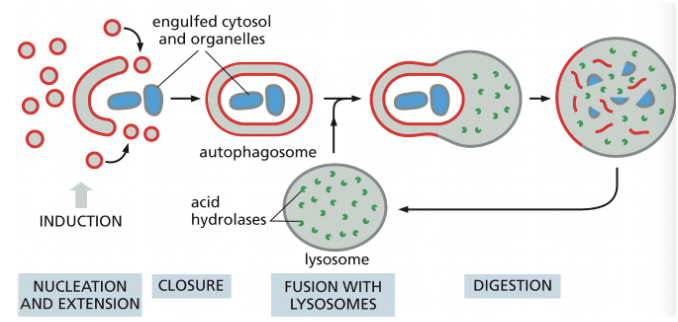
Autophagy
degrades unwanted proteins & organelles
a family of cargo-specific receptors mediates selective autophagy (ex. Ubiquitin)
Cell Signaling
process of detecting, processing, & responding to those signals
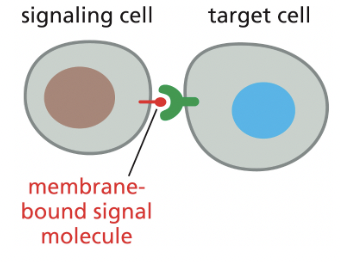
Contact-Dependent Signaling (aka juxtacrine signaling)
requires cells to be in direct membrane-membrane contact
ex) recognition of surface molecules by immune system cells
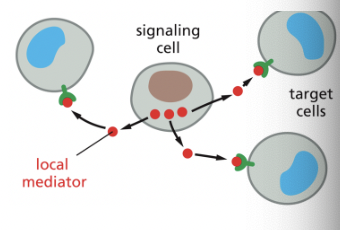
Paracrine Signaling
signaling depends on local mediators that are released into the extracellular space & act on nearby cells
ex) puncture injury to the sin will trigger a local inflammatory response, causing local capillaries to dilate & become leaky
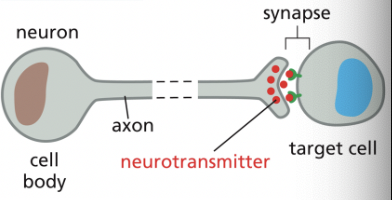
Synaptic Signaling
**a specialized form of paracrine signaling
performed by neurons that transmit signals electrically along their axons & release neurotransmitters at chemical synapses, which are often located far away from the neuronal cell body
Endocrine Signaling
depends on endocrine cells, which secrete hormones into the bloodstream for distribution throughout the body; only cells with the necessary receptor are able to respond
ex) the pancreas releases insulin into the bloodstream & various cells throughout the body respond
Extracellular vs. Intracellular Signal Receptors
when a signal molecules is hydrophilic, it binds to extracellular receptor (most signaling molecules)
when a signal molecule is hydrophobic, they are able to diffuse across the cell membrane & bind to intracellular receptor proteins (inside target cell)
these are typically bound to carrier proteins while in extracellular fluid of blood since they have poor solubility in aqueous solutions
ex) steroid hormones [estrogen & testosterone]
Why do tumors/abnormal growths form?
When cells in a multicellular organism dont integrate lots of incoming info to coordinate activities→ leads to growth without inhibition
Ion-Channel Coupled Receptors (aka transmitter-gated or ligand gated ion channel)
1/3 major classes of cell-surface receptor proteins!
Cells within an animal usually have established a concentration gradient of ions across their membranes; ions cannot easily get through the lipid bilayer
ion-channel coupled receptors open when bound to a specific molecule, allowing a rapid flow of ions across the membrane, changing the membrane potential
VERY important for electrical signaling in neurons & muscle cells
ion channels that open when bound to specific ligand→ utility depends on presence of an ion concentration gradient across the membrane (membrane potential)
when a channel opens, there’s a rapid flow of ions from one side of the membrane to other, changing the membrane potential
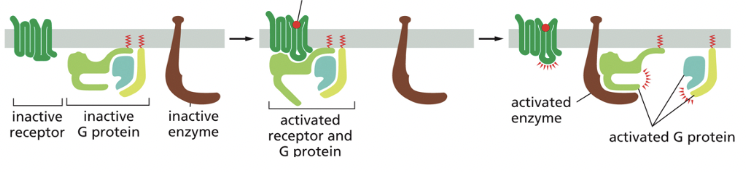
G-Protein-Coupled Receptor
2/3 major classes of cell-surface receptor proteins!
binding of a signaling molecule changes the conformation of the intracellular portion of the receptor, activating a G-protein (GTP binding protein) in the cytoplasm
transmembrane proteins that have a receptor binding site on the extracellular side of the protein
binding of the ligand changes the 3D conformation of the protein, altering the structure on the cytoplasmic side
membrane bound proteins that have several membrane spanning domains
binding of ligand to the receptor causes change in the 3D conformation of the receptor so that it binds to an intracellular membrane associated heterotrimeric G-protein that is bound to GDP
when bound to receptor, the G-protein releases GDP & binds to GTP, becoming acting & leading to various downstream effects
Enzyme-Coupled Receptors
3/3 major classes of cell-surface receptor proteins!
enzyme coupled receptors either become enzymatically active or activate a separate enzyme upon signal molecule binding
in both cases, the enzyme is usually a kinase (enzyme that phosphorylates its substrate) & a dimer, with each monomer phosphorylating the other monomer, leading to various downstream effects
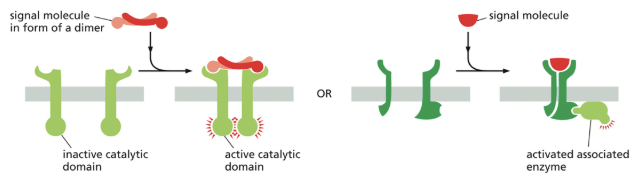
What are the 3 types of intracellular signaling complexes?
**components of signaling cascades are often assembled into a complex or a local region; this increases signal efficiency & reduces the chances of non-specific cross talk btwn different pathways
1) Performed Signaling Complex on a Scaffold Protein
a receptor + intracellular signaling proteins it activates in sequence = pre-assembled into signaling complex on inactive receptor→ interact v quickly & increases local [] w/ respect to each other
2) Assembly of Signaling Complex on an Activated Protein
assembly is transient & only happens when receptor/scaffold is activated
3) Assembly of Signaling Complex on Phosphoinositide Docking Sites
activated receptor phosphorylates local phospholipids which serves as docking sites for intracellular signaling proteins, promoting their interaction
Principles of Cell Signaling
(the relationship btwn signal & response varies in different signaling pathways)
Response timing (ms-days)
Sensitivity (concentrations needed)
Dynamic Range (response is graded or binary [neuron- on or not])
Persistence (short-long lasting)
Signal Processing
Integration (how many inputs?)
Coordination (multiple internal effects from one signal?)
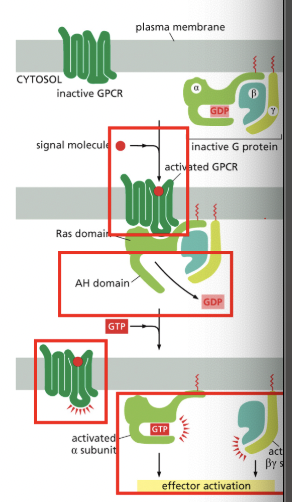
Heteromeric G Proteins
RELAY SIGNALS FROM GPCRs
heterotrimaric = 3 different parts
AH = alpha helical
binding of the receptor changes its conformation so that it binds to the heterotrimeric G-protein → AH domain moves outward, allowing GDP to leave & GTP to enter
the G-protein dissociates into 2 compoents, each of which can regulate downstream molecules
as long as the ligand is bond, receptor can activate more heterotrimeric G-proteins
*some g-proteins regulate production of cAMP in neurons
Why is synthesis & degredation of cAMP so fast?
cAMP is synthesized from ATP via cyclization rxn that removes 2 phosphate groups as PP (pyrophosphate)
cAMP is short-lived/unstable in cell cuz hydrolyzed by specific phosphodiesterases to form 5’-AMP as indicated
Cyclic-AMP-dependent Protein Kinase (PKA)
mediates most effects of cAMP
when cAMP binds to inactive PKA, the inactivator becomes inactivated, releasing the catalytic subunit
activated PKA will enter nucleus & activate CREB (binds to cyclin AMP response element (CRE)
ex) activated PKA enters the nucleus, increasing transcription of the target gene(s) → activating an activator
IP3 Receptors
**part of the ER membrane that are stimulated by low to moderate cytoplasmic Ca²+ concentrations
this Ca²+ induced calcium release (CICR) = positive feedback
inhibited by high [Ca²+] = negative feedback
Calmodulin
multipurpose intracellular Ca2+ receptor, governs many Ca2+ regulated proceses
consists of highly conserved, single polypeptide chain with 4 high-affinity Ca2+ binding sites
when it binds to Ca2+, it undergoes an activating conformational change
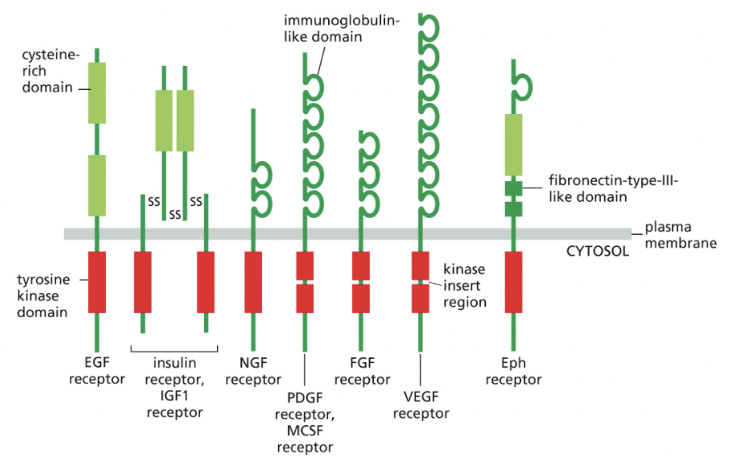
Receptor Tyrosine Kinases
**most common class of enzyme coupled receptor
have one transmembrane domain
in some cases, tyrosine kinase domain is interrupted by a kinase insert region: an extra segment emerging from the folded kinase domain
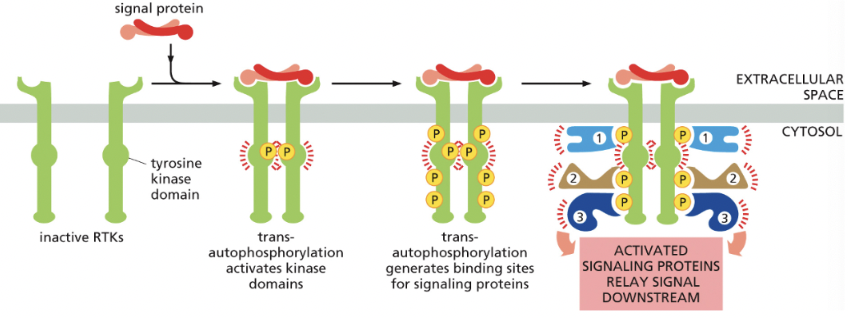
in all cases, binding of signal protein to the ligand-binding domain on extracellular side of the receptor activates the tyrosine kinase domain on cytosolic side
leads to phosphorylation of tyrosine side chains on cytosolic part of receptor = makes phosphotyrosine docking sites for various intracellular signaling proteins that relay the signal/diverge into multiple pathways
Trans-autophosphorylation
case of receptor tyrosine kinases→ when 2 monomers are brought together by binding of a dimerized signal protein
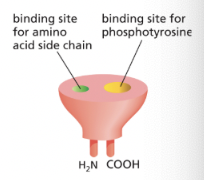
SH2 Domain
protein motif that has a binding site for phosphotyrosine next to a binding site for a specific amino acid, allowing them to bind specific phosphotyrosines
it’s a “plug-in” module→ can be inserted in disordered regions of a protein without disturbing the protein’s folding/function
different SH2 domains recognize phosphotyrosine in the context of different flanking amino acid sequences
**phosphorylated tyrosines on RTKs serve as docking sites for intracellular signaling proteins
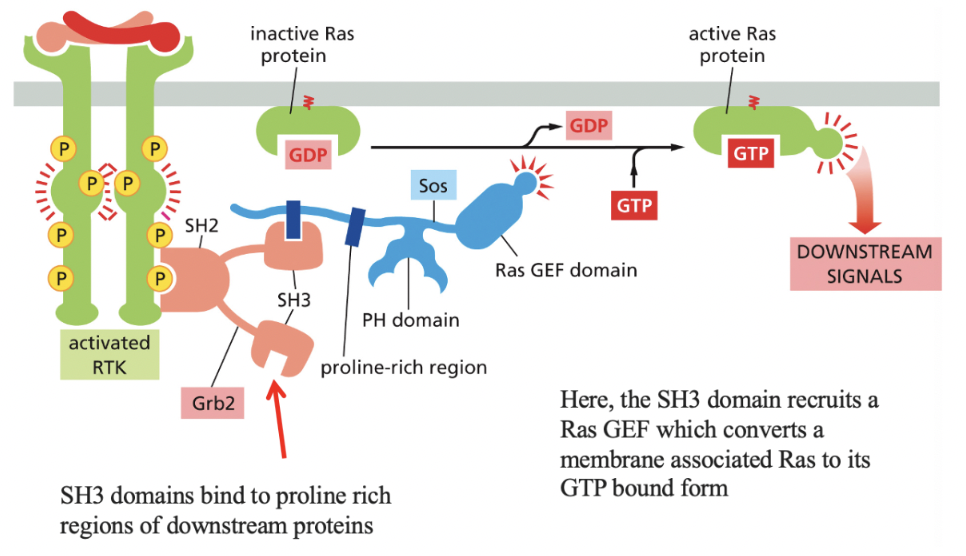
Ras
monomeric GTPase that mediates signaling by most RTKs
next to RTK→ SH2 domains (which has SH3 domains attached) bind to proline rich regions of downstream proteins
then, Sos (contains pH domain that recognizes specific phospholipids) binds to the SH3 domain and activates Ras (gives it GTP)
via recruitment of Ras GEF
Ras activates a kinase cascade via activating MAP Kinase signaling module
this signal can be amplified at each step if the components of the cascade are not bound to a scaffold
Scaffold proteins reduce cross-talk btwn different MAP Kinase modules (ex. binding kinase A onto a scaffold ensures it only acts in pathway initially activated)
PI 3-Kinase
produces lipid docking sites in the plasma membrane by phosphorylating stages of PI (phosphatidylinositol) lipids
ex) phosphorylating stages of PI lipids that become diacylglycerol (prevents this)
the PI-3-Kinase-Akt Signaling Pathway stimulates animal cells to survive & grow
the lipid docking sites are just another way to get components of a signaling pathway in close proximity
Nuclear Receptors
ligand-modulated transcription regulators; some extracellular signal molecules that bind to intracellular receptors
all of them are small & hydrophobic so they can diffuse directly across a lipid bilayer
ex) cortisol, estradiol, testosterone, vitamin D3, thyroxine, retinoic acid
How are nuclear receptors activated?
EXAMPLE
binding of the ligand causes dissociation of the inhibitory proteins, allowing activation of transcription
all nuclear receptors bind to DNA as either homo- or hetero-dimers
Plant Hormones (aka Plant Growth Regulators)
help to coordinate plant development→ small molecules made by most plant cells & diffuse readily through cell walls & can either act locally or be transported in influence cells further away
ex: ethylene (blocks degradation of specific transcription regulatory proteins in nucleus), auxin, cytokinins, gibberllins, abscisic acid, & brassinosteroids
Ehylene-Mediated Triple Response (PLANTS)
occurs when the growing shoot of a germinating seedling encounters an obstacle underground
in the abscense of obstacles, the shoot grows upwards & is long/thin
if the shoot encounters an obstacle (ex. gravel), the seedling responds to encounter in 3 ways
it thickens its stem→ can then exert more force on the obstacle
it shields the top of the shoot by increasing the curvature of a specialized hook structure
it reduces shoot’s tendency to grow away from the direction of gravity as to avoid the obstacle
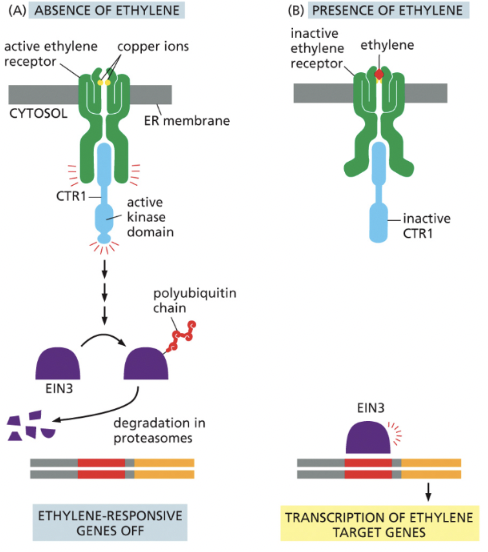
Ethylene Signaling Pathway (PLANTS)
the presence of ethylene prevents the destruction of EIN3, leading to the transcription of ethylene responsive genes
blocks degradation of specific transcription regulatory proteins in nucleus
Auxin Signaling (PLANTS)
in the abscense of auxin, a transcription repressor protein (called Aux/IAA) binds & supresses a transcription regulator (called auxin-response factor [ARF]), which is required for the transcription of auxin-responsive genes
when activated by auxin binding, the receptor-auxin complex recruits a ubiquitin ligase, which ubiquitinates the Aux/IAA protein, marking it for degradation in proteasomes→ ARF is now free to activate the transcription of auxin-responsive genes
![<p>in the abscense of auxin, a transcription repressor protein (called Aux/IAA) binds & supresses a transcription regulator (called <em><u>auxin-response factor</u></em> [<strong>ARF</strong>]), which is required for the transcription of auxin-responsive genes</p><p></p><p>when activated by auxin binding, the receptor-auxin complex recruits a ubiquitin ligase, which ubiquitinates the Aux/IAA protein, marking it for degradation in proteasomes→ ARF is now free to activate the transcription of auxin-responsive genes</p><p></p>](https://knowt-user-attachments.s3.amazonaws.com/16d1c6d4-9075-41ff-a045-ccf16d8aa05d.png)
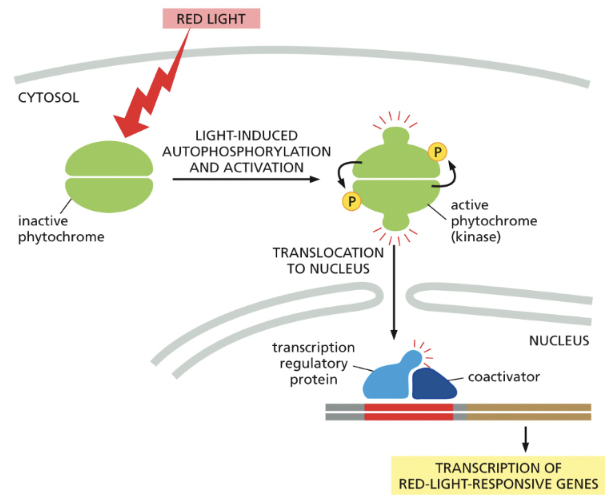
Phytochromes (PLANTS)
detect red light & respond by autophosphorylation
Why is signaling important?
needed for cells to respond to their environments; especially important in multicellular organisms so that the activities of all the cells can be coordinated
Kinase
phosphorylates a substrate
Phosphatase
dephosphorylate a substrate
Cytoskeleton
network of internal filamentous proteins that forms an internal framework which is used for organizing cell components, moving organelles, maintaining cell shape/locomotion
cytoskeletal filaments are dynamic, but can nevertheless form stable structures
the cytoskeleton determines cellular organization & polarity
filaments assemble from protein subunits that impart specific physical & dynamic properties
accessory proteins & motors act on cytoskeletal filaments
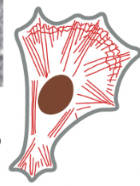
Actin (aka microfilaments)
1/3 major types of protein filaments that form the cytoskeleton
helical polymers of actin (protein) → flexible structures with a diameter of 8 nm that organize into a variety of linear bundles, 2D networks, & 3D gels
Actin filaments are dispersed throughout the cell
most highly concentrated in the cortex, just beneath the plasma membrane
actin subunits assemble head-to-tail to create flexible, polar filaments
plus end (barbed) grows much more & faster than minus end (pointed)
ex) when the actin cytoskeleton is rapidly assembled/reassembled, the cell can be pushed to move from a section of cell
accessory proteins & motors act on this
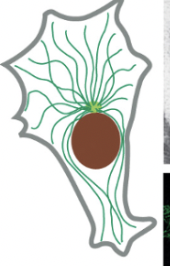
Microtubules
2/3 major types of protein filaments that form the cytoskeleton
long, hollow cylinders made of tubulin (protein)
outer diameter of 25 nm (much more rigid than actin filaments)
long/straight & frequently have one end attached to a microtubule-organizing center (MTOC) = centrosome
part of extending cell in mitosis!
accessory proteins & motors act on this
the addition of GTP-containing tubulin subunits to the end of a protofilament causes end to grow in linear conformation that can readily pack into the cylindrical wall of the microtubule
hydrolysis of GTP after assembly changes the conformation of the subunits & tends to force the protofilament into a curved shape that’s less able to pack into the microtubule wall (happens when GTP cap is removed and causes disruptions)
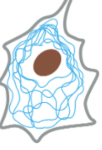
Intermediate Filaments
3/3 major types of protein filaments that form the cytoskeleton
ropelike fibers with diameter of ~10 nm
made of intermediate filament proteins, which constitute a large/heterogeneous family
one type of intermediate filament forms a meshwork called nuclear lamina just beneath the inner nuclear membrane
other types extend across cytoplasm to give cells mechanical strength
ex) in epithelial tissue, they span the cytoplasm from one cell-cell junction to another, thereby strengthening the entire epithelium
accessory proteins & motors DO NOT act on this
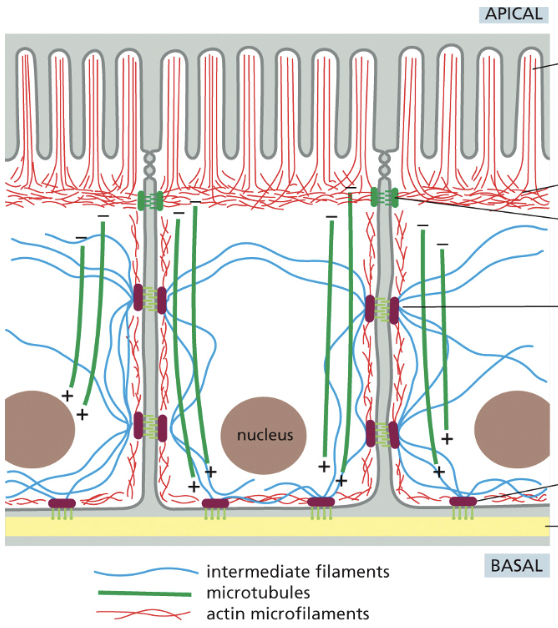
How does the cytoskeleton determine cellular organization & polarity (model is epithelial cells that line the small intestine)
bundled actin filaments form microvilli that increase the cell-surface area available for absorbing nutrients from food
below the microvilli, a circumferential band of actin filaments is connected to cell-cell adherens junction that anchor the cells to each other
intermediate filaments are anchored to other kinds of adhesive structures including desmosomes & hemidesmosomes that connect the epithelial cells into a sturdy sheet & attach them to underlying matrix
microtubules run vertically from the top of the cell to the bottom & provide a global coordinate system that enables the cell to direct newly synthesized components to their proper location
seems to really determine polarity (negative = outside of cell, positive = inside the cell)
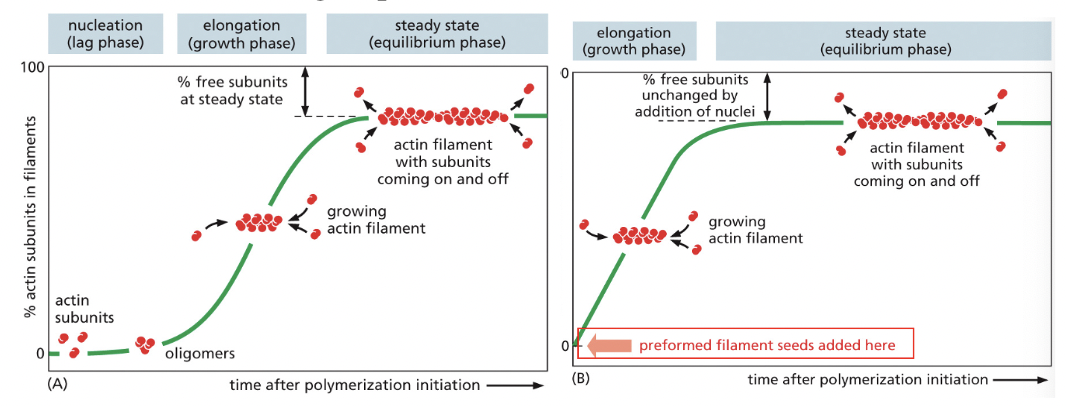
Nucleation
RATE-LIMITING STEP IN FORMATION OF ACTIN FILAMENTS
helical polymer is stabilized by multiple contacts between adjacent subunits
2 actin molecules bind relatively weakly to each other, but, addition of a 3rd actin monomer to form a trimer makes the entire group more stable
further monomer addition can take place onto this trimer, which therefore acts as a nucleus for polymerization
for tubulin, the nucleus is larger & has more complicated structure (13+ molecules) but principle is the same
Critical Concentration (Cc)
the number of monomers that add to the polymer (actin filament or microtubule) per second will be proportional to the concentration of the free subunit (konC)
but, the subunits will leave the polymer & end at a constant rate (koff) but that doesn’t depend on C
as the polymer grows, subunits are used up & C is observed to drop until it reaches a constant value = Cc
EQUILIBRIUM: kon * C = koff
Cc = (koff/kon) = Kd
Kd = dissociation constant
Lag Phase in Polymerization
corresponds to time taken for nucleation
Growth Phase in Polymerization
occurs as monomers add to the exposed ends of the growing filament, causing filament elongation
Equilibrium Phase (aka Steady State) in Polymerization
reached when the growth of the polymer due to monomer addition precisely balances the shrinkage of the polymer due to disassembly back to monomers
Nucleotide Hydrolysis
each actin molecule carries a tightly bound ATP molecule that’s hydrolyzed to a tightly bound ADP molecule soon after its assembly into the polymer
each tubulin molecule carries a tightly bound GTP molecule that’s converted to a tightly bound GDP molecule soon after the molecule assembles into the polymer
hydrolysis of the bound nucleotide reduces the binding affinity of the subunit for neighboring subunits & makes it more likely to dissociate from each end of the filament
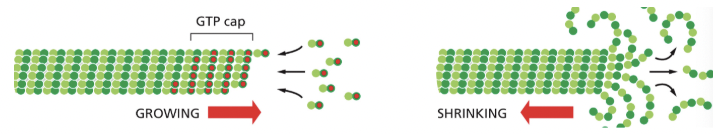
ATP/GTP Caps
the rate of addition of subunits to a growing actin filament or microtubule can be faster than the rate at which their bound nucleotide is hydrolyzed
under such conditions, the end has a “cap” of subunits containing the nucleoside triphosphate→ an ATP cap on the actin filament or a GTP cap on a microtubule
Treadmilling
changing the critical concentration at the two ends of the polymer
because koff & kon refer to different reactions, their ratio koff/kon need not be the same at both ends of the polymer
so… Cc (- end) > Cc (+ end)
if both ends of the polymer are exposed, polymerization proceeds until the concentration of free monomer reaches a value that is above Cc for the plus end but below Cc for minus end
at this steady state, subunits undergo a net assembly at the + end & a net disassembly at the - end at an identical rate→ polymer maintains a constant length
predominates dynamic instability in actin filaments
Dynamic Instability
individual microtubules can alternate between a period of growth & period of rapid disassembly
microtubules depolymerize ~100x faster from an end containing GTP-tubulin
a GTP cap favors growth, but if its lost, then depolymerization ensues
predominates treadmilling in microtubules
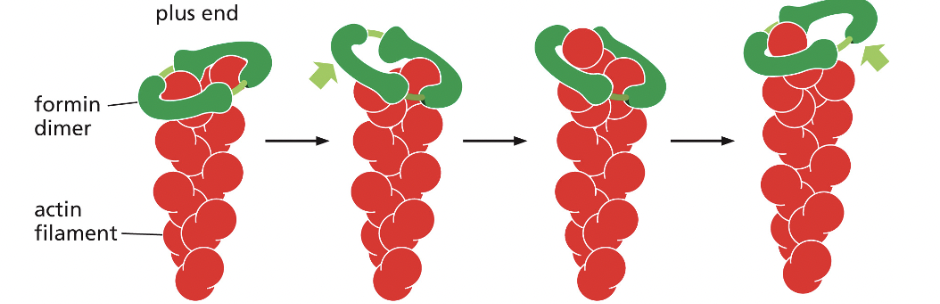
Formin (actin-binding protein)
nucleates assembly and remains associated with the growing plus end
form a dimeric complex that can nucleate the formation of a new actin filament & remain associated with the rapidly growing plus end, increasing the rate of elongation
several formin dimers can nucleate the growth of actin filaments that can be cross-linked by other proteins to form parallel bundles
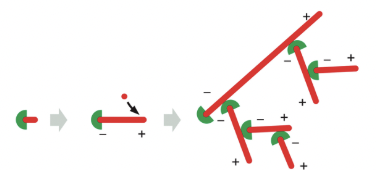
Arp2/3 Complex (actin-binding protein)
nucleates assembly to form a branched network & remains associated with the minus end
Arp = Actin Related Protein
although face of molecule equivalent to the plus end (top) in both Arp2 & Arp3 is very similar to the plus end of actin itself, differences on sides & minus end prevent these Arps from forming filaments on their own or co-assembling into filaments with actin
they can nucleate actin polymerization
Needs Nucleation-Promoting Factor (NPF) to bind to Arp so it can nucleate the actin filament (because now it resembles the plus end of the actin filament)
the Arp2/3 complex nucleates filaments most efficiently when it’s bound to the side of a pre-existing actin filament
filament branches grow at a 70 degree angle relative to the original filament
Thymosin (actin-binding protein)
binds subunits, prevents assembly

Profilin (actin-binding protein)
binds monomers, concentrates them at sites of filament assembly
many nucleation-promoting factors (NPF) contain binding sites for profilin which is bound to actin monomers
this maintains a large pool of actin monomers in the area where actin filaments are growing
Some members of the formin protein family possess whistler-like unstructured domains that contain several binding sites for profilin-actin complexes
This serves to recruit new actin monomers to the growing plus end of the actin filament where formin is bound

Tropomodulin (actin-binding protein)
prevents assembly & disassembly at minus end
caps actin filaments at both ends to prevent depolymerization during muscle contraction

Tropomyosin (actin-binding protein)
stabilizes filament, modulates binding of other accessory proteins
held in place by troponin complex
in muscle contraction, blocks myosin binding sites on actin subunits at low [Ca2+]
At high [Ca2+], tropomyosin shifts position, revealing the myosin binding sites (this forms a cross bridge & starts contraction cycle)
![<p>stabilizes filament, modulates binding of other accessory proteins</p><ul><li><p>held in place by troponin complex</p></li></ul><ul><li><p>in muscle contraction, blocks myosin binding sites on actin subunits at low [Ca2+]</p><ul><li><p>At high [Ca2+], tropomyosin shifts position, revealing the myosin binding sites (this forms a cross bridge & starts contraction cycle)</p></li></ul></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/b773bc51-35a9-4b67-9e22-216290a79496.png)
Cofilin (actin-binding protein)
ds ADP-actin filaments, accelerates disassembly by inducing actin filament twisting
the energy of cofilin binding serves to deform the actin filament, twisting it more tightly & reducing the distance spanned by each twist of the helix
this puts strain on the actin/actin monomer bonds, destabilizing the filament & promoting depolymerization

Gelsolin (actin-binding protein)
severs filaments & binds to plus end
Capping Protein (actin-binding protein)
prevents assembly & disassembly at plus end

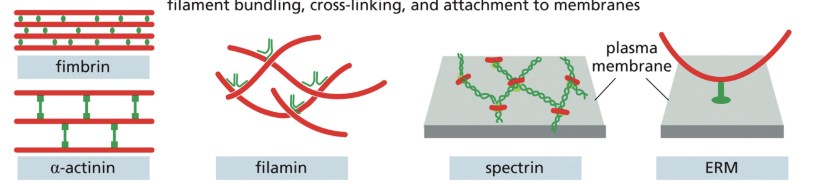
What actin-binding proteins are involved with filament bundling, cross-linking, & attachment to membranes?
Fimbrin, alpha-actenin, filamin, spectrin, myosin motor proteins & ERM
How does severing proteins regulate actin filament depolymerization?
breaking actin filaments into pieces can have two effects, depending on conditions:
exposed ends rapidly depolymerize
fragments act as nucleation sites, so there is growth of many new filaments