Lecture 1 - Structure and Organization of Nucleic Acids and Protein
1/42
Earn XP
Description and Tags
Dr. Natoya Peart, Fall 2025
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
43 Terms
DNA and RNA, formerly thymus and yeast nucleic acid, are ____ ______ of repeating subunits, with end-to-end directionality.
long polymers; subunits are nucleotides made up 5C sugar, nitrogenous base, and phosphate group
Differences Between RNA and DNA
DNA has no 2’ OH on sugar group, just H
DNA is more stable
DNA has a more rigid structure
RNA comes first
RNA has U, DNA has T
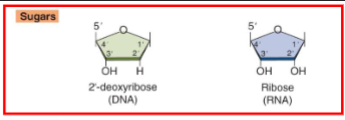
Subunits of DNA and RNA
bases: proton (H+) acceptors
purines are double C-N ring (A, G)
pyrimidines are single C-N ring (C, T, U)
phosphate group: proton donor that will form the phosphodiester bond
A nucleoside is made of:
the sugar and nitrogenous base only; nucleotides also contain a phosphate group
The phosphodiester bond between adjacent nucleotides in DNA or RNA is formed between ______ and _______.
3’ C OH group on nitrogenous base; 5’ C PO4- group
this maintains directionality and polarity in the strand
Phoebus Levene
studied carbohydrates and nucleic acids
thought nucleic acids were made of phosphoric acid and nitrogenous and non-nitrogenous substances
studied composition of nucleic acids in yeast and cattle (thus yeast and thymus) and determined that they are in equal proportion (1:1:1:1) → INCORRECT
Edwin Chargaff (1940s)
studied organism specificity of nucleic acids and observed that ratios of nucleotides and composition of bases varies amongst species
gave us Chargaff’s rules:
[A] = [T]
[G] = [C]
[G] + [A] = [T] + [C]
There are _____ H-bonds between adenine and thymine and ___ H-bonds between guanine and cytosine.
2; 3
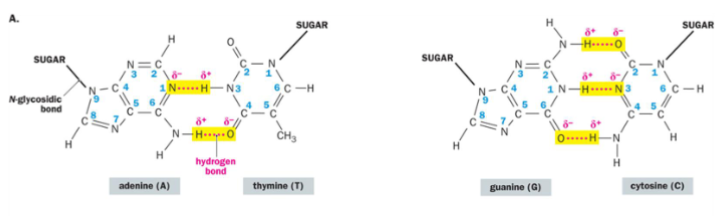
Watson-Crick Base Pairing
maximizes the number of H-bonds and confers the antiparallel
AT with 2 H-bonds; CG with 3 H-bonds
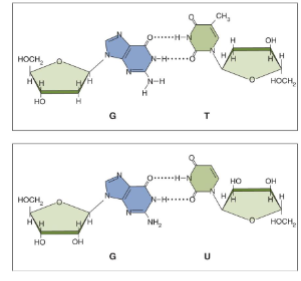
Non-Watson Crick Base Pairs
not typically found in dsDNA helix but are common in RNA - wobble bond because they can flip around
Structural Features of the Double Helix
secondary structure of DNA
has a hydrophobic core that is stabilized by base stacking (to keep water molecules out)
bases are also hydrophobic
major and minor grooves have different abilities to H-bond
more exposed DNA in major groove means it can do more H-bonding
also allows for interactions with proteins
Strand Separation
denaturing DNA by melting can be induced with higher temperature from increase of thermal energy → breaks H-bonds and other stabilizing forces in the helix
hyperchromicity: absorbance of 260nm UV light increases when DNA denatures
renaturation happens with slow cooling to allow reannealing or hybridization
Can DNA be unwound under physiological conditions?
yes; primarily by helicase
What affects the melting temperature (Tm) of DNA?
length and G:C content
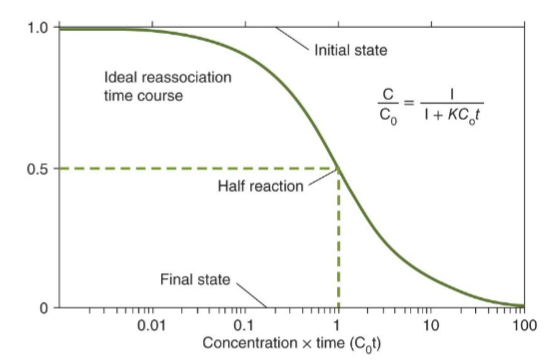
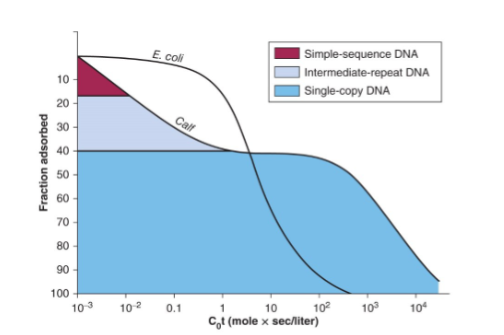
DNA Reassociation Kinetics
rate of renaturation of depends on:
length
concentration
repeated sequences
the rate that a sequence reassociates is proportional to the number of times it is found in the genome
Cot Curve: plot of % DNA reassociation vs. Cot value that reflects the complexity of the genome
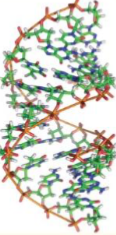
A-DNA
right-handed
deep and narrow
shallow and broad (superficial)
11 BP per turn
low humidity, high salt
not usually adopted by DNA; can happen in RNA
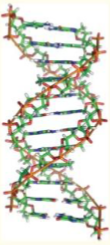
B-DNA
Watson-Crick DNA (regular double helix)
right-handed
moderate depth, wide
moderate depth, narrow
10.5 BP per turn
high humidity, low salt
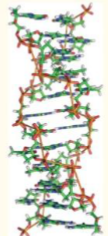
Z-DNA
left-handed (increased potential energy from torsional stress)
very shallow, basically a single groove
very deep and narrow
12 BP per turn
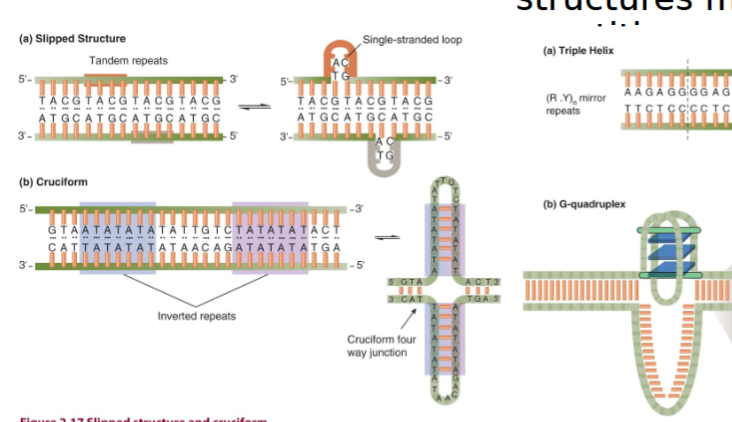
Non-B DNA Forms
most often occurs due to repetitive sequences to make unexpected structures
triplex: ssDNA comes back to H-bond with double helix
cruciform: resembles a cross; from inverted repeat sequences
RNA-DNA hybrid: three-stranded structure when a short RNA molecule base-pairs with dsDNA
G-tetrad: 4 G bases make a square
slipped DNA structures in tandem repeats
Hoogsteen Bonds
alternate H-bonds found in triplex and G-tetrad DNA
occur when purine rotates 180 degrees to the helix axis
RNA Secondary Structure
depends on various H-bonds
AU reverse Hoogsteen
GU wobble
GA sheared
GA imino
DNA Tertiary Structure
occurs when ends of double helix are constrained by circularizing or by DNA binding proteins underwinding or overwinding
virtually all DNA exists as supercoil
Positive Supercoiling
right-handed turn that decreases the # of bases per turn
double helix gets wound more tightly in the same direction
Negative Supercoiling
left-handed turn that increases the # of bases per turn
opposite of normal DNA direction which basically straightens it
Types of RNA
mRNA - messenger
tRNA - transfer
snRNA - small nuclear
snoRNA - small nuceolar
rRNA - ribosomal
miRNA - micro
Why is RNA more versatile than DNA?
it is a single-stranded ribose sugar structure which allows it to take on more 3D structures
Components of RNA
polymer of NTPs (ribonucleotides) linked by phosphodiester bonds
RNA Secondary Structures
uses both Watson-Crick and non-Watson Crick base pairs to facilitate structure formation 2
interactions with 2’ hydroxyl facilitates long range interactions - changes the shape of minor and major grooves
nucleosides are often highly modified that allow for more H-bonding
large RNA molecules require protein chaperones to properly form their structure and prevent random folding
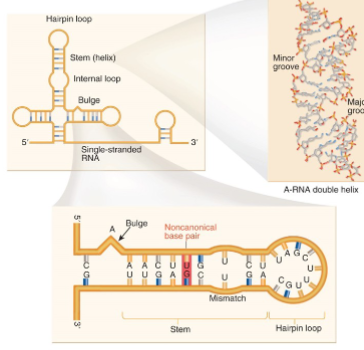
Is structure or sequence more important for function?
structure; it is more conserved than sequence to protect the hydrophobic region
tRNA model structure features non-Watson Crick base pairs, base stacking, modifications, and intramolecular forces
Amino Acids
monomer of proteins of which 22 are encoded
amino group and carboxyl group are charged at physiological pH
side chain confers properties to the amino acids (polarity for water solubility, basicity/acidity for charge, bulk)'
all except glycine are chiral and exist as enantiomers (non-superimposable mirror images); mostly L-amino acids
peptide bond is made between carboxyl group and amino group
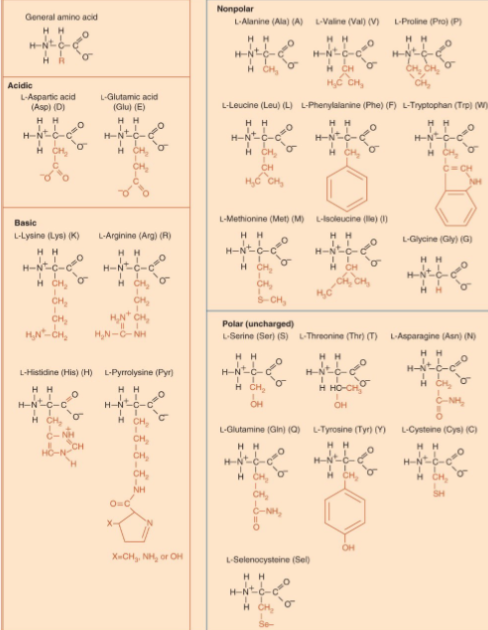
Polypeptide
made of amino acids linked by peptide bonds between carboxyl and amino groups
peptide bond is rigid; cis and trans configurations are fixed and are mostly trans in protein to minimize steric hindrance
polypeptide is read from N-terminus to C-terminus since it has polarity
Translating the Genetic Code
codon: 3 base sequence in the mRNA that specifies a single amino acid BUT multiple codons may code for the same amino acid (degeneracy)
mRNA is synthesized from the non-coding/antisense/template strand; the strand will have the same sequence as coding strand
codon on the sense strand of DNA → codon of mRNA
start codon: AUG, stop codons: UGA, UAA, UAG
The difference in amino acids usually lies in the ____ base because it _______.
third; doesn’t participate in Watson-Crick base pairs rather non-Watson Crick
this is known as the wobble base
mRNA codons are read by the ____ in tRNA.
anticodon; tRNA is charged with amino acid
_____ and _____ are the building blocks of life.
nucleic acids (DNA and RNA); protein
Adenine (A)
DNA nucleoside: deoxyadenosine
RNA nucleoside: adenosine
Guanine
DNA nucleoside: deoxyguanosine
RNA nucleoside: guanosine
Cytosine
DNA nucleoside: deoxycytidine
RNA nucleoside: cytidine
Thymine
DNA nucleoside: deoxythymidine
RNA nucleoside: N/A since RNA
Non-B Form DNA Structures
most often occurs due to repetitive sequences
slipped structure from tandem repeats (repeated in head-to-tail copies)
cruciform structure from inverted repeats (read the same forward on one strand and backward on complementary strand H-bonds to self)
non-Watson Crick base pairing makes:
triple helix from R, Y mirror repeats
G-quadruplex
Cot Curve
graphical representation of genome complexity that plots ratio of ssDNA against the log scale of the product of initial concentration with time
shows the rate of DNA reassociation to determine the complexity of a genome → affected by concentration, temperature, repetitive sequences, GC content
Does RNA need help to fold?
yes and no; it can fold on its own or with aid from a chaperone protein
DNA, RNA, and proteins can all take on _______ structures.
higher order