GCC550 Exam 4 Part III
1/67
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
68 Terms
The cell cycle
Features: cellular growth, DNA replication, chromosome segregation, an division of cell and contents
single cell accumulating mass
half of DNA segregation
all components need to be segregated
Highly ordered series of events, highly conserved process
needs to be unregulated cell division process
Necessary for Development replacement of adult tissue that has high cell turnover
need to be able to make copies, organ system needs to be able to be replaced
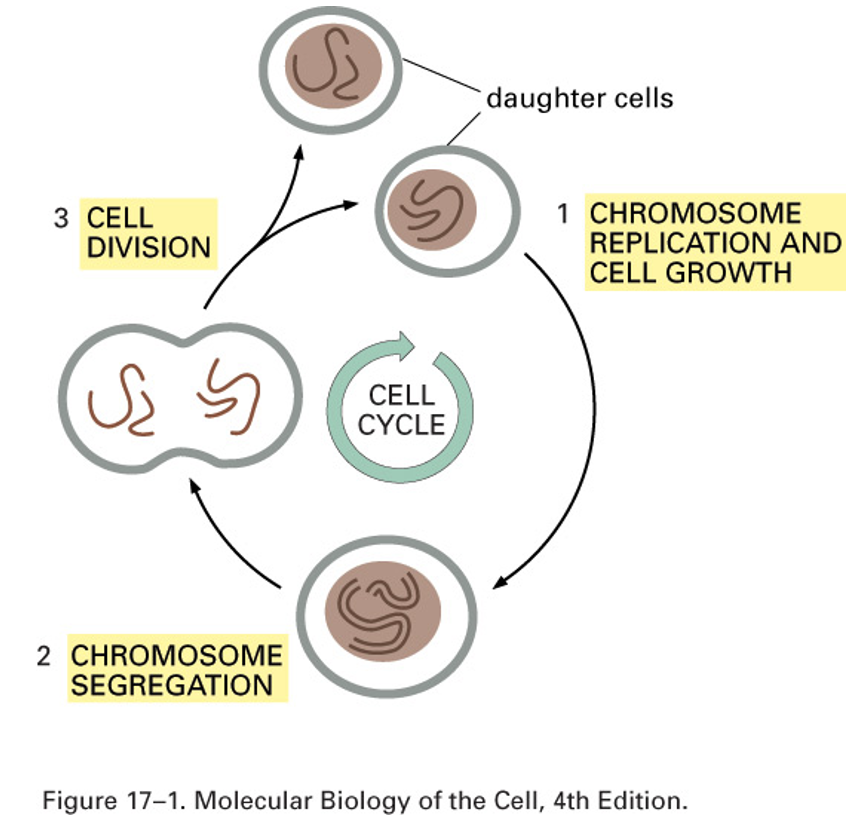
Cell Cycle Phases
G1 - Gap phase 1, growth of cell
S - DNA synthesis
G2 - Gap phase 2, growth and preparation for mitosis
M - mitosis, cell division
Each step has different checkpoints, and or cues
problems in regulators leads to cancer
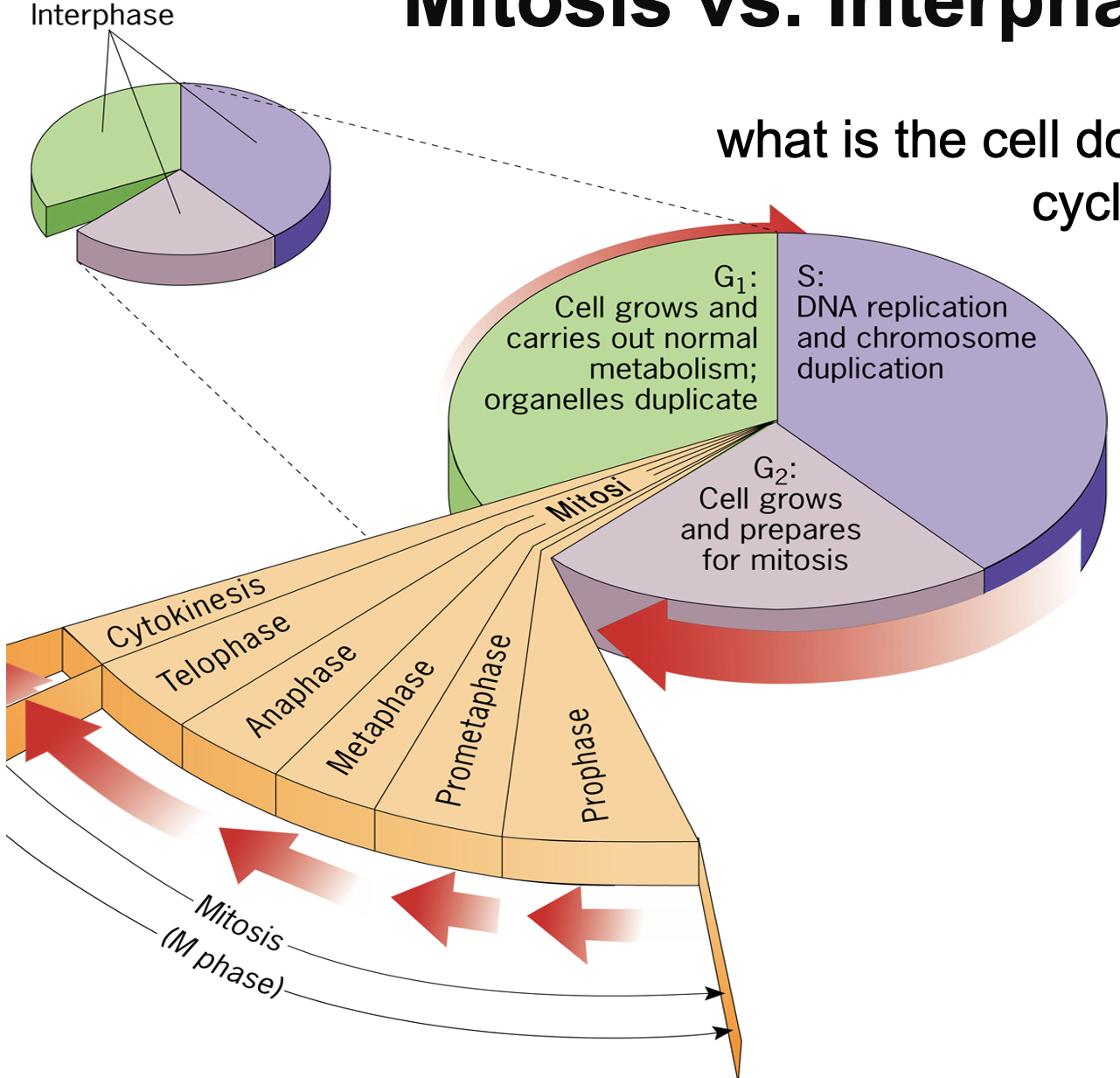
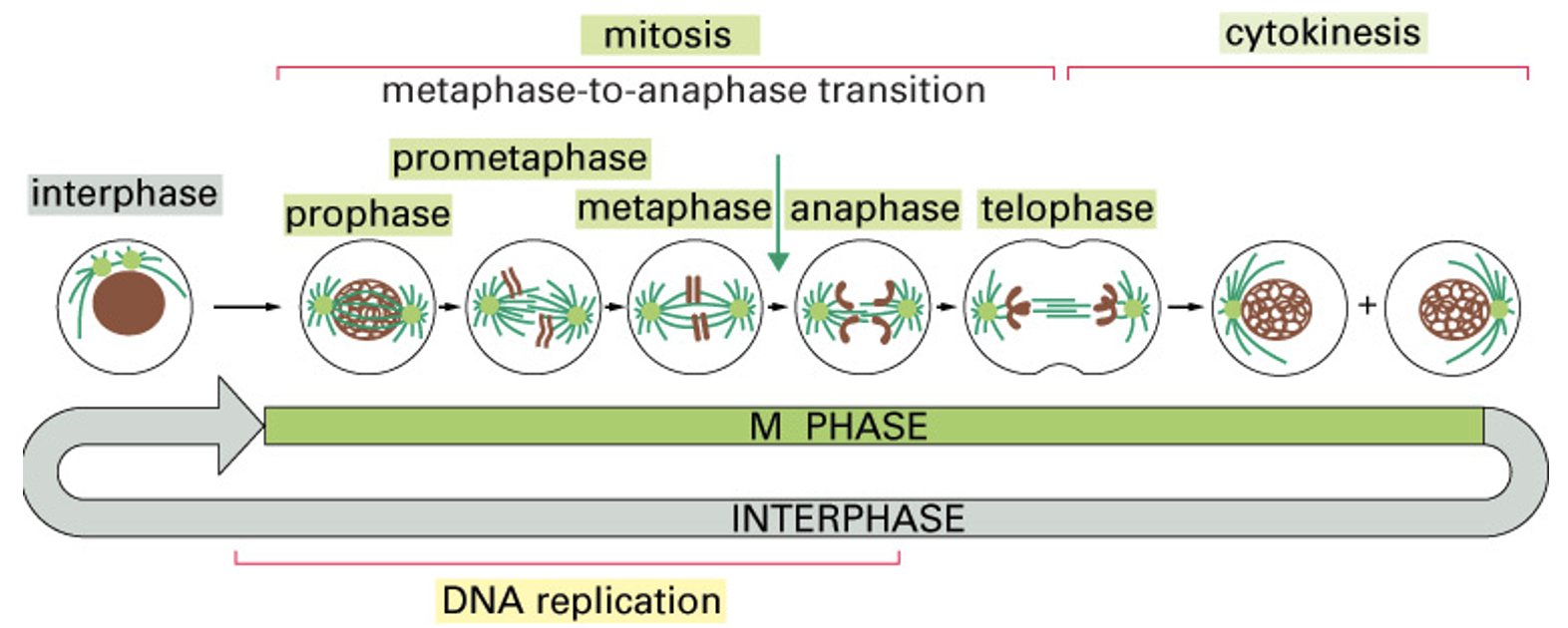
Mitosis vs Interphase
Interphase:
G1: Cell grows and carriers out normal metabolism; organelles duplicate - is also carrying out cue, sensing environment for prep to divide
S: DNA replication and chromosome duplication - once DNA fully replicated
G2: Cells grows and prepares for mitosis - replicated organelles, sense DNA synthesis, replication/ quality of replication.
Mitosis: Small steps for cell separation - Prophase, prometaphase, metaphase, anaphase, telophase, cytokinesis
Mitosis and Meiosis
M phase - 5% of the total cell cycle extensively studied because of mitotic chromosomes and their behavior
G0
Reversible cell cycle exit (post-mitotic cells)
has all characteristics of G1
cells reversibly enter
neurons are typically in this state
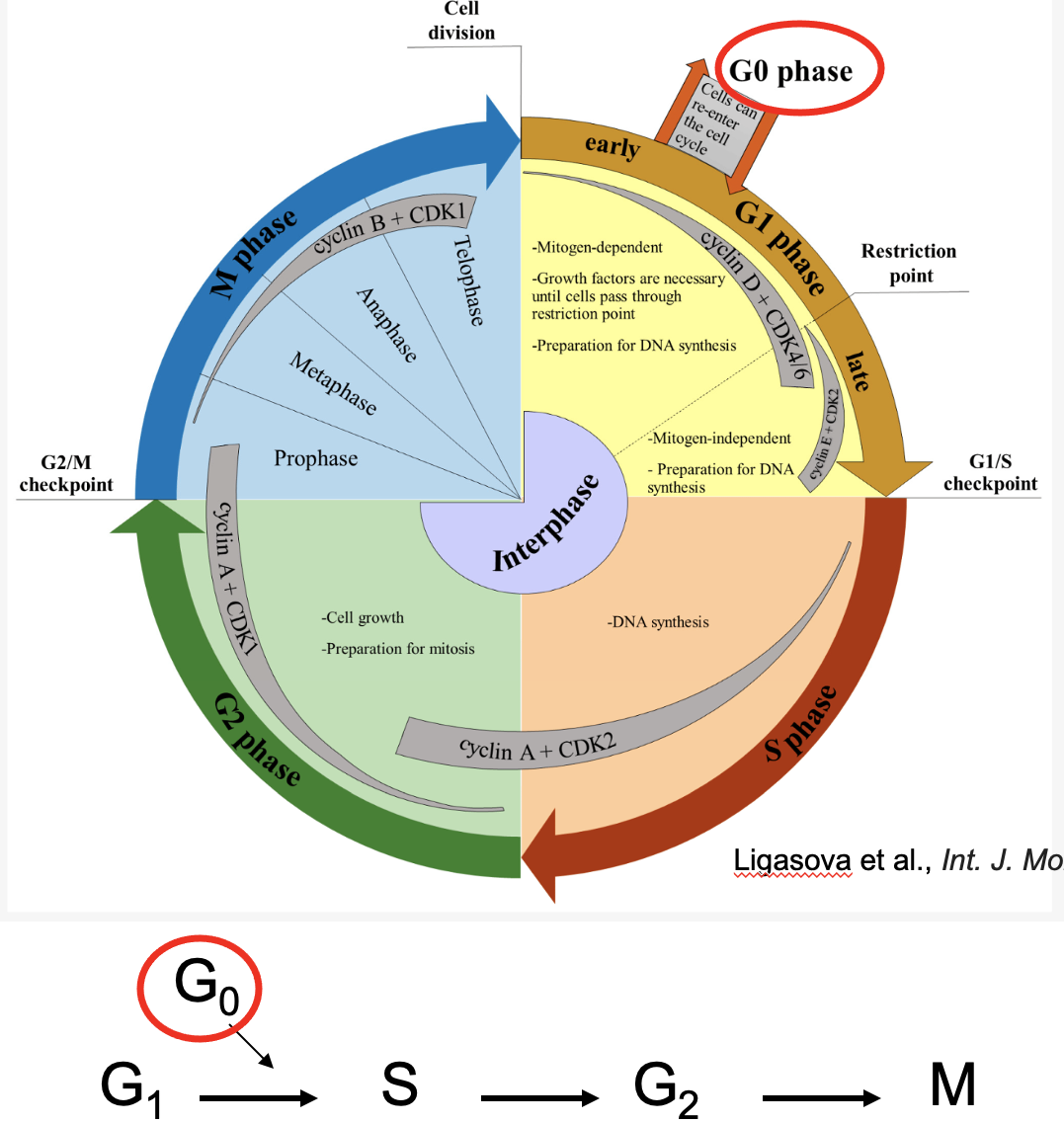
G0 Cell cycle length vary dramatically
Non-dividing cells - nerve cells, muscle cells, RBCs. Cell that can be induced to divide-liver cells, B and T cells.
Rapidly dividing cells - hematopoietic stem cells, epithelial cells and embryonic cells
G0 time varies in cells
Cell cycle length of typical mammalian cell cycle (S through Mitosis) 18-24 hours
How can we determine which stage of the cell cycle a cell is in?
DNA content:
G → S → G2 → M
2N-2N-4N-4N—4N-2N
In s phase, DNA not fully duplicated
N - varies by species, but is equal to 1 haploid genome
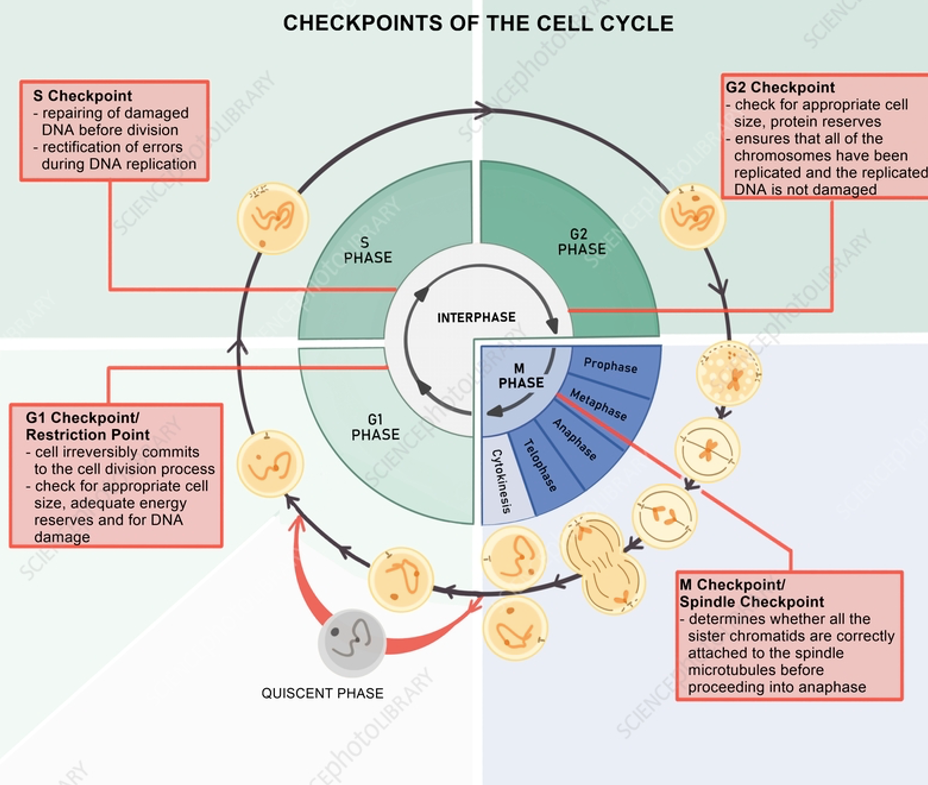
Decisions, Checkpoints, and Transitions
G1 Checkpoints:
Is environment good?
Growth factors present?
DNA damage?
S checkpoints:
DNA damage?
DNA replicated?
G2 Checkpoint:
DNA Damage?
DNA replication errors?
Mitotic spindle properly formed?
M checkpoint:
DNA replication errors?
Mitotic spindle properly formed?
Checkpoints are enforced by regulation of CDK activities
Biochemical events in Cell Cycle Regulation
Proteins involved in checkpoints and regulating phases
Kinases: cyclin-dependent kinases (cdks)
cell cycle specific
specific cyclins for each cycle
specific ones for each cycle
Active kinases phosphorylates substrates, which in turn allows the cell to go through different stages of cell cycle
There are multiple cdks for each cell cycle transition - multiple substrates known
Cyclins bind CDK to activate
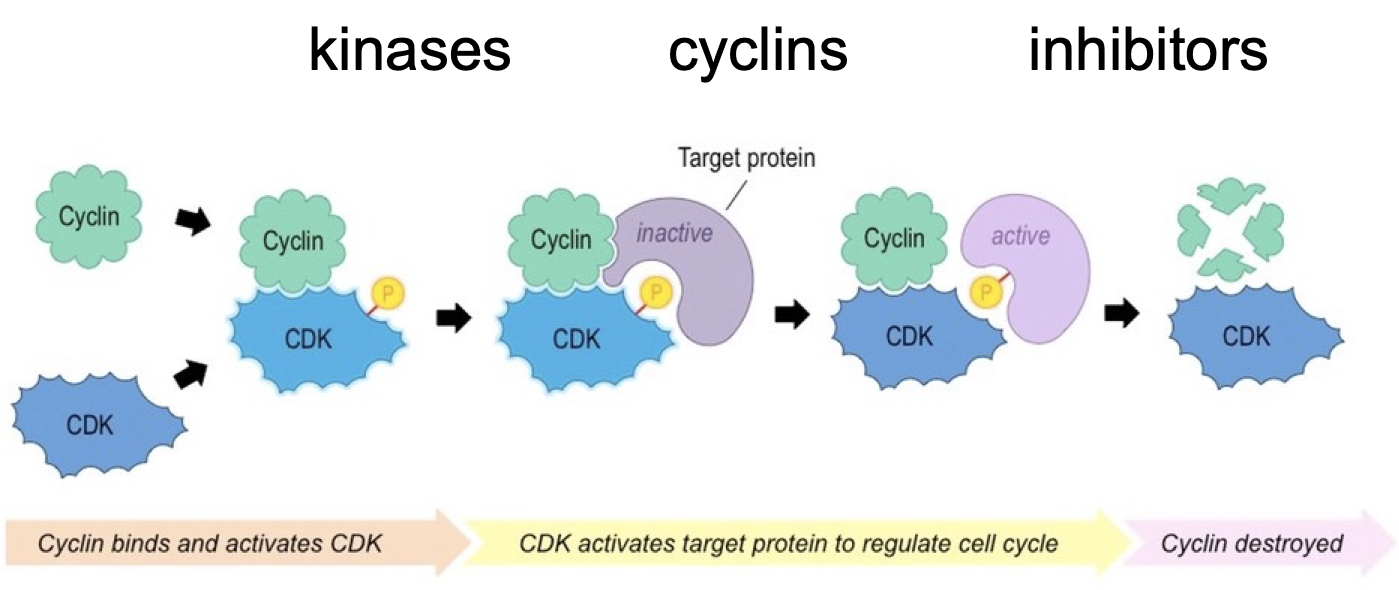
Active enzyme
A complex of cdk and cyclin
set of machines made to regulate cell cycle
CDKs - always expressed
cyclins - phase dependent expression
distinct from enzymes that replicate cell components a dedicated cell cycle control machine
Regulatory mechanisms: expression, activity, interaction, degradation
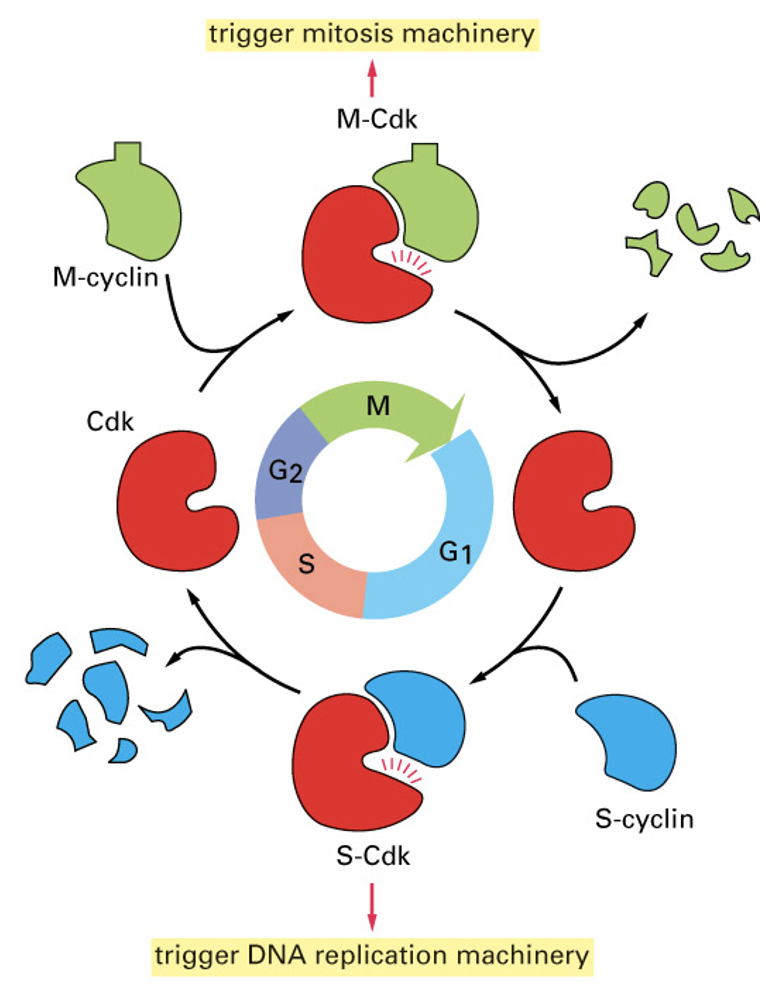
Specific cell cycle transitions
Controlled by specific cyclin/cdk molecules
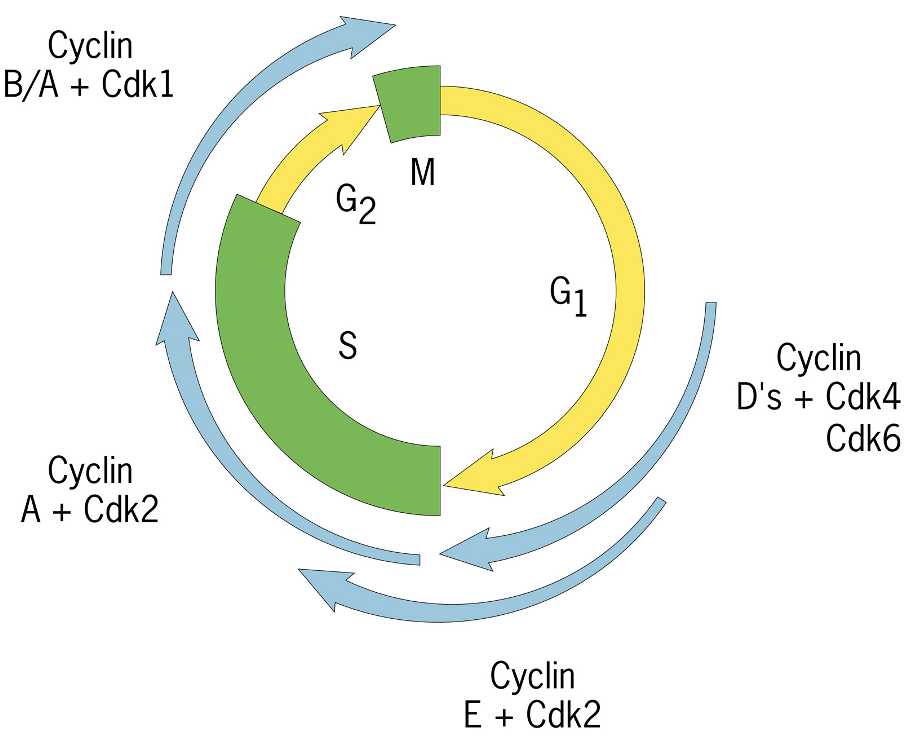
G1: Cyclin D with CDK4/CDK6 - expressed late in G1
G1/S: Cyclin E with CDK2
swaps cyclin partners
S: Cyclin A with CDK2
M: Cyclin B/A with CDK1
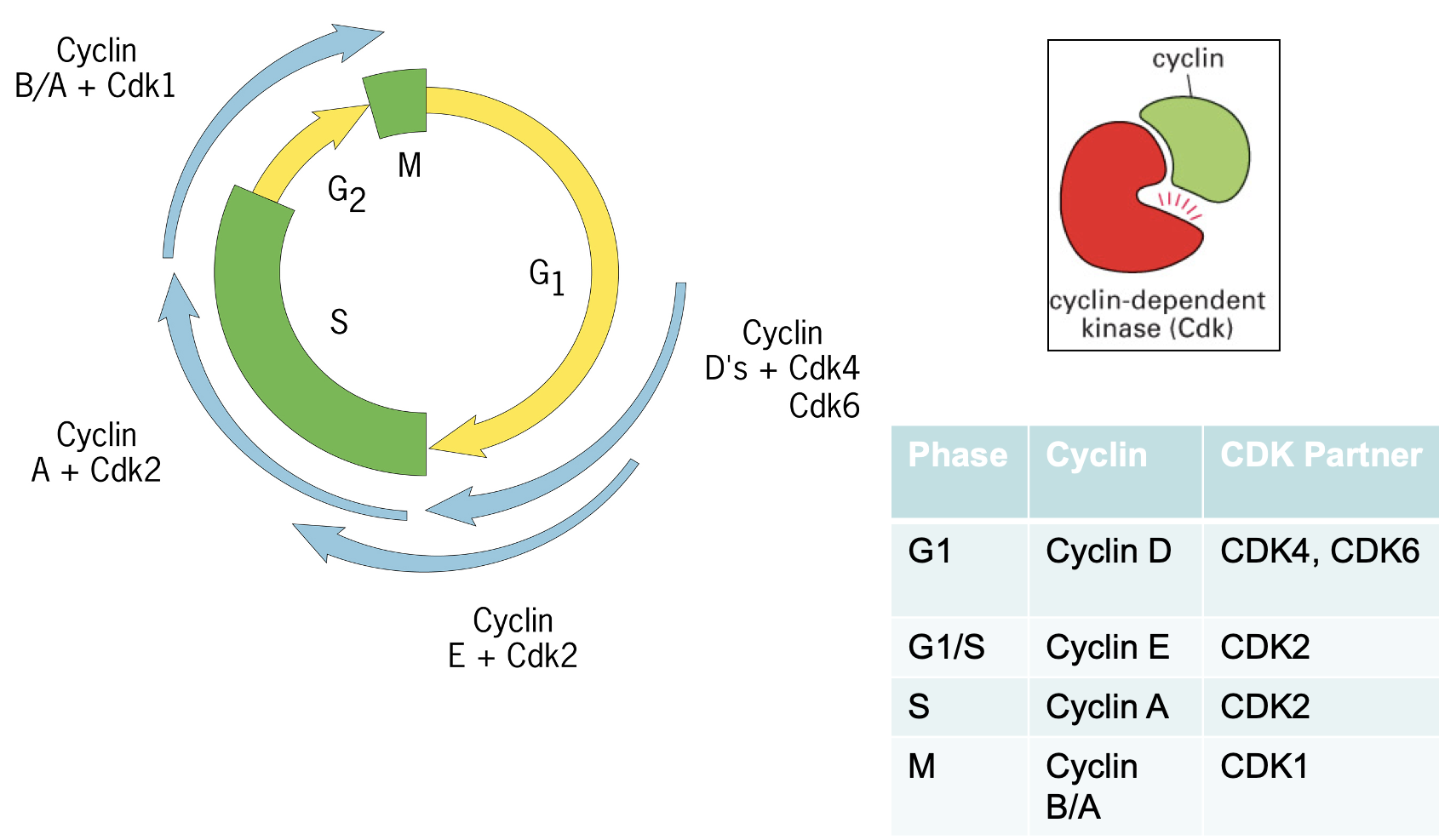
Regulation of Cyclin proteins
Cyclin levels can be regulated to determine if CDKs are active or not
several ways to regulate:
Cyclin protein levels
Phosphorylation of cdks themselves
Inhibitory molecules
Cyclin protein accumulation
Regulates Cyclins
Affects of cyclin mRNA transcription
cyclin mRNA translation
Other steps
CDKs are always present but cyclins switch
Cyclin D1 transcription
Linked to many growth factor signaling pathways
c-JUN, c-FOS, c-MYC
closely connected to signaling pathways
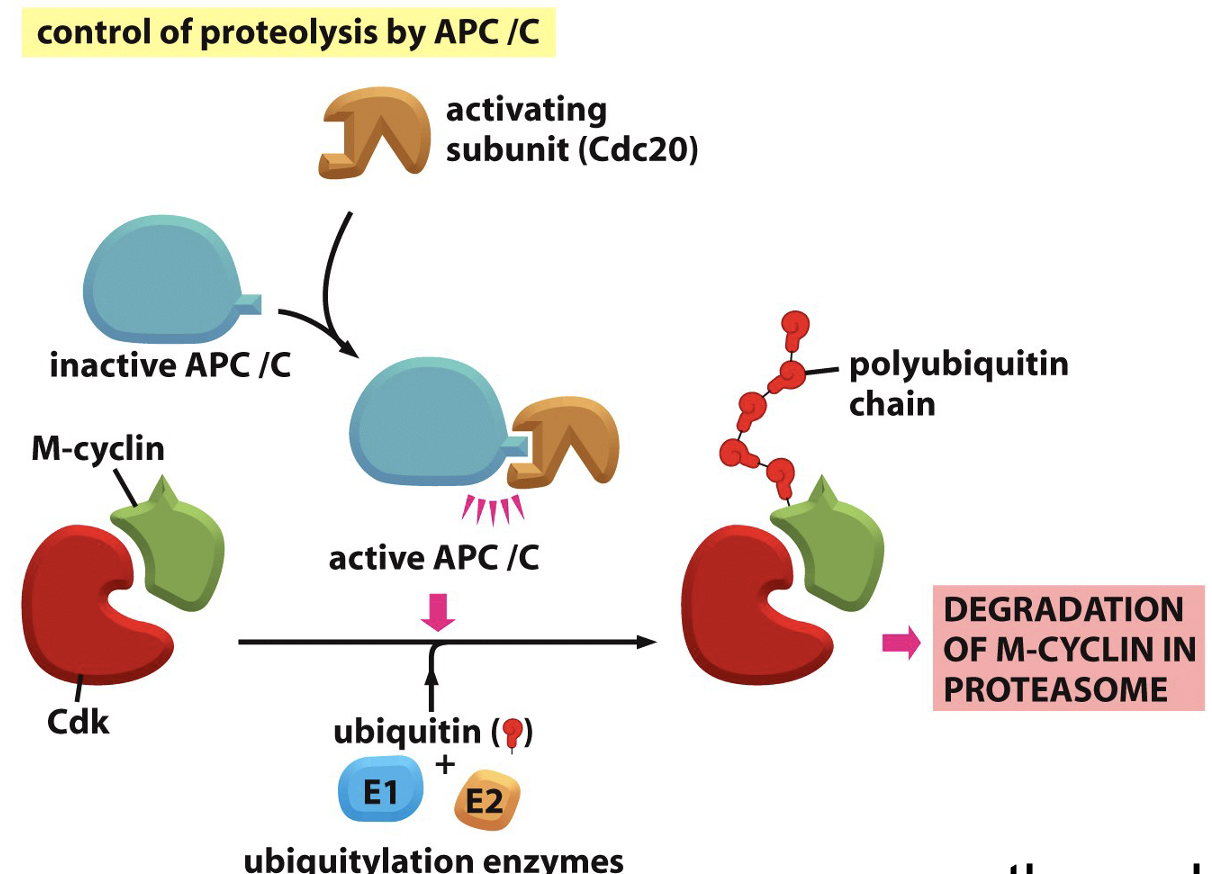
Cyclin protein degradation
Protein stability is regulated, multiple protein structures
Cyclin is ubiquityated by APC/C
Forms polyubiquitin chain on cyclin
spend energy to get rid of cyclin
gets degrated by proteasome
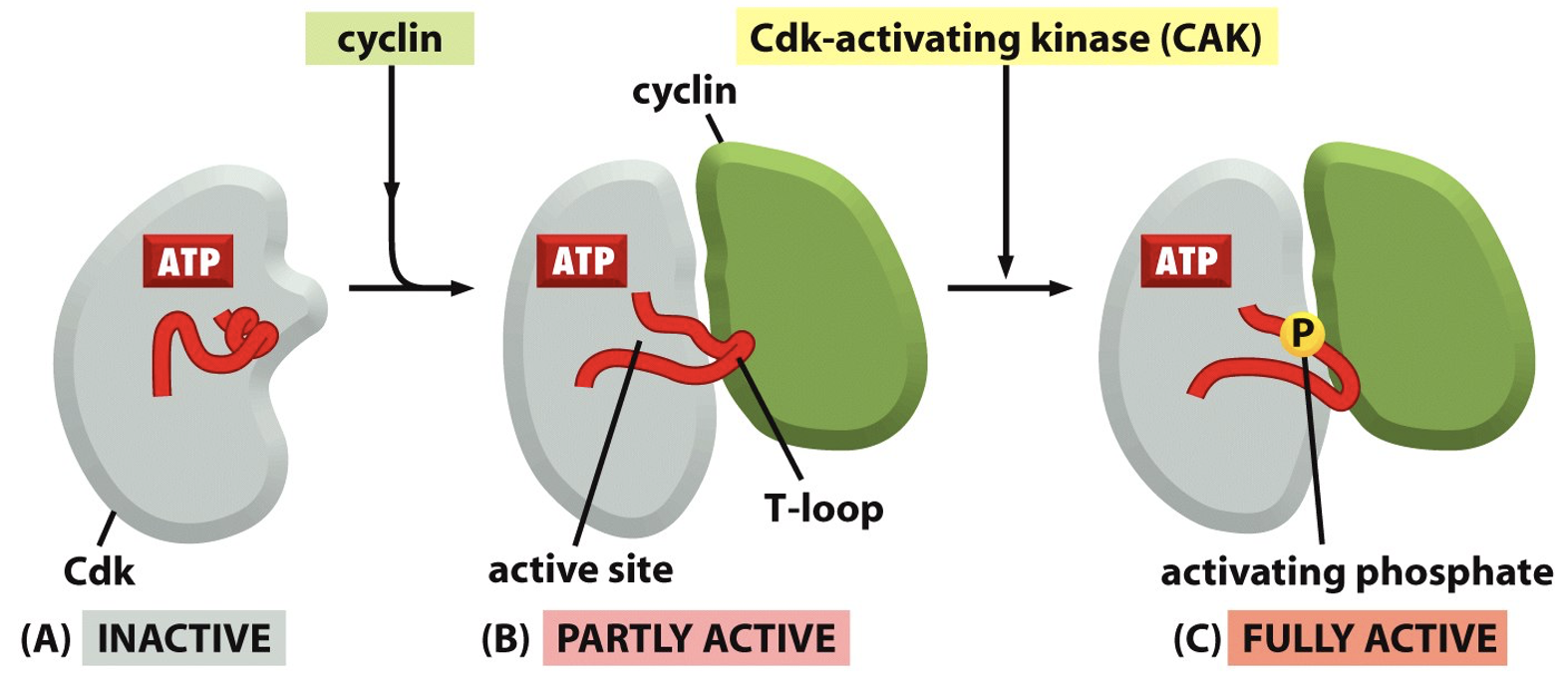
Cdk phosphorylation
Cdk is an enzyme can itself be post-translationally modified by phosphorylation
there are 2 types of phosphorylation events inhibitory [Tyr 15] and stimulatory [Thr 161]
de-phosphorylation of tyrosine 15 is able to phosphorylation important components for M phase
Inhibitory phosphorylation “wins” - active kinase must have stimulatory phosphorylation and NOT have inhibitory phosphorylation
Results in conformational change
![<p>Cdk is an enzyme can itself be post-translationally modified by phosphorylation</p><ul><li><p>there are 2 types of phosphorylation events inhibitory [Tyr 15] and stimulatory [Thr 161]</p></li><li><p>de-phosphorylation of tyrosine 15 is able to phosphorylation important components for M phase</p><ul><li><p>Inhibitory phosphorylation “wins” - active kinase must have stimulatory phosphorylation and NOT have inhibitory phosphorylation</p></li><li><p>Results in <strong>conformational change</strong></p></li></ul></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/3f0b32f6-a7f9-4e75-acba-edf4a185da80.png)
Multiple steps in enzyme activation
A) Inactive (no cyclin bond)
B) Cyclin binds, Partly active - forms T loop (no longer blocking active site)
C) T-loop is phosphorylates - fully active form
Confirmational changes in cdk not cyclin
Inhibitory Molecules that Blocks Cdks
These proteins interact with cdk molecules and inhibit their activity
Ex. p21, p27, p16 (CDKN1A, 1B…)
Currently there are 16 known inhibitors
Each cdk has its own group of inhibitory proteins
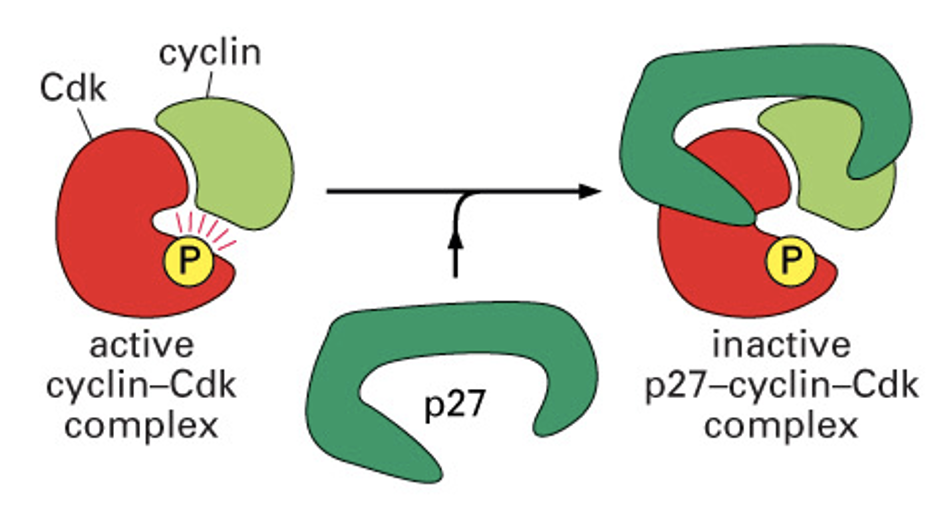
Inhibitors bind CDk/cyclin complex
Alter shape of pocket as a result
ex: p27 Kip1 - cdk inhibitor forms c shape across proteins - blocks substrate
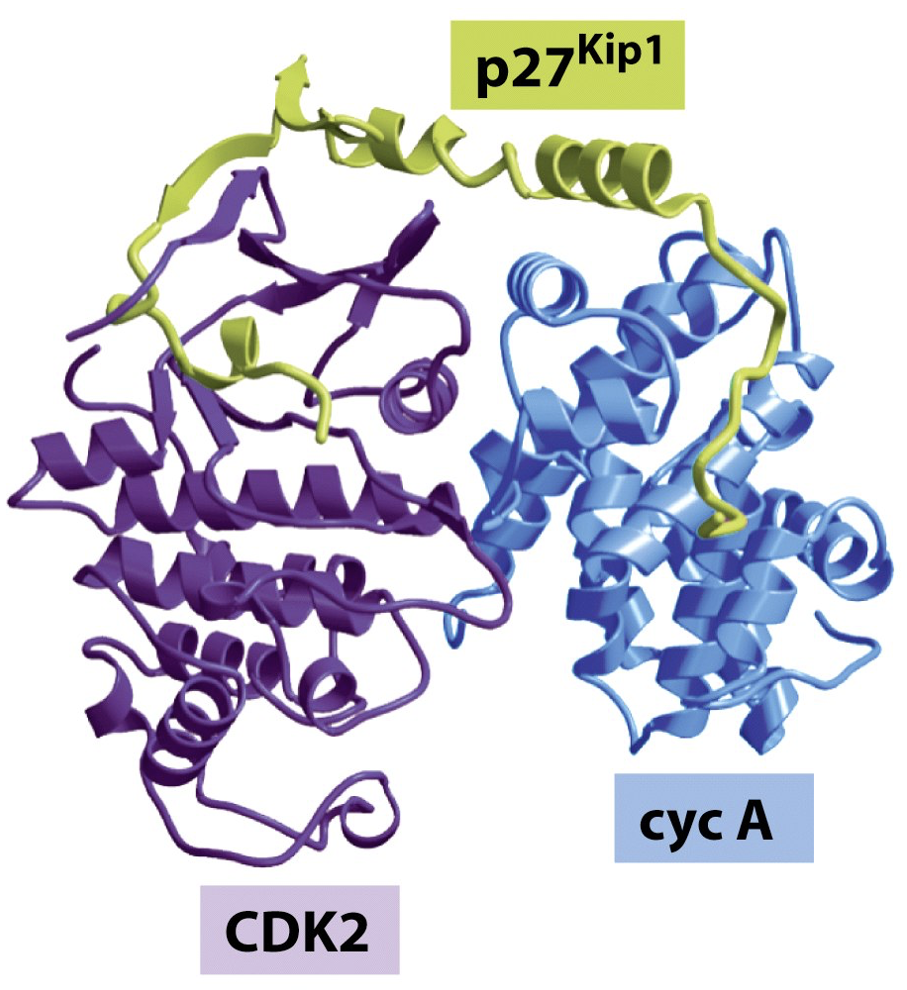
Inhibitors have structure based specificity
p16 INK4A binds specifically to CDK6
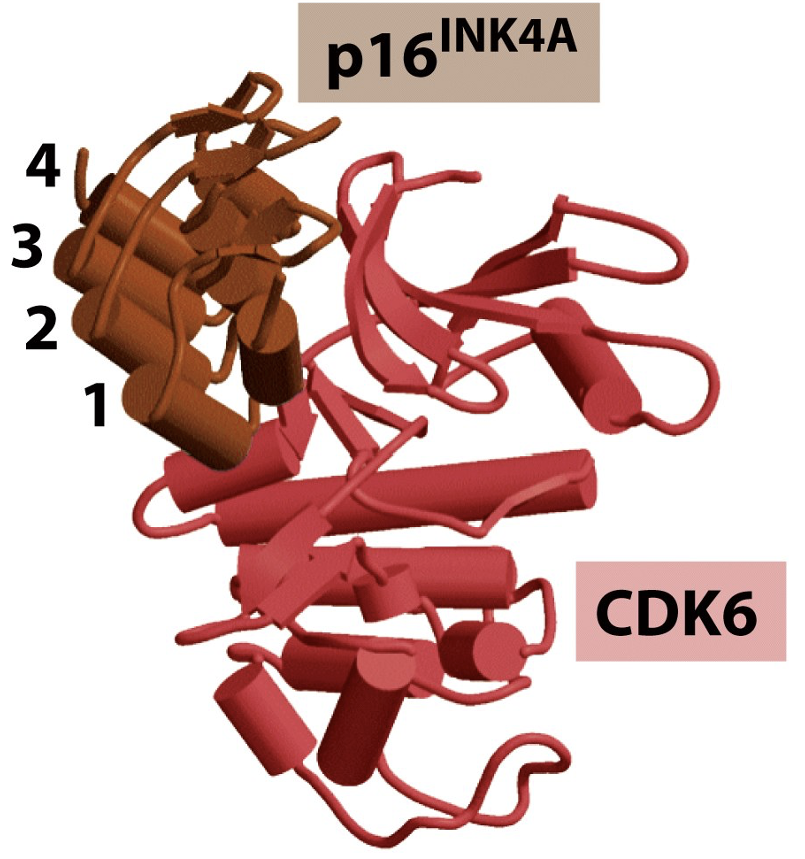
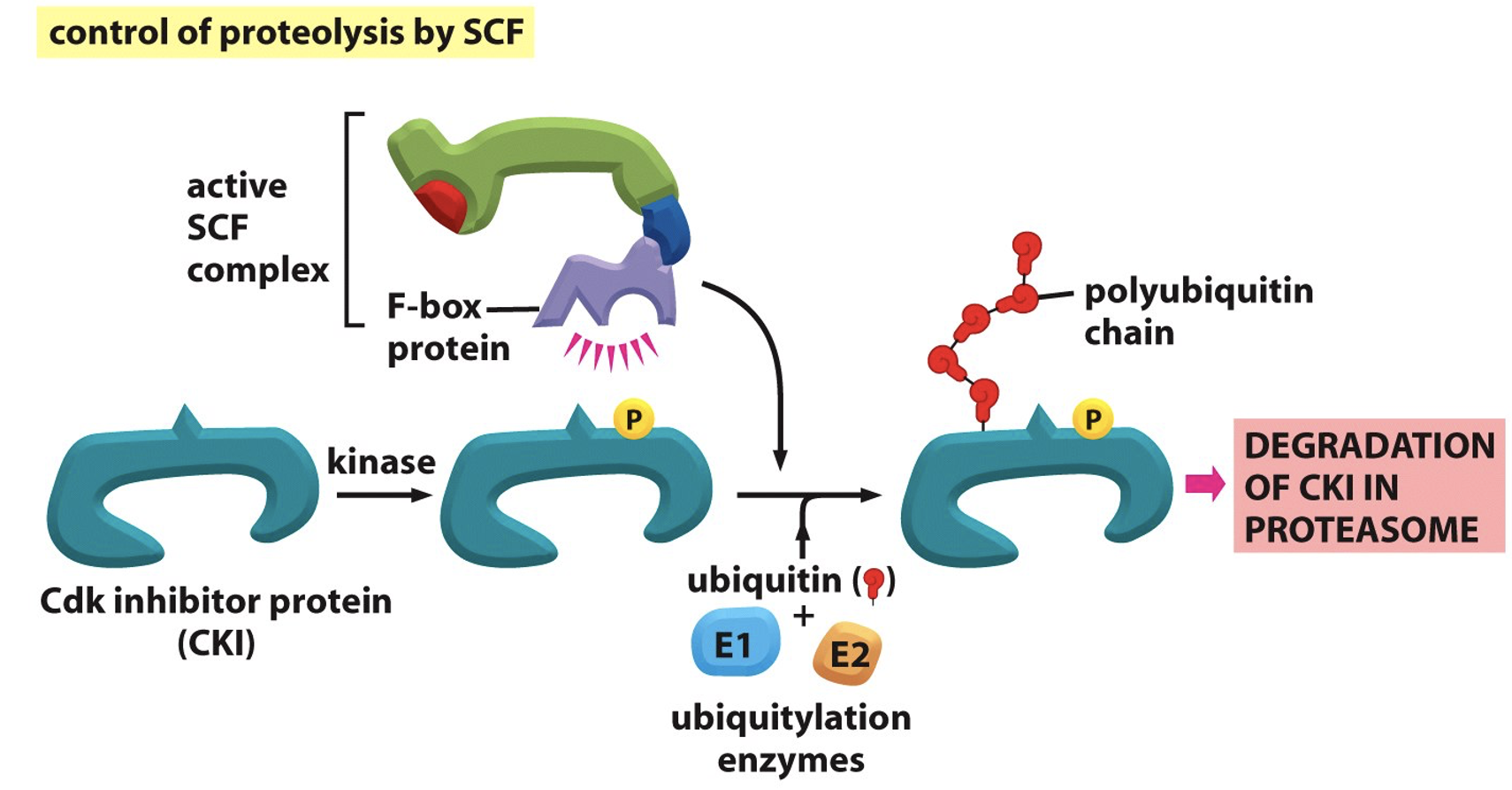
Inhibitors are tightly regulated
Active SCF complex can’t bind Cdk inhibitor protein for degradation unless there is a specific phosphorylation event
Focus on the G1 to S transition
Early in G1 - CycD/cdk4 becomes activated:
phosphorylate substrates
substrate phosphorylation permits cell cycle progression
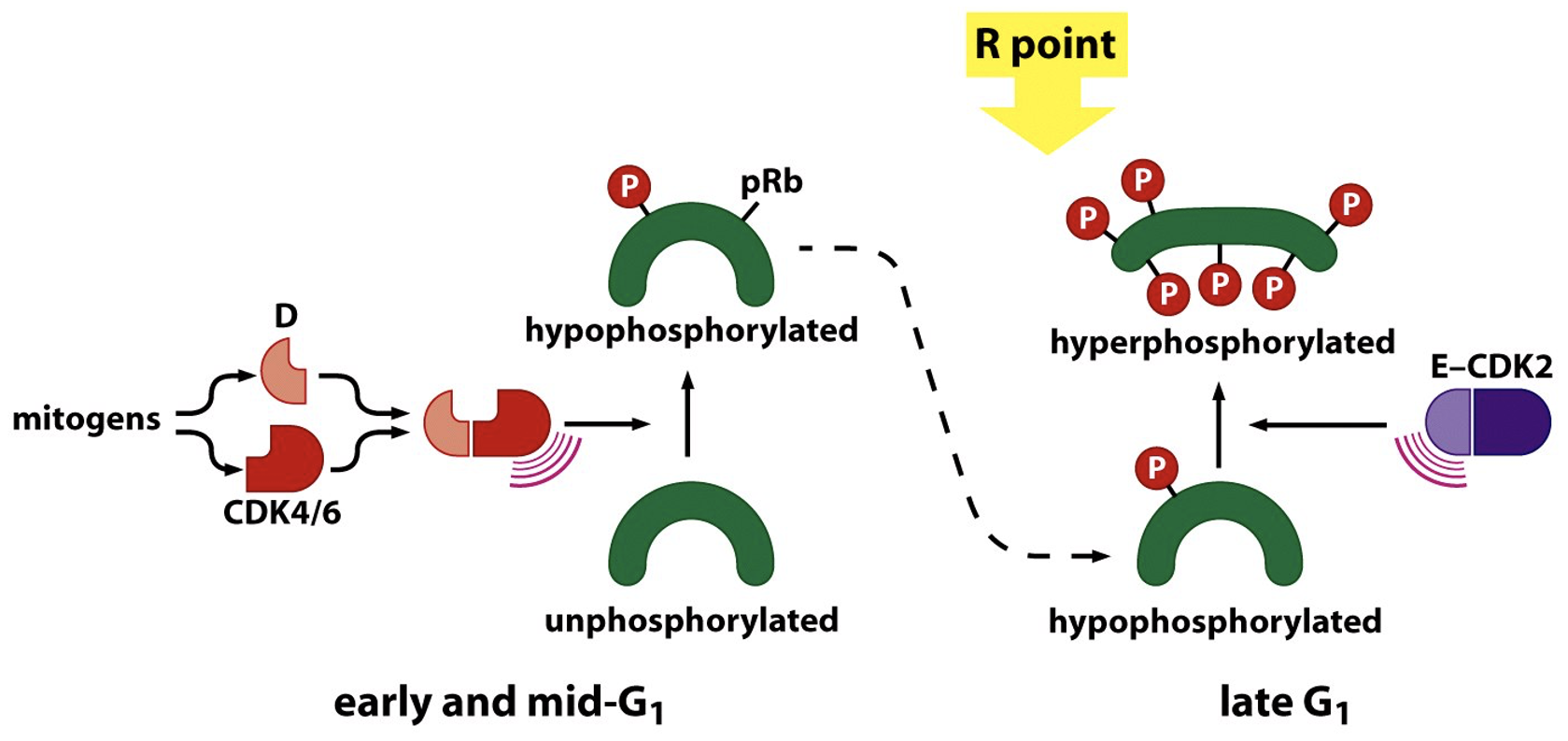
One of the Substrates is Rb protein
pRb inhibit E2F - activation leads to pRb phosphorylation and dissociation from E2F
outcome is turning on sets of genes that allow cell cycle progression
Cyclin E
DNA polymerase
thymidine kinase
E2F drives transcription of genes in S phase
nucleotide biosynthesis
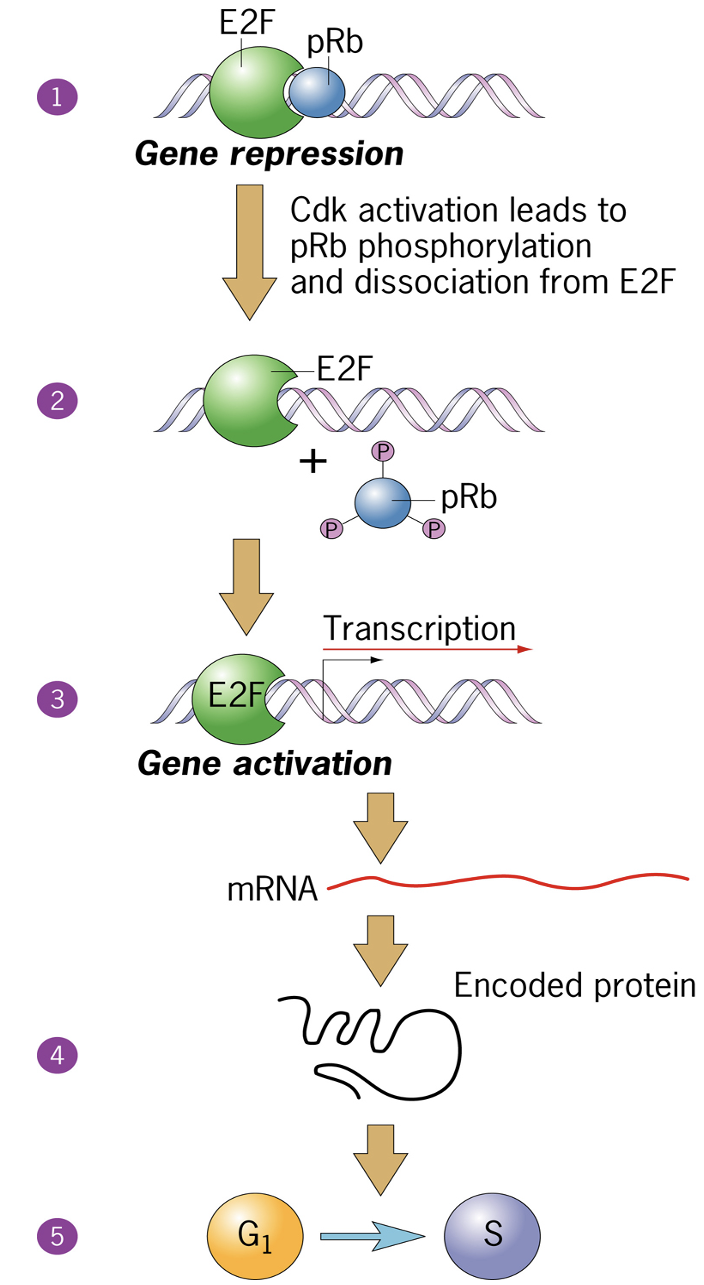
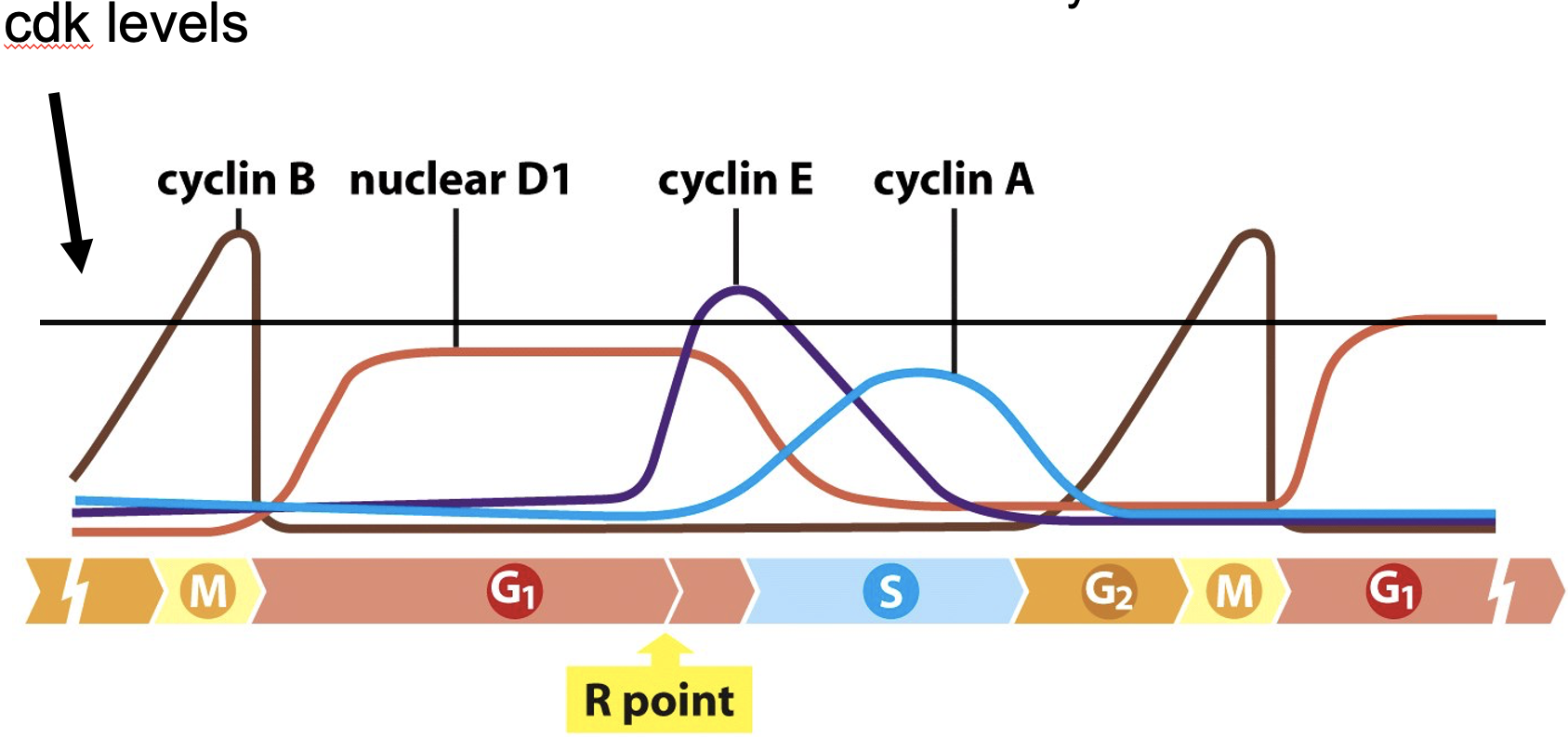
Regulation of G1/S phase transition by the RB protein
When hyper-phosphorylated, Rb is inactivated
at R point
multiple sites of phosphorylation
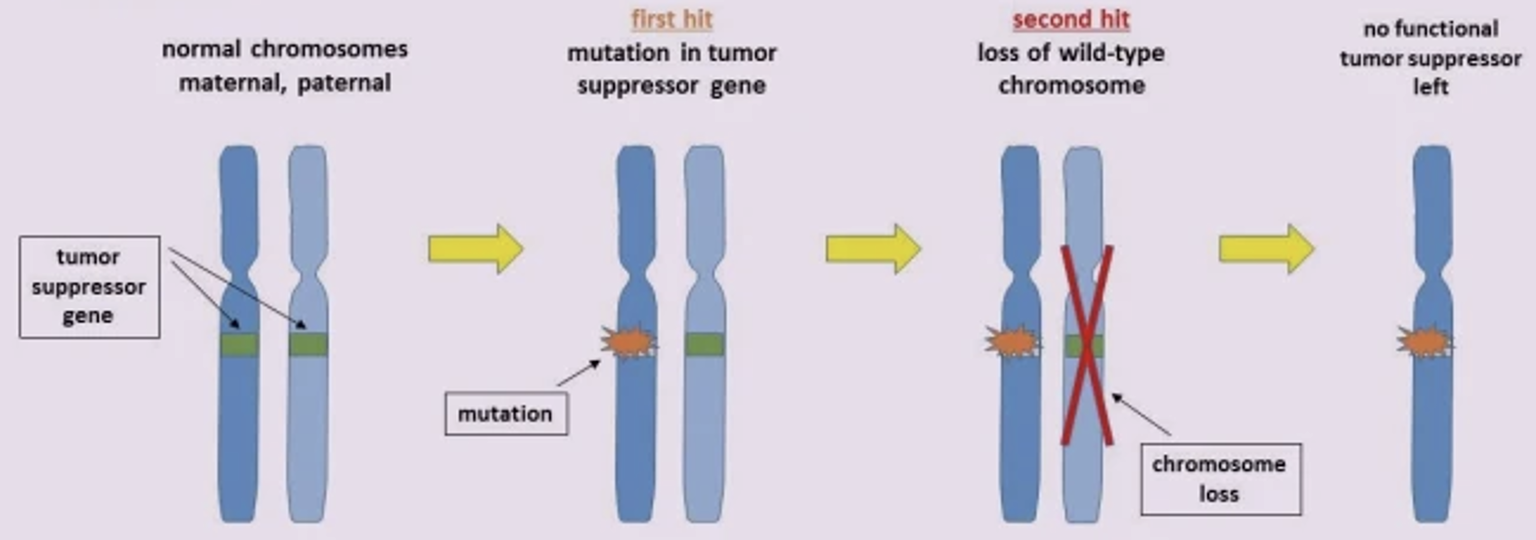
Alfred Knudson (1971) the two-hit hypothesis
Of tumor suppressors
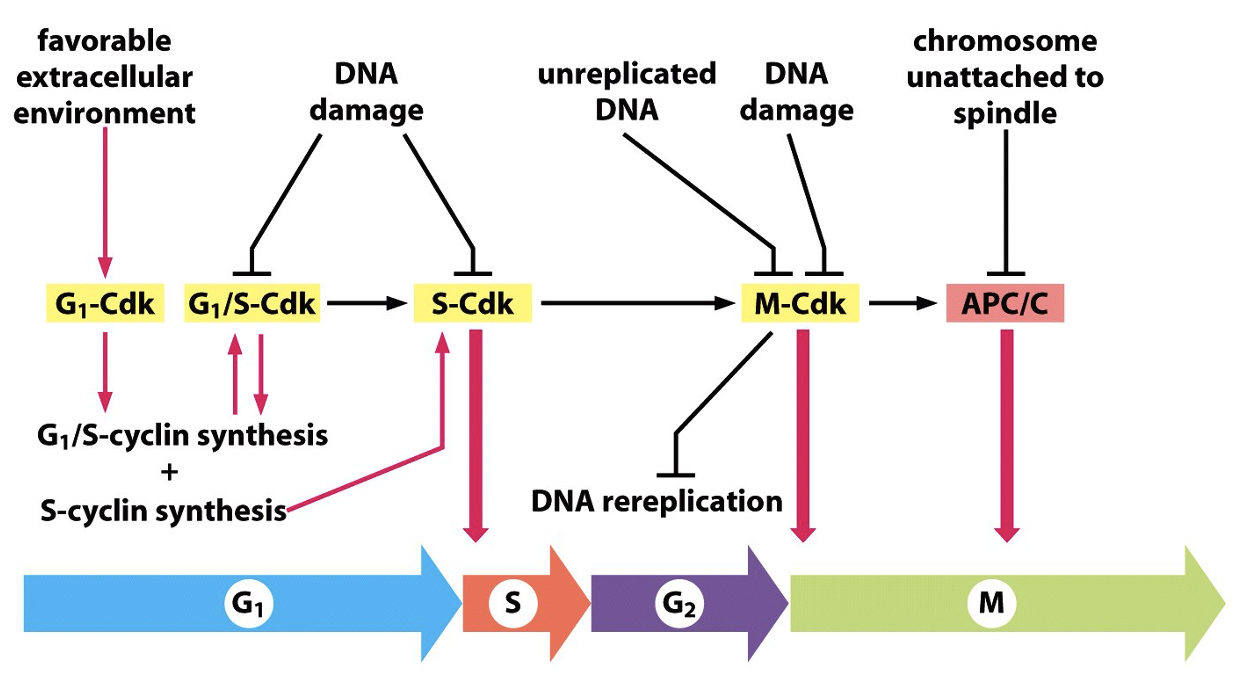
3 Major regulatory transitions (checkpoints)
1) Start (G1 to S)
2) G2 to M
3) Metaphase Anaphase
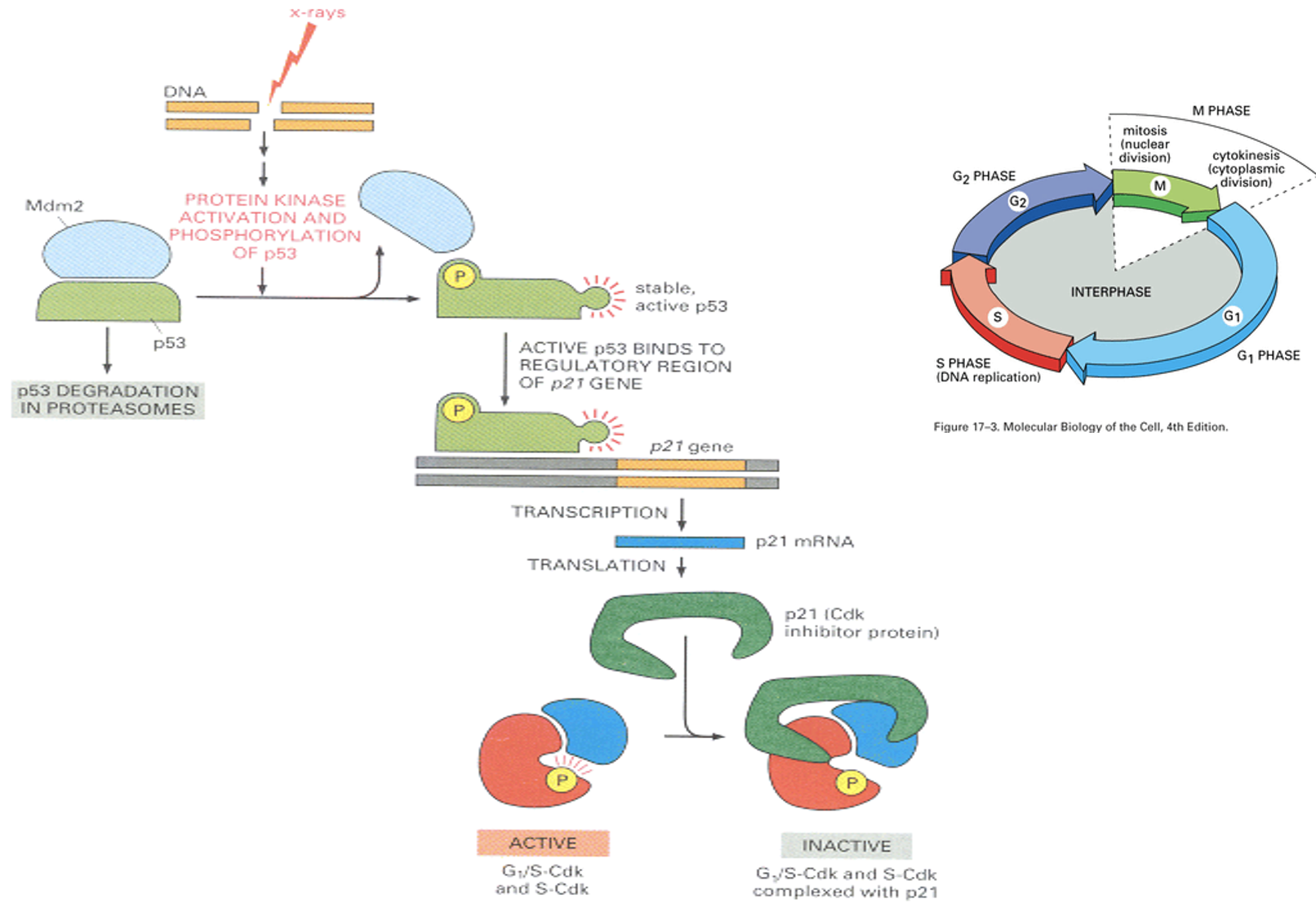
p53-dependent checkpoint after DNA damage
Transcription factor, drivers cyclin inhibitor transcription, and complexes and inhibits CDK-cyclin complex
Cdk/cyclin complexes regulate RNA Pol II-based transcription
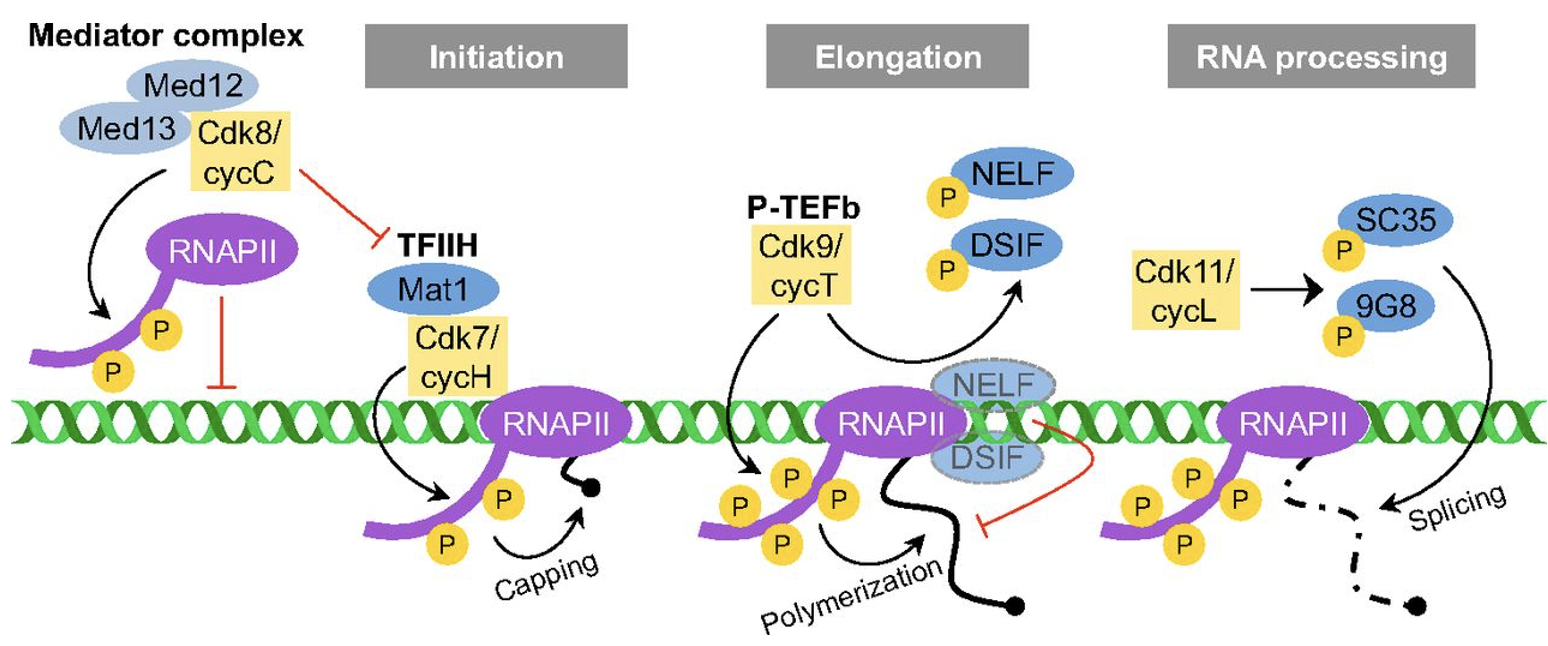
Cell cycle regulators influence DNA damage repair
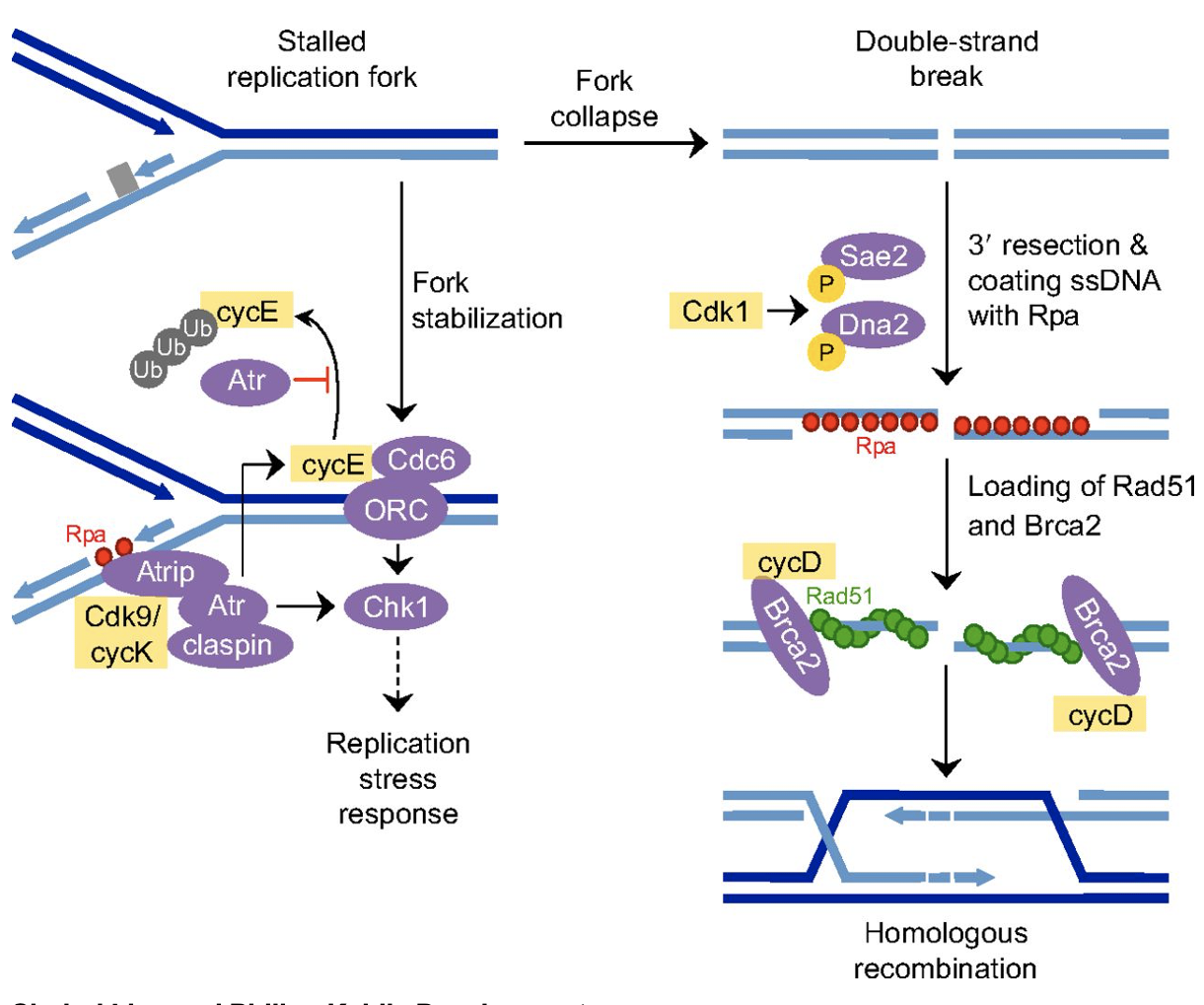
Cell cycle regulators and epigenetic regulation
modify histones
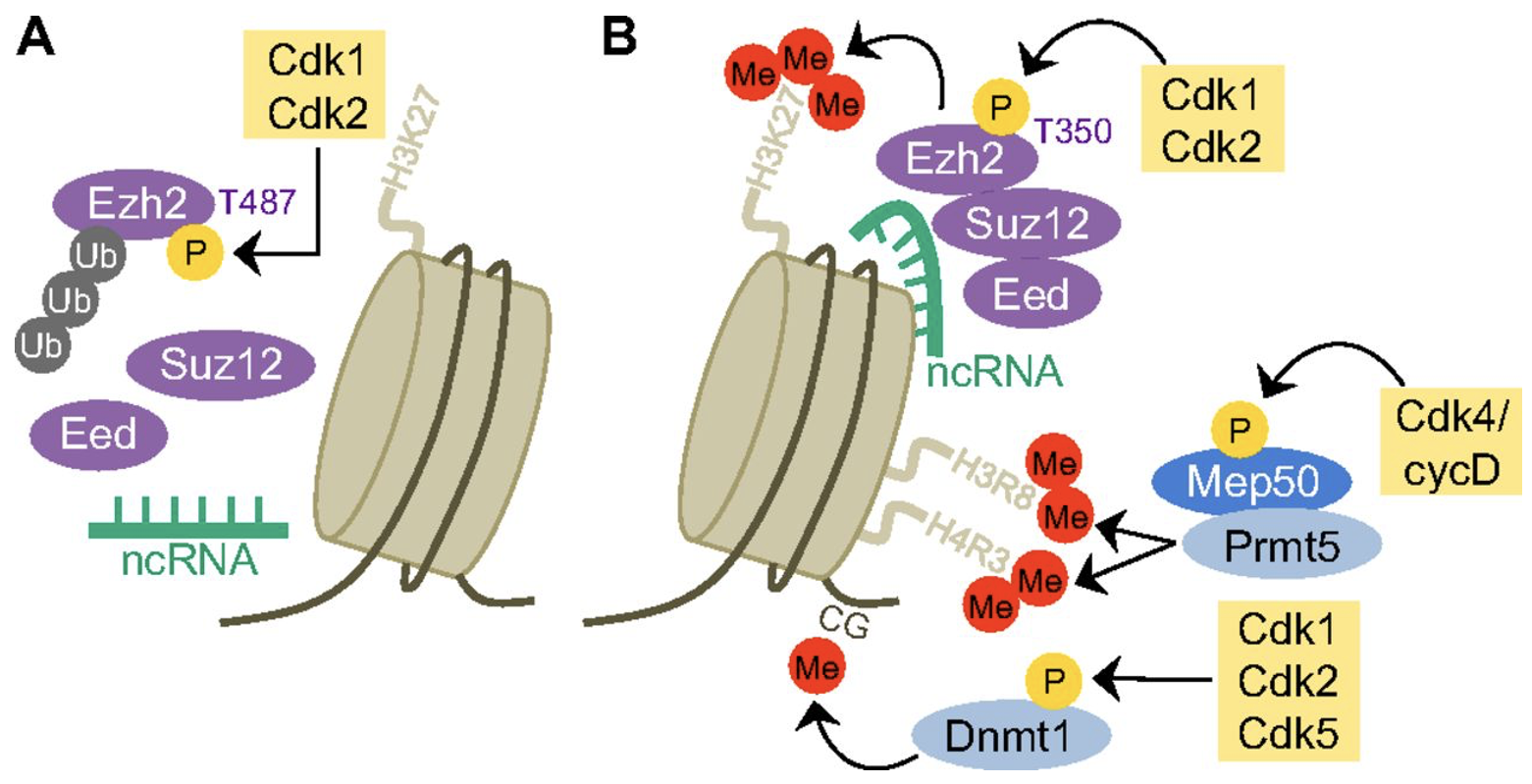
Cell cycle regulators controlling stem cell self-renewal
stem cell function
Why choose a mouse as a model organism?
Similar to humans, have placenta, small, short generation time
has human genetic and physiologic similarities
are well documented, established tools, deep literature records
Genetic Modifications
Progression of available tools: Find a mutant or make a mutant
Either look for a visible different gene
Or use radiation, chemical, or transposon insertion to damage DNA and result in repair → cause changes in DNA sequence, required for a lot of screening
Genetic Modifications - Target a mutant
Knock-out: use homologous recombination to introduce a disruptive sequence into a locus - target specific genes - make a null allele
Knock-in: insert foreign DNA into a locus of choice
distinct from transgenesis where the insertion point is random, and does not disrupt a targeted locus
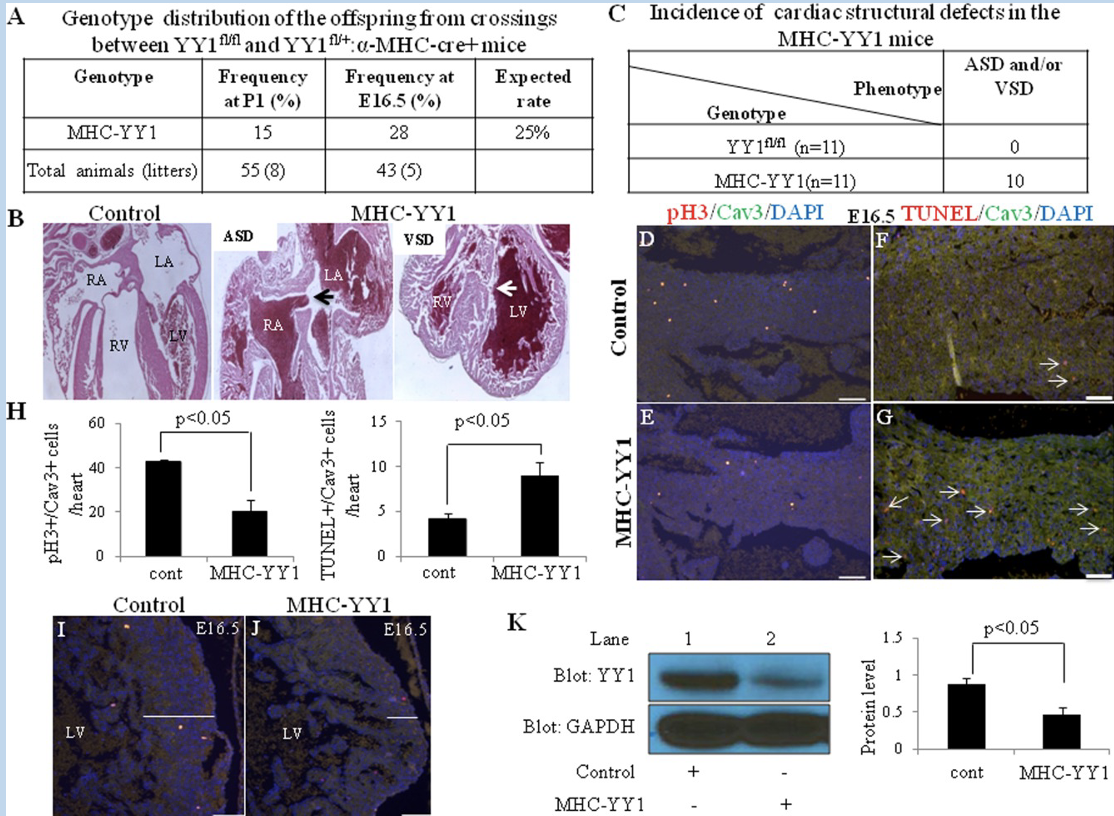
Conditional Mutagenesis: selective knockout in a tissue, cell population, and/or developmental stage of choice
compare tissue-specific vs whole organism - YYI
Whole organism mutagenesis
YYI (and other gene) knock-out mice die near implantation
YYI is an essential gene
mice with mutant alleles where pari implantation lethal
Conditional mutagenesis
Cell population and or developmental stage restriction allow other tissues to function
tissues to function
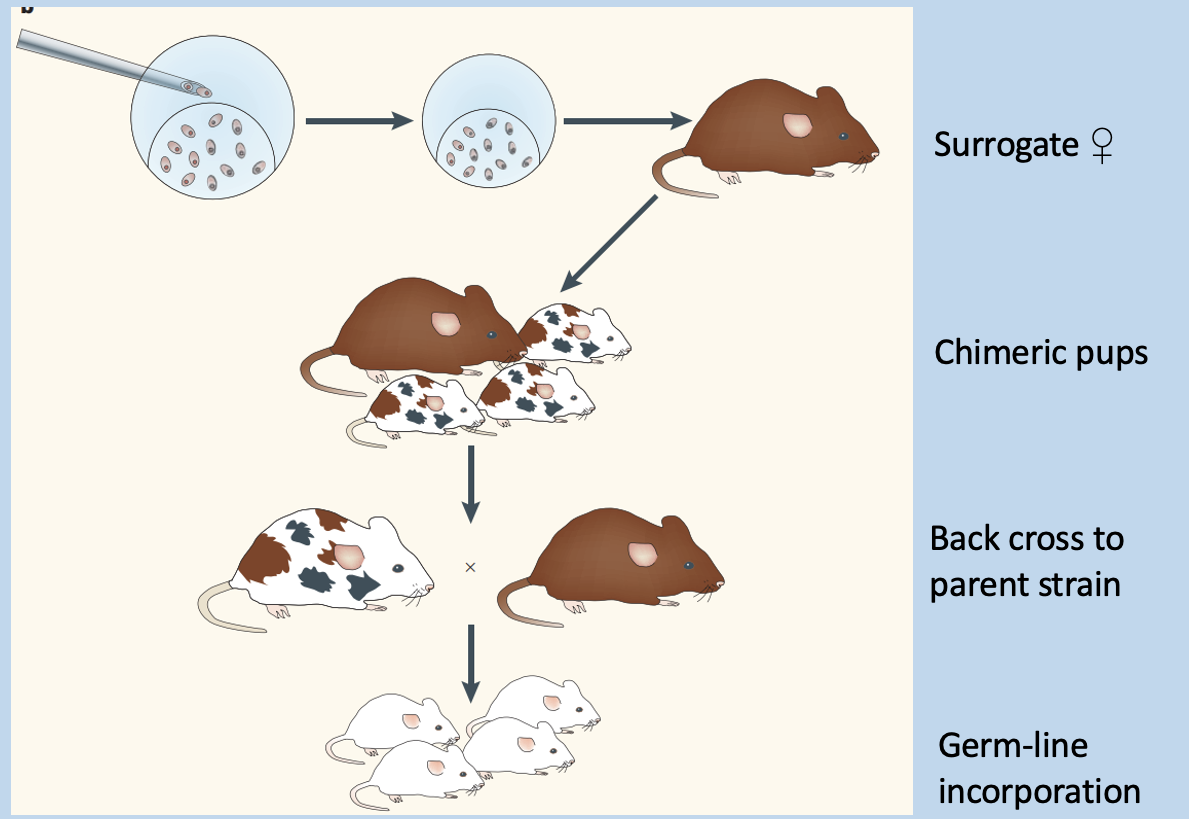
Embryonic Stem (ES) manipulation
Requires drug selection, chimeric embryos, and back crossing
embryos develop in uetero
form chimeric mice
cross with wildtype to obtain stain with expression in all cells
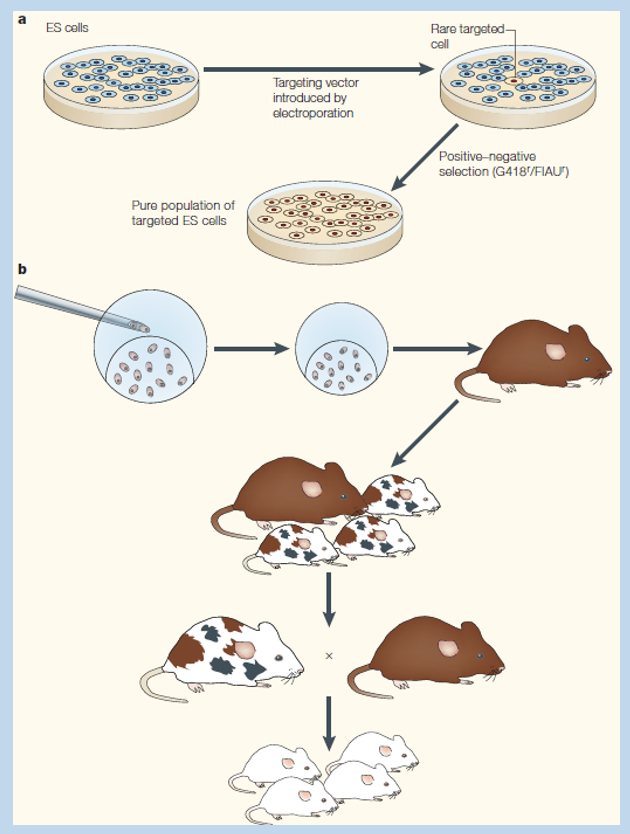
ES cells can be manipulated in vitro
Selected through drug resistance
explain ES manipulation and husbandry to produce targeted KP mice
explain role of G418 an FIAU selection to select for correctly targeted mutation
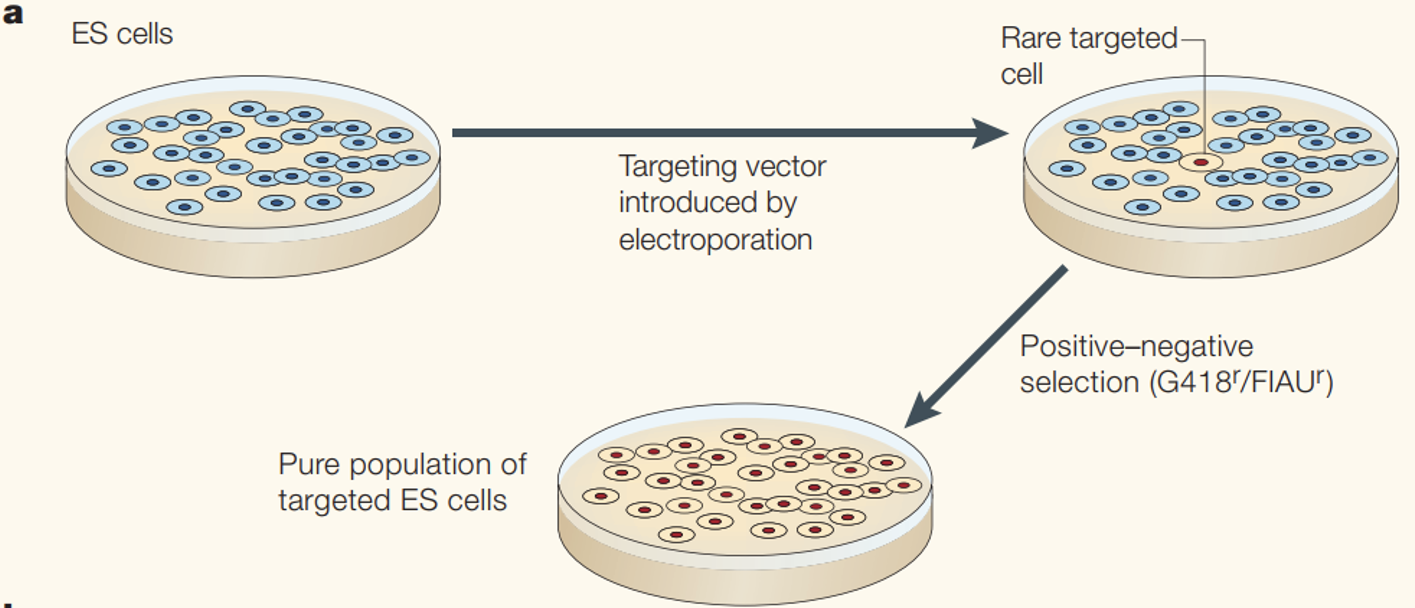
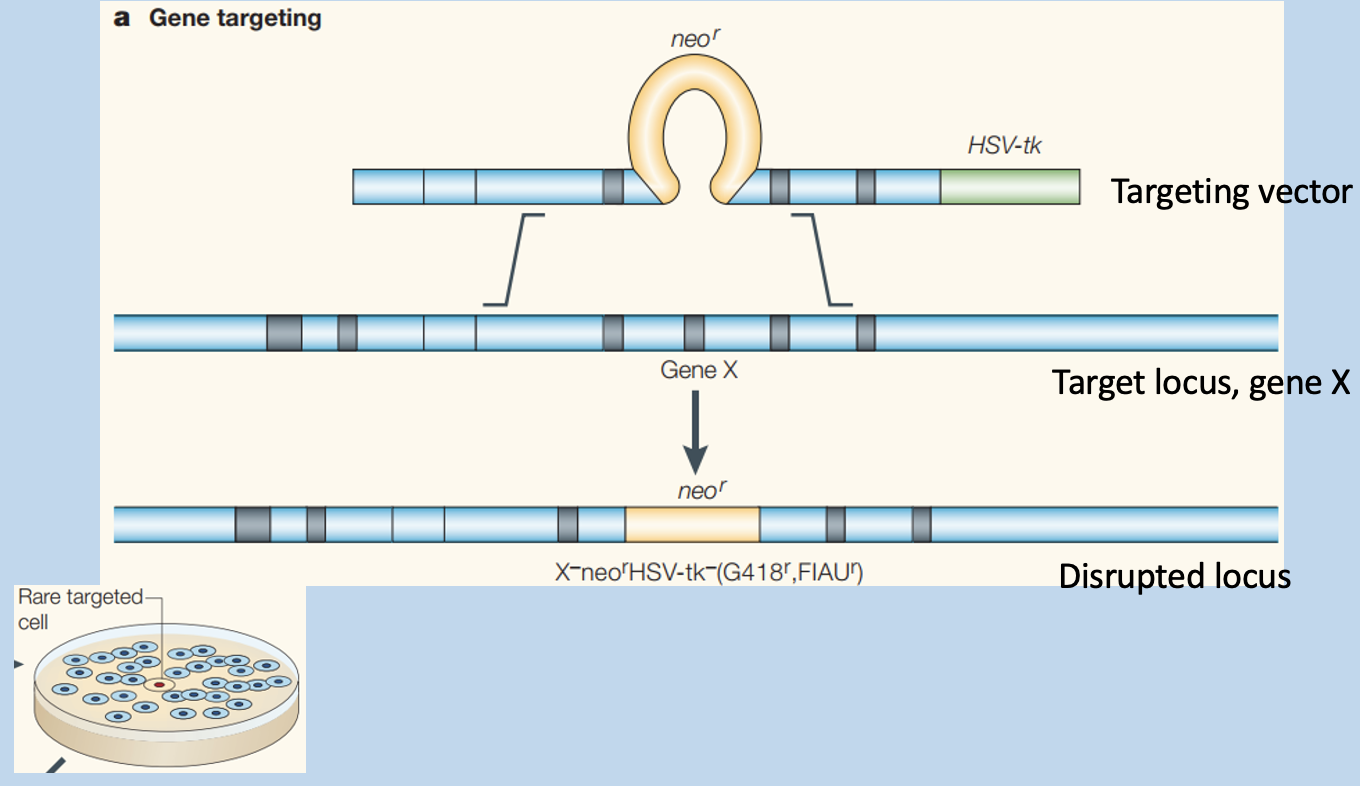
The gene targeting vector has 2 selection markers and homology to the target locus
Results in outside genes getting lost
Targeting vector is inserted into target locus, disrupting the locus
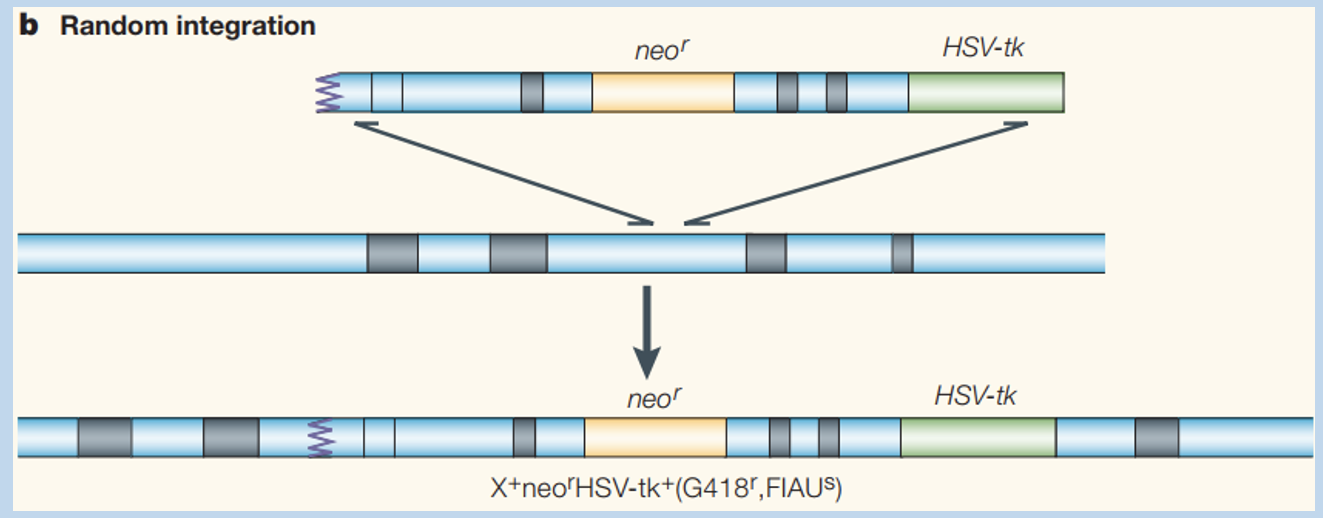
Gene targeting vector has 2 selection markers and homology to target locus
Randomly integrated
Compare the incorporation of cassettes and predict consequences of selection
low efficiency
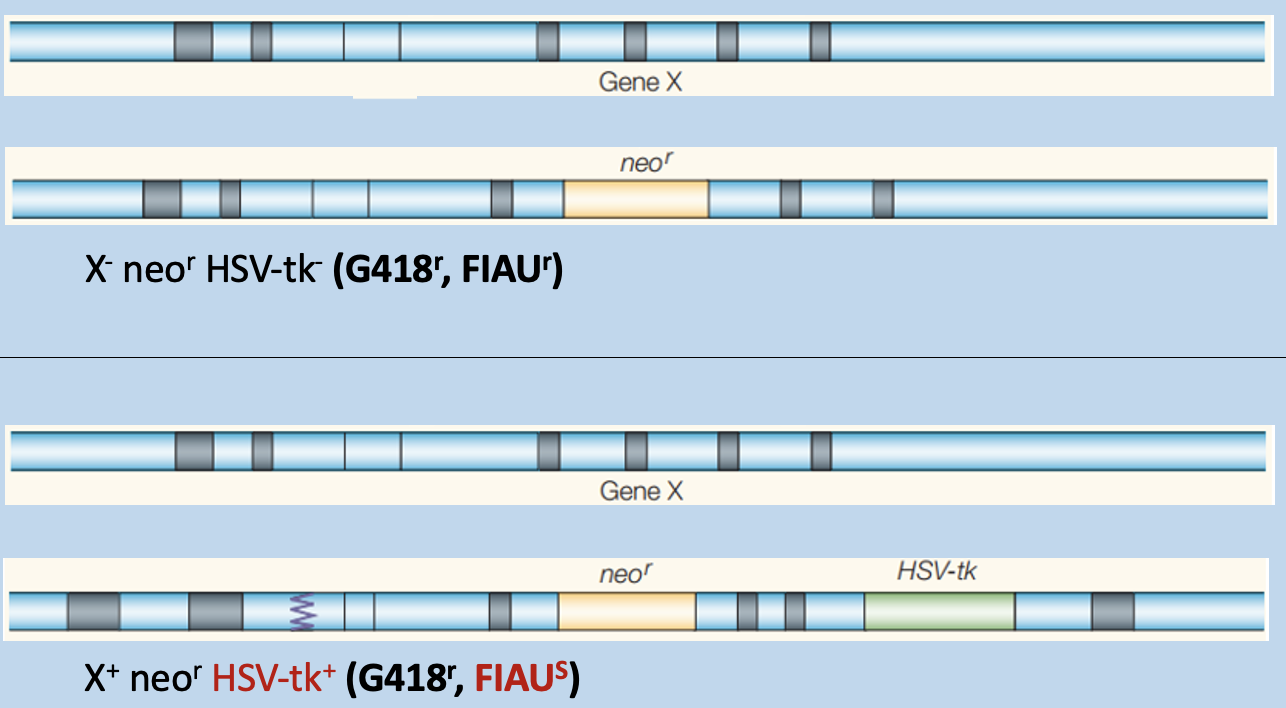
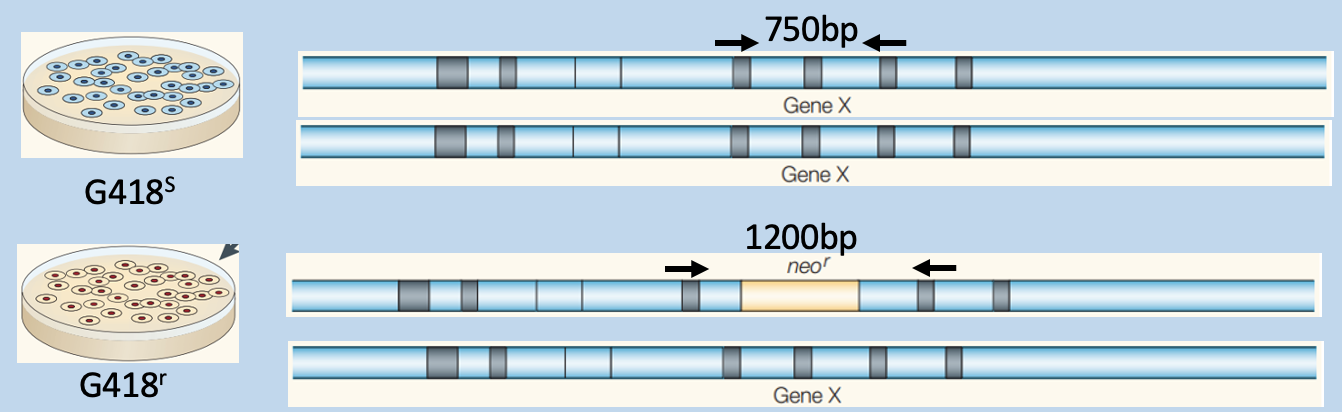
Predict outcome of genotyping
G418s has only 750bp
G418r has 1200bp (recombinant locus) - modified population
has a mix of to fragments
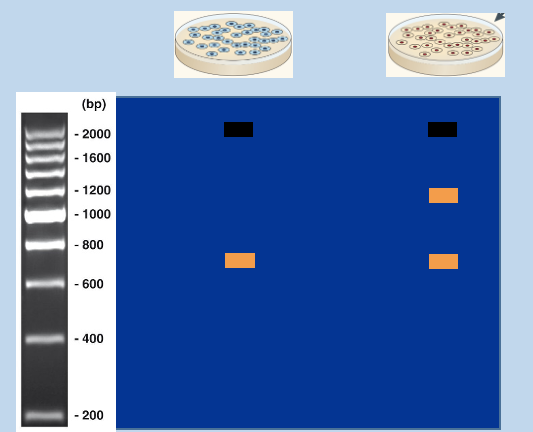
Embryos bearing modified ES cells are cultivated in vivo
Pups requires back-crossing - genotyping mice
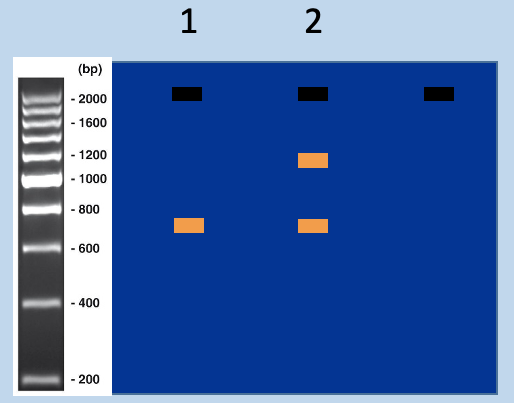
Match genotyping PCR outcome to mouse strains - 2
has amplification at 1200bp and 750bp
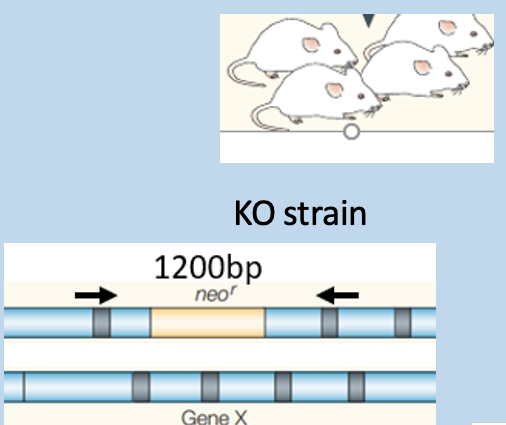
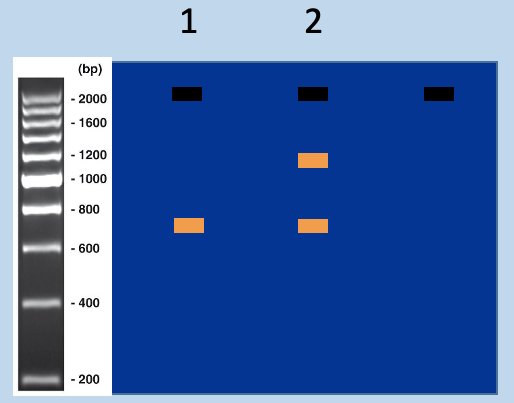
Match genotyping PCR outcome to mouse strains - 1
has amplification at 750bp
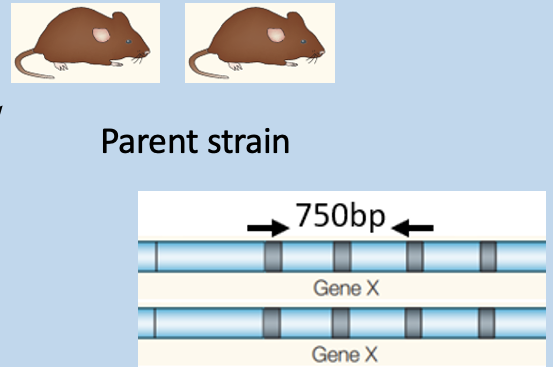
Determine the genotypes of these targeted mice
Using genotyping PCR to distinguish targeted and untargeted mice
assign genotypes from sizes of PCR products
Wildtype (+), recombinant (-)
heterozygous (has 2 bands)
WT homozygous' (has one band in between 800/600bp)
recombinant (has on band at 1300bp)
Recombinant
How does targeting vector disrupt a genome locus?
homology arms flank neo gene can be separately recombined with the genome locus through homologous recombination (HR, crossing over in meiosis)
leads to neo incorporation of neo into genomic locus
excised region is chosen to remove critical structural aspects of genomic locus gene product
Why does HR lead to disruption of the target locus, but integration to other genomic sites occurs through distinct mechanisms?
HR requires a significant amount of homology between the exchanging DNA molecules
other genomic loci are not predicted to have significant homology with the vector
integration of the targeting vector at these sits cannot use the HR mechanism
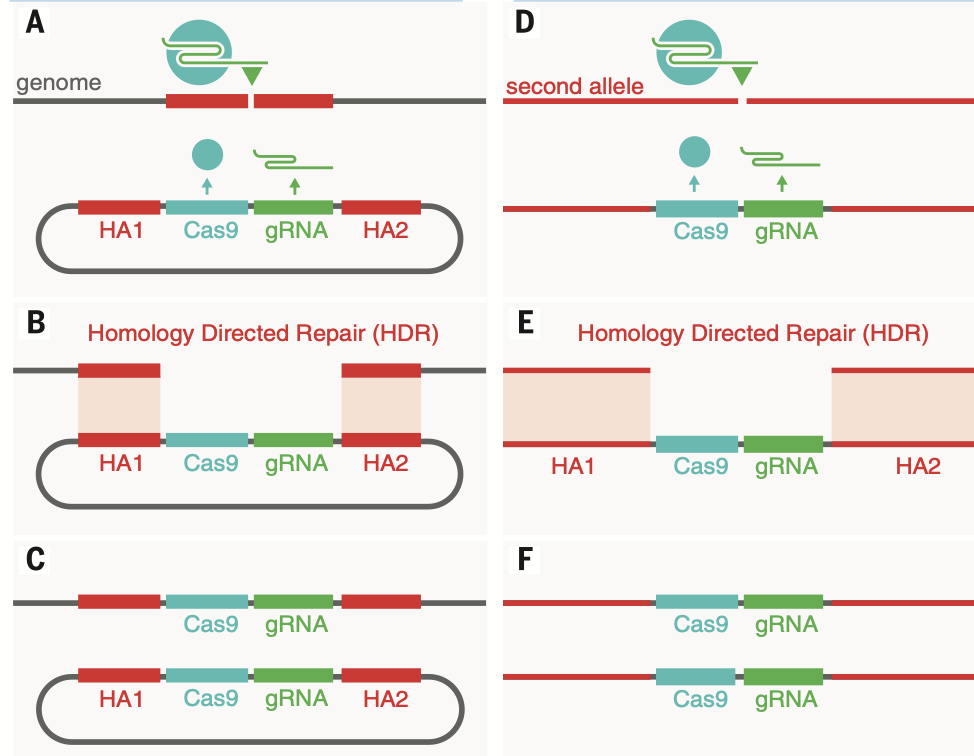
Mutagenesis using CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)
CRISPR uses guide RNA and PAM to recognize target sites - Cas9 performs enzymatic activity
guide RNA stitched together - directs where we want Cas9 to go
pairing of protospacer DNA with the guide RNA leads to activation of Cas9
PAM (protospacer adjacent motif) and complementary region lead to activation of Cas9 endonuclease activites
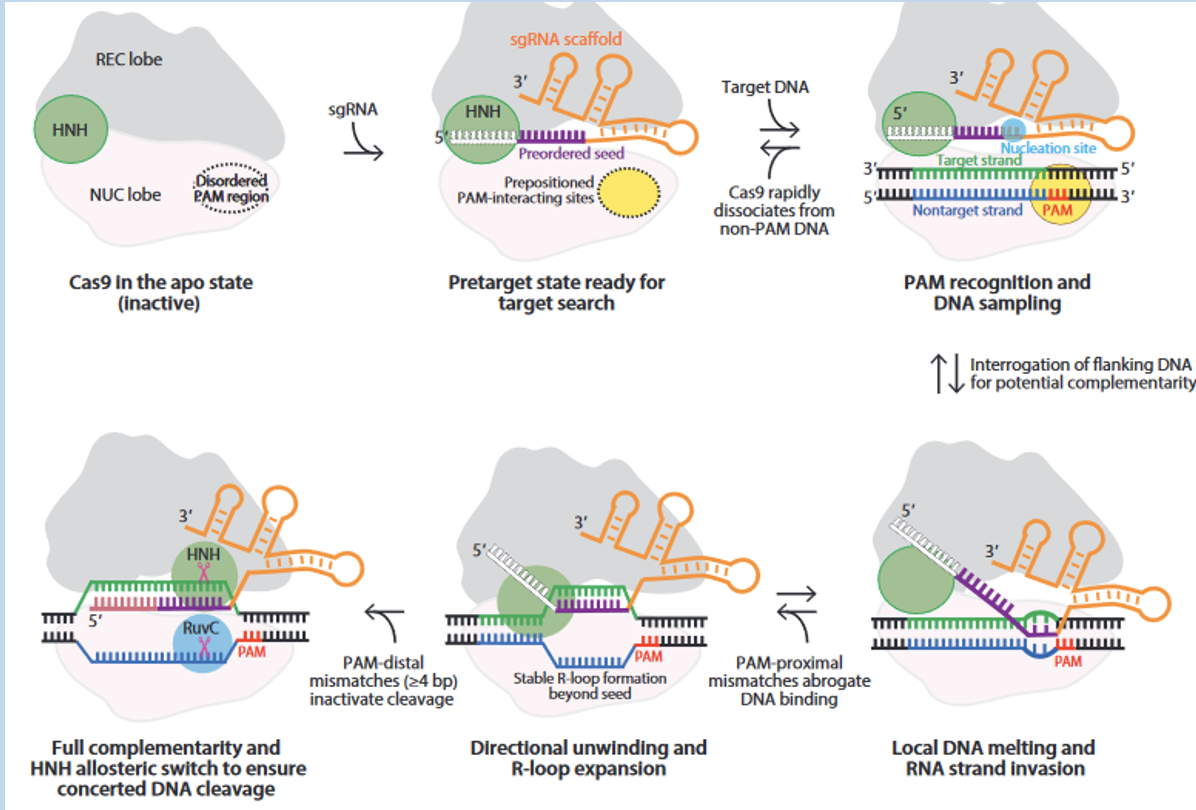
Repair Pathways introduce changes at the target genomic loci
Works for knock-in strategies, can go down two different pathways:
NHEJ: stitch together - results in mutagenesis
HDR: move access to repair template to stear towards it
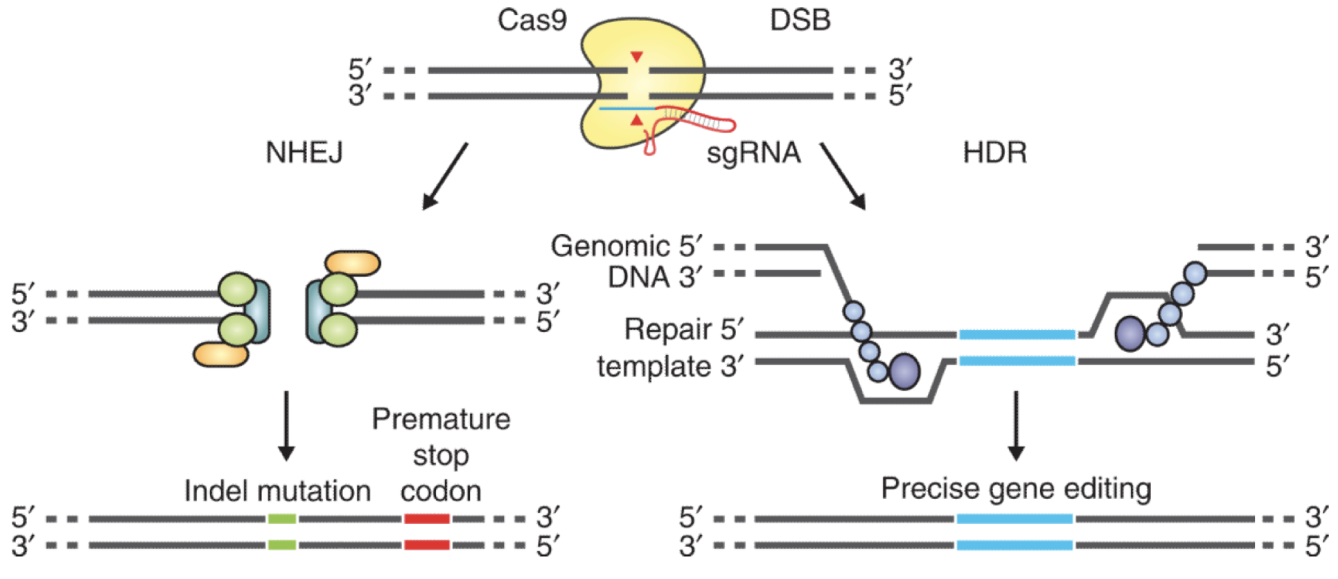
Homology-directed repair using a co-delivered template
Used to make precise alterations
introduces HM1 cut
get high modification of fibroblast
Off-target Effects are a Danger
CRISPR is great - except genome is full of a lot of repeats
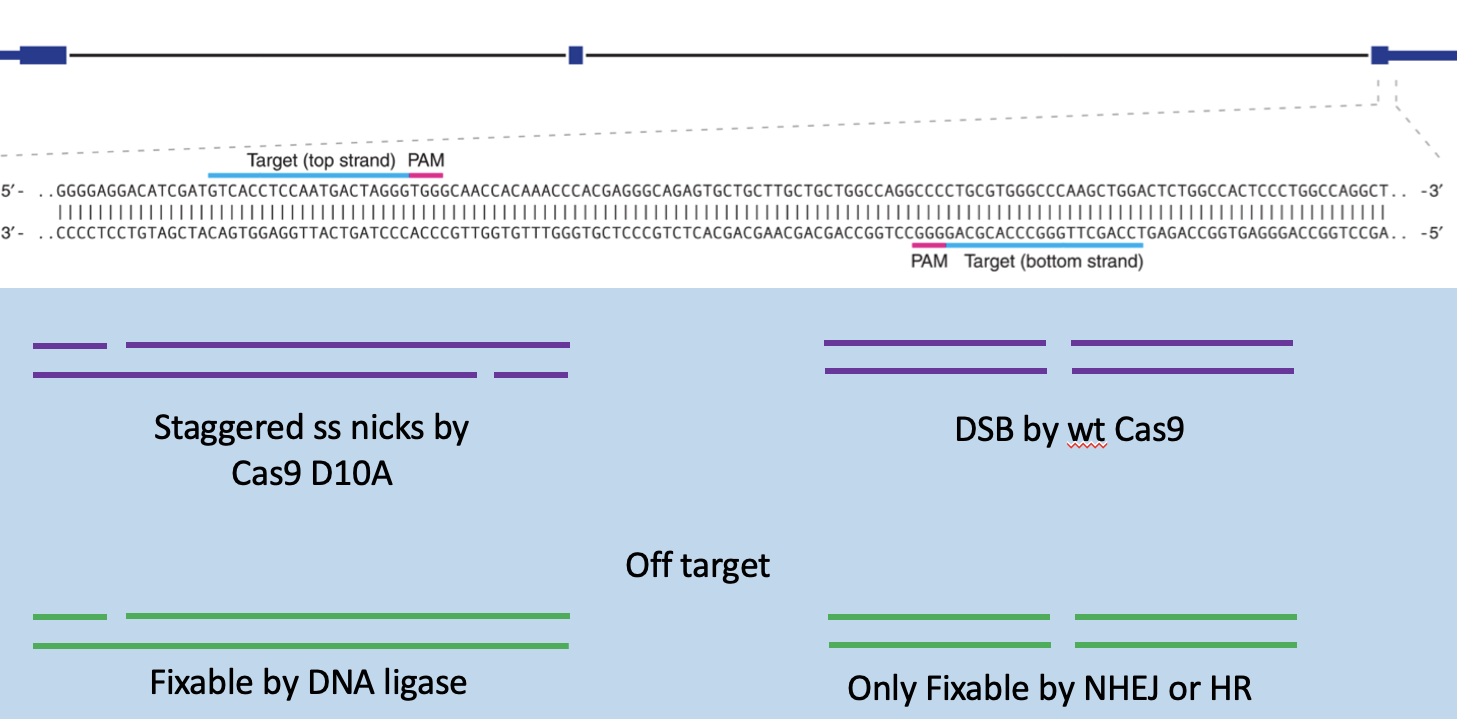
5bp closet to PAM must match exactly but mismatches closer to the 5’ end of the sgRNA are tolerated
Chromatin accessibility (less efficient when sequences within condensed chromatin are targeted)
lays chucks of repeats → predicts off target sites
“Nickase” Cas 9 mutations
inactivate a nuclease site and only cut one strand
eliminates where cas9 cleaves
cleaves only target strand
Individual point mutation in Cas9 lead to loss of nuclease activity and prevents double strand bearks
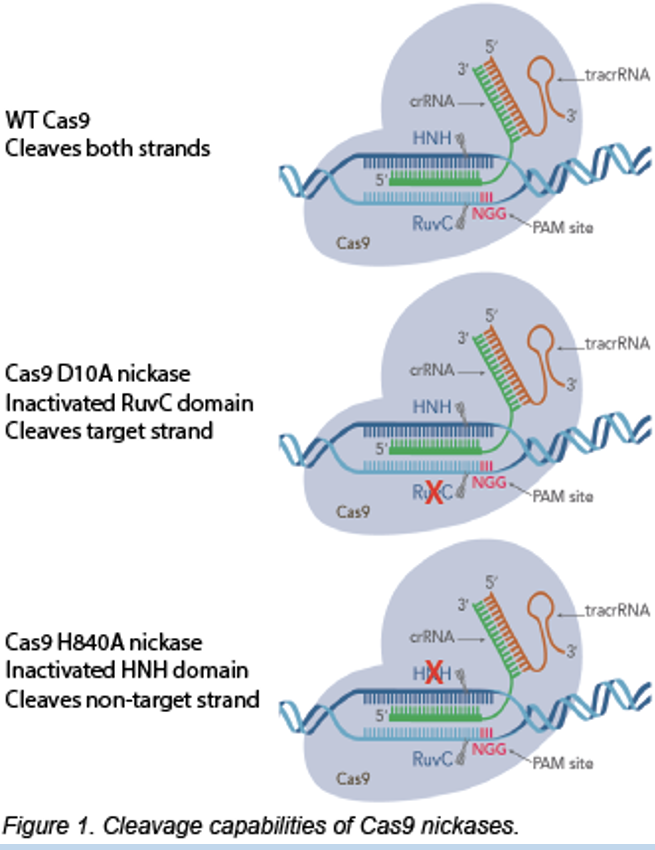
Staggered, single-stranded nicks
Nickase Cas 9 mutant, 2 sgRNAs and D10A
restains off target effects
off target is fixable by DNA ligase
CRISPR modification of pro-nuclear embryos
Can be used for germ-line incorporation
these modifications can be adapted for mouse embryo genome engineering
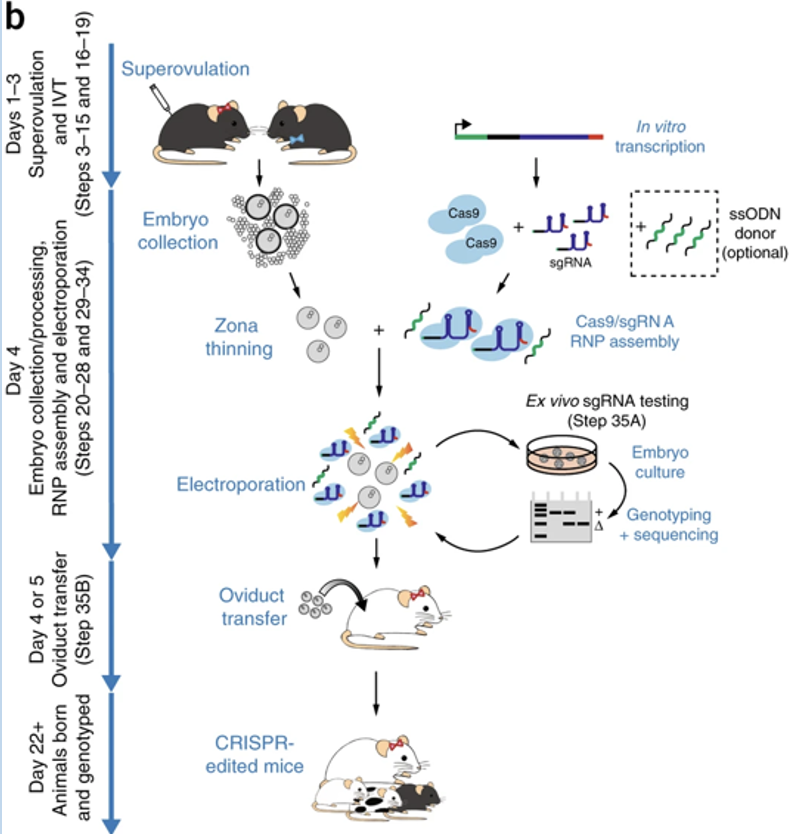
How does guide RNA direct Cas9 endonuclease to genomic targets
Complementary base pairing between the RNA and the genome locus to high selectively for targeting
Cas9 is ineffective in the absence of these interactions
How does Cas9 activity lead to mutations?
Double stranded break introduced by Cas9 will recruit either non-homologous end joining (NHEJ) or homology-directed repair (HDR)
NHEJ is usually mutagenic since bases are removed in the repair
HDR can be supplemented with a repair template that can be introduce specific mutations
Conditional mutagenesis
About 30% of knock-outs are lethal, genes often have functions in different tissues or at developmental stages that are required for viability
limitations of knock-out strategy to desired tissue or developmental stage, does not cause ablation in other essential functions
uses site-specific recombinase enzymes, that can be expressed in select tissues or stages
alleles, containing recombinase recognition sites
Site-specific recombinases
Their cognate recognition sites, set the stage for conditional mutagenesis
harvest from DNA phases
setup recombination event for us
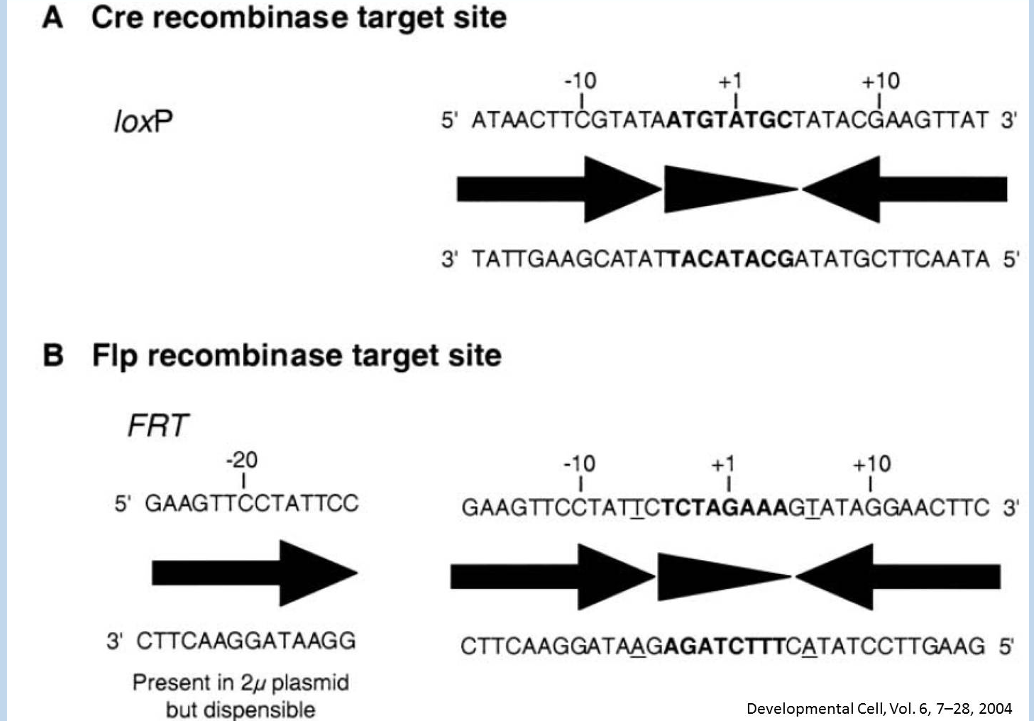
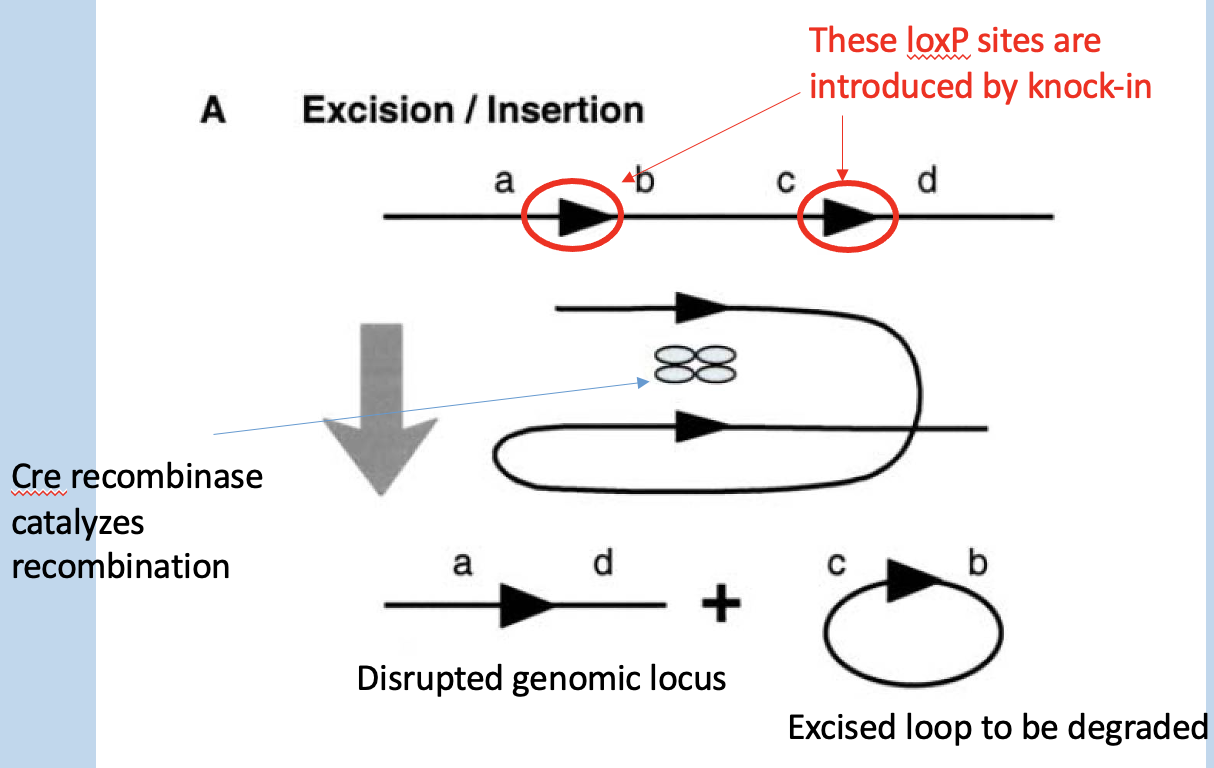
Following knock-in of a “flozed-allele” to a genomic locus
Cre recombinase can excise DNA between the loxP sites, disrupting the allele
loxP sites are introduced by knock-in
intro - loose loops or large groups
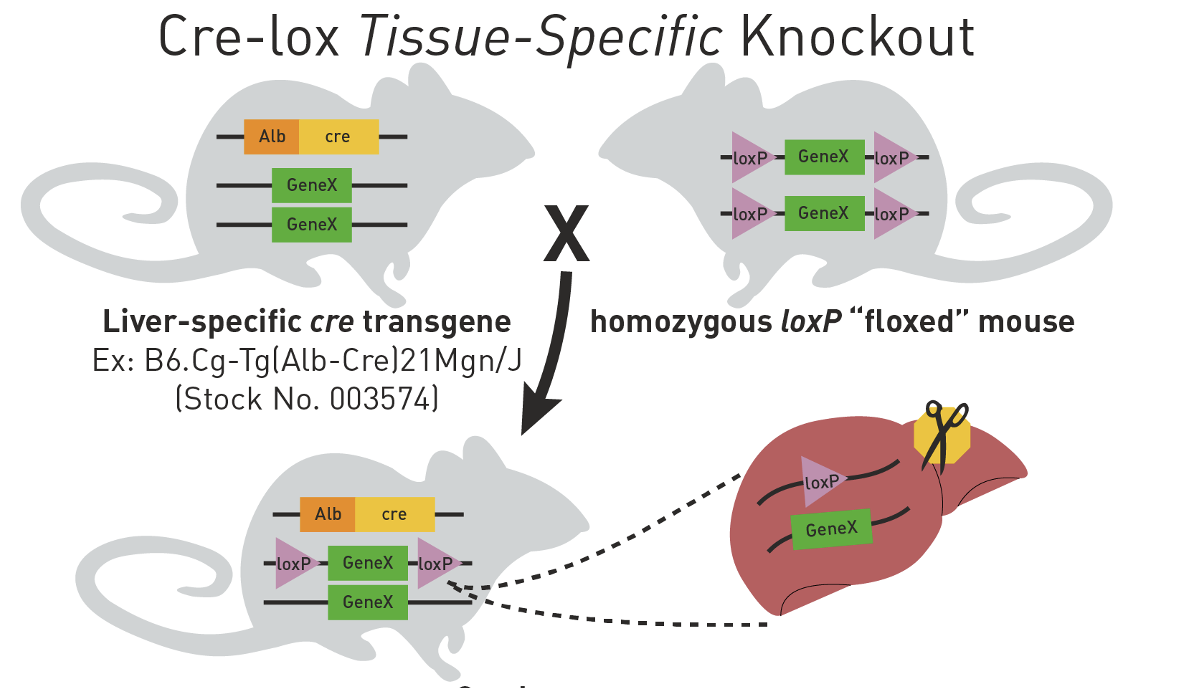
Tissue-specific expression of Cre recombinase leads to tissue-specific disruption of targeted locus
Cre-lox tissue-specific knockout
heterozygous for GeneX conditional knockout after 1 generation
how husbandry can be used to prepare unique targeting of floxed locus in mice
Generate homozygous knockouts
Additional crosses are needed to make a mouse that has two floxed alleles and Cre expression transgene
results in null effect in liver
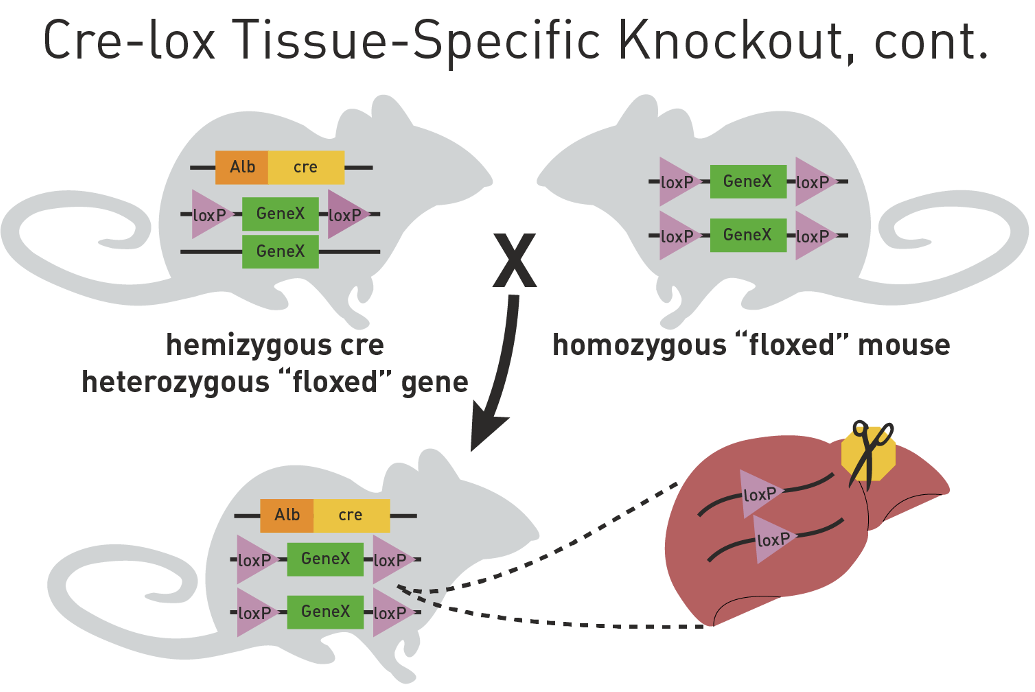
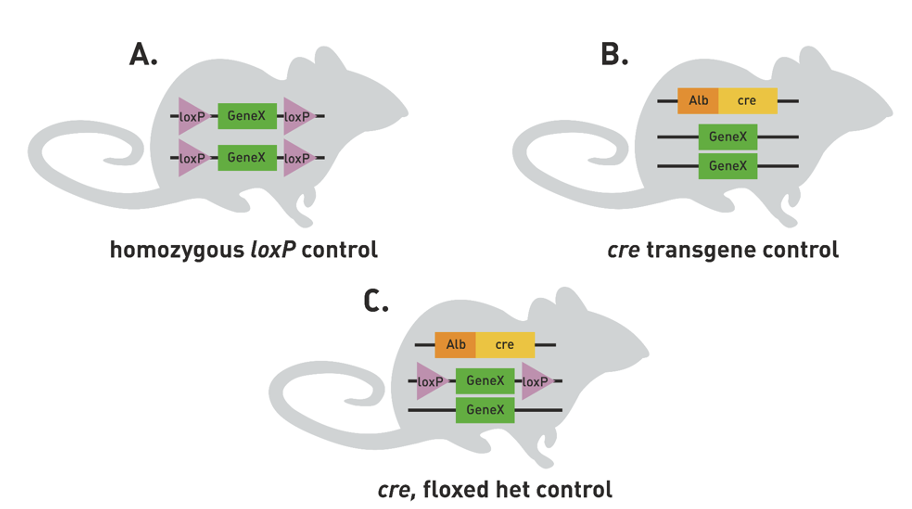
Control mice are needed to determine that transgenic cre cassete and the loxP sites
Do not cause any phenotypic effects on their own
A) homozygous loxP control - check they don’t have strange livers before hand
B) cre transgene control - make sure cre is effective
C) cre, floxed het control
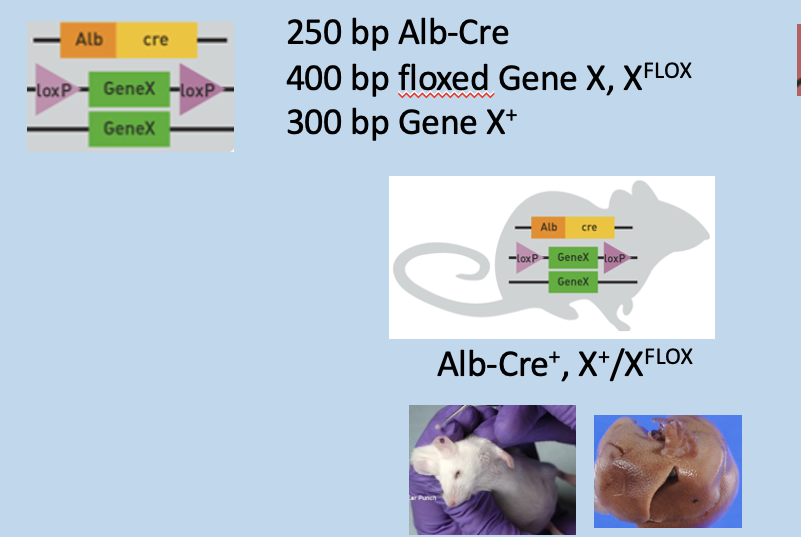
One Copy of floxed gene in genotyping PCR (heterozygous for loxp gene)
Ear punch) has 3 bands: 250bp Alb-Cre, 300bp Gene X, and 400bp floxed gene X
Liver) has 3 bands: 100bp disrupted locus, 250bp Alb-Cre, and 300bp Gene X
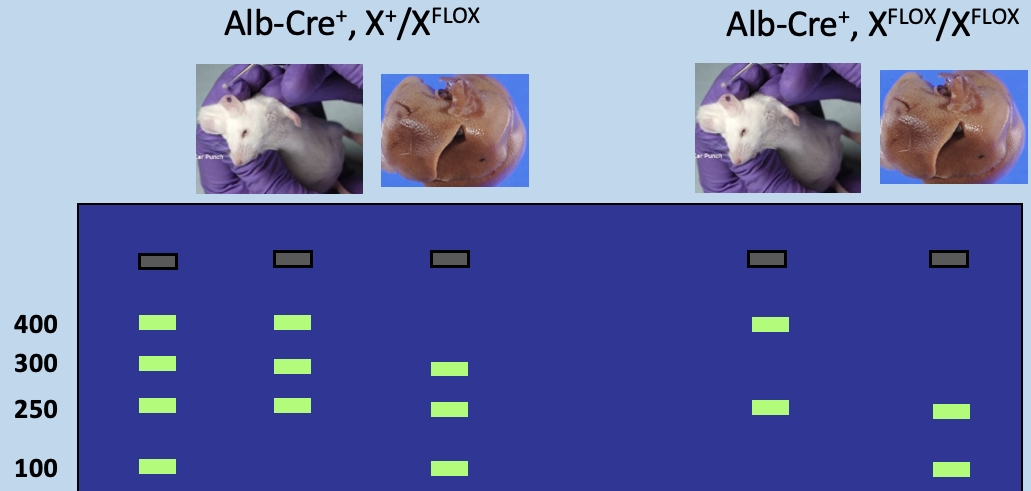
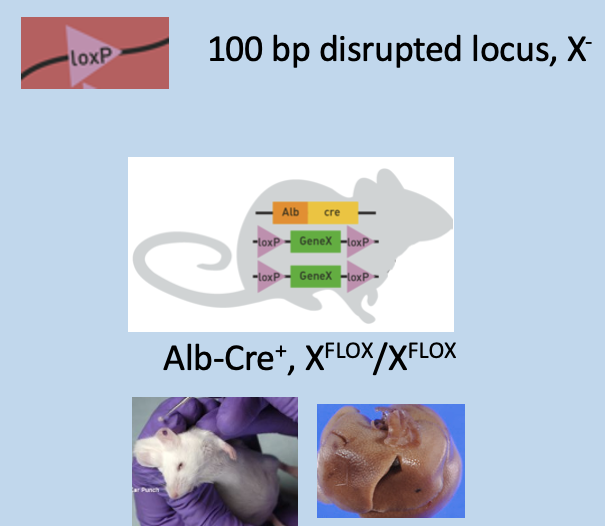
Two copies of floxed gene in genotyping PCR
Ear punch) has 2 bands: 250bp Alb-Cre and 400bp floxed gene X
Liver) has 2 bands: 100bp disrupted locus and 250bp Alb-Cre
Why are Floxed alleles generally non-disruptive in the absence of recombinase activity?
Modest size of loxP sites permit integration into intronic or intergenic regions such that coding information or expression of the target gene is not altered
How is Cre expression limited to select tissues?
Cre expression can be controlled by tissue-specific promoter (or some regulatory element)
to limit its expression to desired regions
Gene Drive
Converting heterozygous to homozygous mutations
can disrupt locus
knockout homologous allele (ensure all alleles)
Good for targeting reproductive system in misquito to kill them off