BIOL 3000 Proteins
1/44
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
45 Terms
Proteins
Large biological molecules (macromolecules) consisting of one or more polypeptide chains that perform a wide variety of functions in living organisms.
Structure gives rise to?
Function
Functional diversity and versatility of a protein derives from
-Chemical diversity of the constituent amino acid side chains
-Flexibility of the polypeptide chain
-Large number of ways in which polypeptide chains interact with different amino acids and fold
Overview of protein function
Binding, catalysis, switching, structural
Chemical diversity of the constituent amino acid side chains
There are 20 unique amino acid side chains
They each have different tendencies to interact with one another and other molecules
Non-polar (hydrophobic)
The tendency to repel water and pack closely against each other
They are on the inside
Non-polar examples
Alanine, valine, leucine, isoleucine, proline, methionine, phenylalanine, tryptophan
Polar (hydrophilic)
The tendency to form hydrogen bonds with one another, to the peptide backbone, to other molecules and to water
Polar examples
Glycine, serine, threonine, cysteine, tyrosine, asparagine, glutamine
Charged
The tendency to reside on the outside of a globular protein and interact with the other side chains or macromolecules to give structure.
There are repulsion and attraction forces that alter the structure of the protein
The functional structure of protein folds onto each other
Positively charged (+) examples
Lysine, arginine, histidine
Negatively charged (-) examples
Aspartic acid, glutamic acid
Non-covalent bonds
Hydrogen bond, ionic bond, hydrophobic interactions
Covalent bonds
Disulfide bond between amino acids
Backbone of all amino acids
An amino group at the front, an alpha carbon and side chains, and a carboxylic group at the back
Where do interactions occur?
At the side chains (R-group)
Peptide bond
A covalent bond between the carboxyl carbon and the amide nitrogen of two adjacent amino acid residues
Forms water
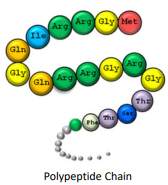
Polypeptide chain
Globular 3-dimensional protein
Flexibility of the polypeptide chain
The carboxyl carbon, carboxyl oxygen, and the amide nitrogen of the next amino acid are on a peptide plane which is coplanar
N-alpha carbon and the alpha carbon-carbon bonds are single bonds allowing free rotation provided there is no interference from the side chains
Covalent bonds
The sharing of electron pairs creating a very stable interaction
Stronger interactions
Ex: amide bonds of the amino acid backbone, disulfide bonds in some excreted or exterior surface proteins between the side chains of cysteine residues
Electrostatic bonds
The interaction of amino acids based on their charge
Weaker interactions
Ex: hydrogen bonds which occur when hydrogen has a significant positive charge and attracts significantly negatively charged atoms; Van der Walls Interactions due to the fluctuating electron clouds causing interactions between two atoms
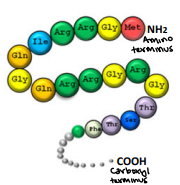
Primary structure
A linear sequence of amino acids in a polypeptide
Held together by covalent bonds of the peptide backbone
The two ends of the structure are referred to as the Amino Terminus (N-Terminus) which is the start and the Carboxyl Terminus (C-terminus) which is the end
Determined directly from the gene corresponding to the protein
Nothing really going on here
Folding leads to secondary structure
Secondary structure
Highly regular local substructures of a protein
Three types: beta turn, beta sheet, and alpha helix
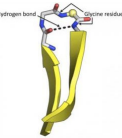
Beta turn
“u-turn”
The simplest secondary structure
Usually involving 3-4 residues
Polypeptide chain is forced to reverse direction making compact folding of the chain possible
Can make an incredibly small, sharp turn because of the hydrogen
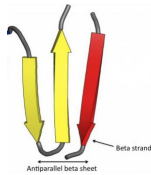
Beta sheets
Two or more strands widely separated in the primary sequence orient side-by-side with hydrogen bonds between the strands
Held together by R groups interacting laterally
Forms an opening in the membrane, “porin”
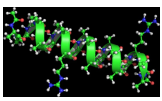
Alpha helix
The most common secondary structural element
Can be right or left-handed
The R groups points out, so they can interact with each other
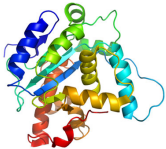
Tertiary structure
The folding of the secondary structural elements into a compact and nearly solid object stabilized by chemical bonding interactions (mostly electrostatic)
3-dimensional structure of a protein in space
ALL PROTEINS HAVE TERTIARY STRUCTURES
Quaternary structure
A three-dimensional structure of multi-subunit proteins and how those subunits stick together
Potassium ion channel protein
Made up of 4 polypeptide chains, but can be different number structures
NOT ALL PROTEINS HAVE QUATERNARY STRUCTURES
It can change its structure to interact

Three types of post-transcriptional modifications
Disulfide bridges, metal binding, bind of effector molecules
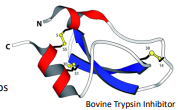
Disulfide bridge
Helps to stabilize and forms depending on the structure
-S-S oxidation of two sulfhydryl groups
Highly sensitive to the environment and is reversible
Not commonly found in intracellular proteins due to the reducing nature of the cytoplasm but very common in secreted proteins
Metal binding
Coordinated binding of a metal ion to several amino acid side chains in a single protein forming an internal metal chelate
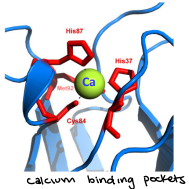
Binding of effector molecules
Most important for stability is glycosylation
Other modifications can affect the conformation and function of the protein
Glycosylation
The addition of carbohydrates to specific amino acids, imagine a candied apple
Reversible modification
Turns proteins off or on
Ex: Phosphorylation and acetylation
Phosphorylation
Adds a phosphate group
Acetylation
Adds an acetyl group
Irreversible modifications
Ubiquitination and methylation
Ubiquitination
Adds a ubiquitin to a protein which tells the cell to degrade the protein
Methylation
Adds methyl group
Size-wise, where are motifs and domains
They’re in between secondary and tertiary
Types of protein motifs
Sequence, functional
Sequence motifs
Generally consist of a few structural elements relating primary structure to tertiary structure. Very small areas of 3D shape. So small that if you were to cut out a motif, then the protein falls apart
Functional/structural motif
A set of contiguous secondary structural elements that have a specific function.
Protein domains
A compact region of protein structure capable of stable folding independent of a large protein and having a specific function
Maintains its shape if it is cut out
Not up to tertiary structure
~40-~900 amino acids
Averaging ~200 amino acids
Have multiple domains