Midterm 2 (10-19) - BIS 2A
5.0(2)
Card Sorting
1/345
Earn XP
Description and Tags
Last updated 5:09 AM on 12/7/22
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
346 Terms
1
New cards
How long did it take for O2 to build up in the atmosphere and oceans?
About a billion years to get to 10% of today's levels
2
New cards
methanogens
Archaea that release methane, prospered on anoxic Earth
3
New cards
Why was oxygen considered toxic waste?
O2 is a powerful oxidant, would immediately oxidize susceptible metals
4
New cards
What else happened to the O2 generated by cyanobacteria, besides reacting with iron?
Reacted a lot with methane, might have caused "snowball Earth"
5
New cards
Why did CO2 levels go down?
A lot of available electrons for photosynthesis, so CO2 used up
6
New cards
What else happened when O2 accumulated in Earth's atmosphere and oceans?
Once 0.0002 of present levels reached, started creating a UV-protective ozone layer
7
New cards
A consequence of ozone layer:
UV-driven reaction of O2 with methane slowed, = increased rate of O2 and ozone accumulation
8
New cards
Another consequence of ozone layer:
UV-induced water splitting (to O2 and H2) slowed, preventing escape of H2 (= oceans) to outer space
9
New cards
Yet another consequence of ozone layer is:
life could move onto land.
10
New cards
enzymes
- catalysts for living things
- made of protein and other things (prosthetic groups and cofactors)
- made of protein and other things (prosthetic groups and cofactors)
11
New cards
Some proteins are:
structural, pumps, machines, carriers - not always catalysts
12
New cards
Why does life use a catalyst for every reaction?
a) To make it go faster
b) To emphasize rate useful reactions and avoid toxic alternatives
c) To optimize rates for current conditions
d) To allow one species to complete
a) To make it go faster
b) To emphasize rate useful reactions and avoid toxic alternatives
c) To optimize rates for current conditions
d) To allow one species to complete
All of the above
13
New cards
The active site of enzymes:
fit their substrates
14
New cards
What are proteins made of?
Several units of amino acids attached in chains which are folded up
15
New cards
What holds proteins together?
Hydrogen, disulfide, ionic, hydrophobic, peptide bonds
16
New cards
How do the various levels of organization affect protein structure?
The levels allow different shapes and different types/places of bonding - each contributes to different function
17
New cards
How does the cell generate very specific 3D shapes?
Sometimes linear forms fold themselves up into their desired shape spontaneously (2', 3', 4') - the code is in the linear sequence of amino acids. (1')
18
New cards
primary structure
- precise series of individual amino acids linked with peptide bonds
- long, flexible chain
- long, flexible chain
19
New cards
peptide linkage
bond between amino group and carboxyl group in amino acids
20
New cards
N-C-C backbone
makes up amino acids
21
New cards
The R group is:
used to represent one of 20 possible side chains, each for a different amino acid.
22
New cards
protein size
can vary from a dozen amino acids to a few thousand
23
New cards
protein
a specific polymer of amino acids
24
New cards
The protein backbone:
helps the protein fold into secondary structures.
25
New cards
secondary structure
- flat planes = beta sheets
- rigid rods = alpha helices
- rigid rods = alpha helices
26
New cards
The secondary structure is stabilized by:
several hydrogen bonds between N-H and C=O (from backbone).
27
New cards
beta barrel
- created from beta pleated sheets
- forms a pore through bacterial outer membrane, allowing larger molecules to pass through
- forms a pore through bacterial outer membrane, allowing larger molecules to pass through
28
New cards
rhodopsin
- transmembrane protein that carries retinal molecule at core
- made of alpha helices
- made of alpha helices
29
New cards
The likelihood of folding up into an alpha helix or beta sheet is determined by:
the sequence of R groups.
30
New cards
Glycine's R group is too entropic:
the backbone can form too many shapes.
31
New cards
Proline's R group is too rigid, so:
the N-C bond can't rotate.
32
New cards
Amino acids with positively charged R groups:
Arginine, Histidine, Lysine
33
New cards
Amino acids with negatively charged R groups:
Aspartic Acid, Glutamic Acid
34
New cards
Amino acids with polar, uncharged side chains
Serine, Threonine, Asparagine, Glutamine
35
New cards
Special case amino acid R groups:
Cysteine, Glycine, Proline
36
New cards
Amino acids with hydrophobic side chains:
Alanine, Valine, Isoleucine, Leucine, Methionine, Phenylalanine, Tyrosine, Tryptophan
37
New cards
hydrophobic R groups
- oily
- tend to interact only with other oily R groups or membranes and exclude water and charger or polar R groups
- tend to interact only with other oily R groups or membranes and exclude water and charger or polar R groups
38
New cards
Tertiary and quaternary structures stabilized by:
a variety of weak interactions - ionic bonds, hydrogen bonds, hydrophobic bonds.
39
New cards
Denaturing agents disrupt:
the tertiary and secondary structure of proteins.
40
New cards
denatured proteins
unfolded proteins, often won't fold correctly again
41
New cards
Denatured proteins can be caused by:
heat, high salt, etc.
42
New cards
Very stable proteins are generally:
small and stabilized by disulfide bonds.
43
New cards
Cysteine R-groups covalently pair up to form Cystine, which is more stable because:
the covalent bond 'tacks' them together, giving fewer options.
44
New cards
quaternary structure
multiple subunits - different proteins working together as one complex
45
New cards
An example of the influence of local environment on reduction potential of a cofactor:
46
New cards
What is the difference between a cofactor and a substrate or product?
Cofactors come out of a reaction unchanged, substrates are used up.
47
New cards
Why might binding a regulatory molecule change the accessibility or functionality of the active site?
48
New cards
allostery, in terms of protein structure
as regulatory molecules bind, change protein shape - active site is more or less available (activator protein or inhibitor)
49
New cards
An example of logical regulation of an enzyme by a molecule that is neither a substrate or product:
50
New cards
Aside from enzymes, proteins also:
act as motors, act transporters or pores, define cell shape, regulate processes (gene expression), and determine cell-cell interactions.
51
New cards
3D structure and pattern of charges and/or hydrophobic patches allows the enzyme to:
bind the correct substrate(s) and cofactor group(s) in the right orientation.
52
New cards
metal-binding cofactors
iron-sulfur clusters, heme, chlorophyll...
53
New cards
non-metallic cofactors
NAD, FAD, coA, ... (coenzymes)
54
New cards
Substrates are:
used up in the reaction.
55
New cards
Cofactors are:
regenerated and part of catalysis.
56
New cards
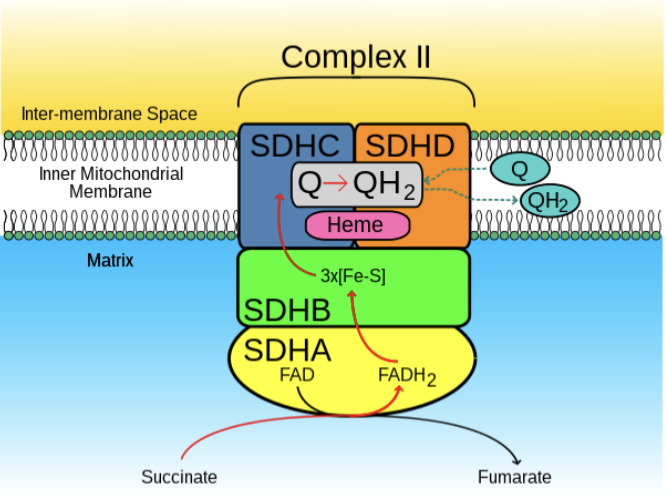
This diagram of Complex II (succinate dehydrogenase) of the respiratory chain includes many non-protein factors. Which is not a cofactor?
a) FAD
b) Succinate
c) Fe-S
d) Heme
a) FAD
b) Succinate
c) Fe-S
d) Heme
b) Succinate
57
New cards
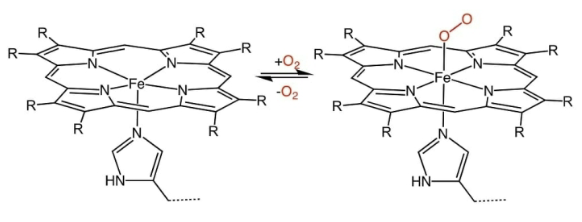
This heme from hemoglobin is not covalently bound to methionine. Why?
It cannot have another covalent bond, the space is left for oxygen, since it's a oxygen carrier.
58
New cards
How might local environment of active site affect electronics of an e- donor? (1)
Negative charges near a potential donor would encourage loss of en electron (smaller reduction potential = stronger electron donor/weaker electron acceptor).
59
New cards
How might local environment of active site affect electronics of an e- donor? (2)
Water or polar R groups might rearrange to partially neutralize the affects of nearby charges.
60
New cards
Not all enzymes require cofactors, instead enzymes: (1)
' R-groups can participate in chemical reactions with the substrate.
61
New cards
Not all enzymes require cofactors, instead enzymes: (2)
can simply orient/bend substrates.
62
New cards
Not all enzymes require cofactors, instead enzymes: (3)
can provide local changes in environmental conditions (concentration/pH), affecting only their trapped substrate(s).
63
New cards
Many minerals and cofactors critical to catalysis may be clues to how biological catalysis arose:
some may have been adapters, enhancing or restricting catalytic activity.
64
New cards
What early life nucleotide based?
Amino acids (... proteins) could be newer developments that further enhanced specificity for particular substrates.
65
New cards
What is the difference between FAD/FADH2 acting as a cofactor and FADH2 being a product of a reaction?
When the FAD/FADH2 is cyclic and covalently bound/permanently embedded to the protein, it's likely a cofactor
66
New cards
The intermediates and products of metabolism are:
maintained at ideal levels and shared between a variety of pathways.
67
New cards
How does an enzyme stay informed?
68
New cards
feedback inhibition
product of a long pathway inhibits the enzyme that catalyzes the first dedicated step of the pathway
69
New cards
Why make enzymes that shut down unnecessary pathways?
Making more of something that isn't needed is wasteful
70
New cards
Why not just allow mass action (accumulation of product) shut down the pathway?
Mass action isn't reliable, sometimes can require massive amounts - over constrained
71
New cards
How can an enzyme perceive conditions in the cell?
Through allosteric interactions
72
New cards
When the enzyme is in inactive form:
it cannot accept substrate.
73
New cards
Phosphofructokinase catalyzes the first dedicated step in glycolysis (F6P to FBP) and is regulated allosterically by high concentrations of ADP. Does high [ADP] switch the enzyme on or off?
It switches it on - the point of glycolysis is to make ATP, high [ADP] is low [ATP]
74
New cards
Would high [PEP] inhibit or activate phosphofructokinase?
75
New cards
competitive inhibitor
races against substrate to active site, more substrate can reverse it
76
New cards
noncompetitive inhibitor
binds allosterically to a regulatory site to disable active site, more substrate won't help
77
New cards
Penicillin G is a noncompetitive inhibitor - it binds to bacterial enzyme required for cell wall synthesis, this:
irreversibly modifies active site.
78
New cards
irreversible inhibition
inhibitor covalently (or otherwise permanently) binds to or modifies the allosteric or active site
79
New cards
signal metabolites
can switch protein on or off by allosterically affecting its affinity for its substrate
80
New cards
Controlling the concentration of an active catalyst will:
control the reaction rate.
81
New cards
Allosteric regulation of enzyme activity is faster than:
turning on synthesis of a protein.
82
New cards
What kind of reaction joins two amino acids?
A condensation reaction
83
New cards
What two functional groups react in a condensation reaction of two amino acids?
The amino group and the carboxyl group
84
New cards
What is a cell?
A high organized compartment with thin, flexible membrane, concentrated chemicals (aq) - capable of metabolism and autonomous replication
85
New cards
simplest cell
cell lacking nucleus and often other membrane bound organelles
86
New cards
Bacteria and archaea have a _____ cell type.
simple (prokaryotic)
87
New cards
How can cells keep functional molecules from floating away?
Developing a selectively permeable membrane, using lipids
88
New cards
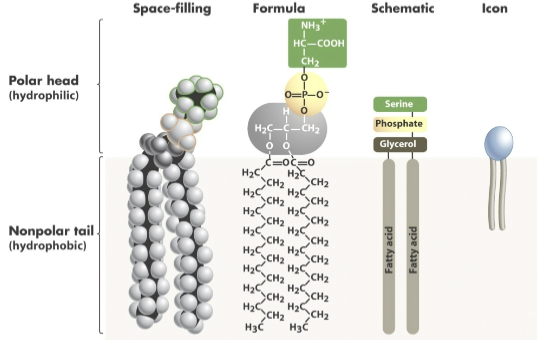
phospholipids
charged, phosphate-containing head + glycerol + two fatty acid tails
89
New cards
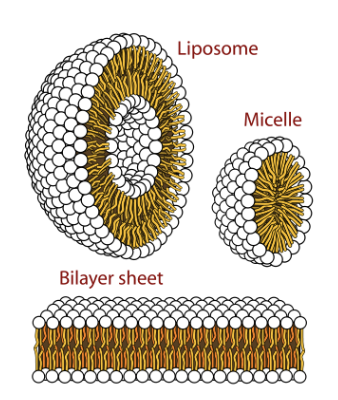
Self-ordered shapes of phospholipids result from:
water maximizing hydrogen bonding with other polar molecules - mostly water, plus the polar head groups of lipids.
90
New cards
diffusion
different solutes on opposite sides of lipid bilayer, molecules of each diffuse freely across until equilibrium established - solutes continue to move but now at equal rates
91
New cards
What can diffuse across a membrane?
Non polar molecules (gases, alcohol, benzene...) - small polar molecules can but slowly and large polar molecules rarely can
92
New cards
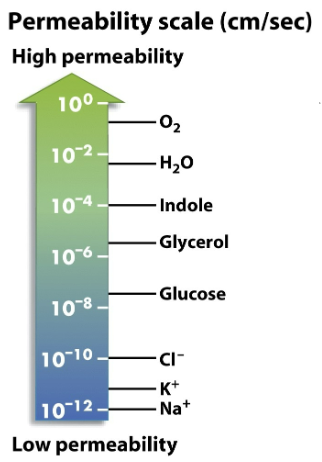
Which chemical is a million-fold more likely than Na+ to cross a lipid bilayer?
a) Cl-
b) Glucose
c) Glycerol
d) Indole
a) Cl-
b) Glucose
c) Glycerol
d) Indole
c) Glycerol
93
New cards
Double bonds in fatty acids cause:
kinks in phospholipid tails, meaning more permeability - kinked = unsaturated.
94
New cards
saturated fatty acid
has as many Hs as possible - many or all saturated fatty acids cause lower permeability
95
New cards
How do lipids move in a membrane?
They can spin and exchange places but cannot flip
96
New cards
Membrane permeability and fluidity are related to: (1)
percentage of unsaturated fatty acid tails - more saturated, more viscous.
97
New cards
Membrane permeability and fluidity are related to: (2)
the length of fatty acid tails - longer tails = thicker membrane = less permeable.
98
New cards
Cold-adapted organism have:
more kinks, preventing the membrane from solidifying.
99
New cards
osmotic issues in the membrane
if a cell has a higher concentration than its environment, water will leak into cell - can potentially blow up cell
100
New cards
How is osmotic pressure prevented in the cell?
A cell wall limits the amount of water that can be taken up