Week 4B: DNA Synthesis
1/31
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
32 Terms
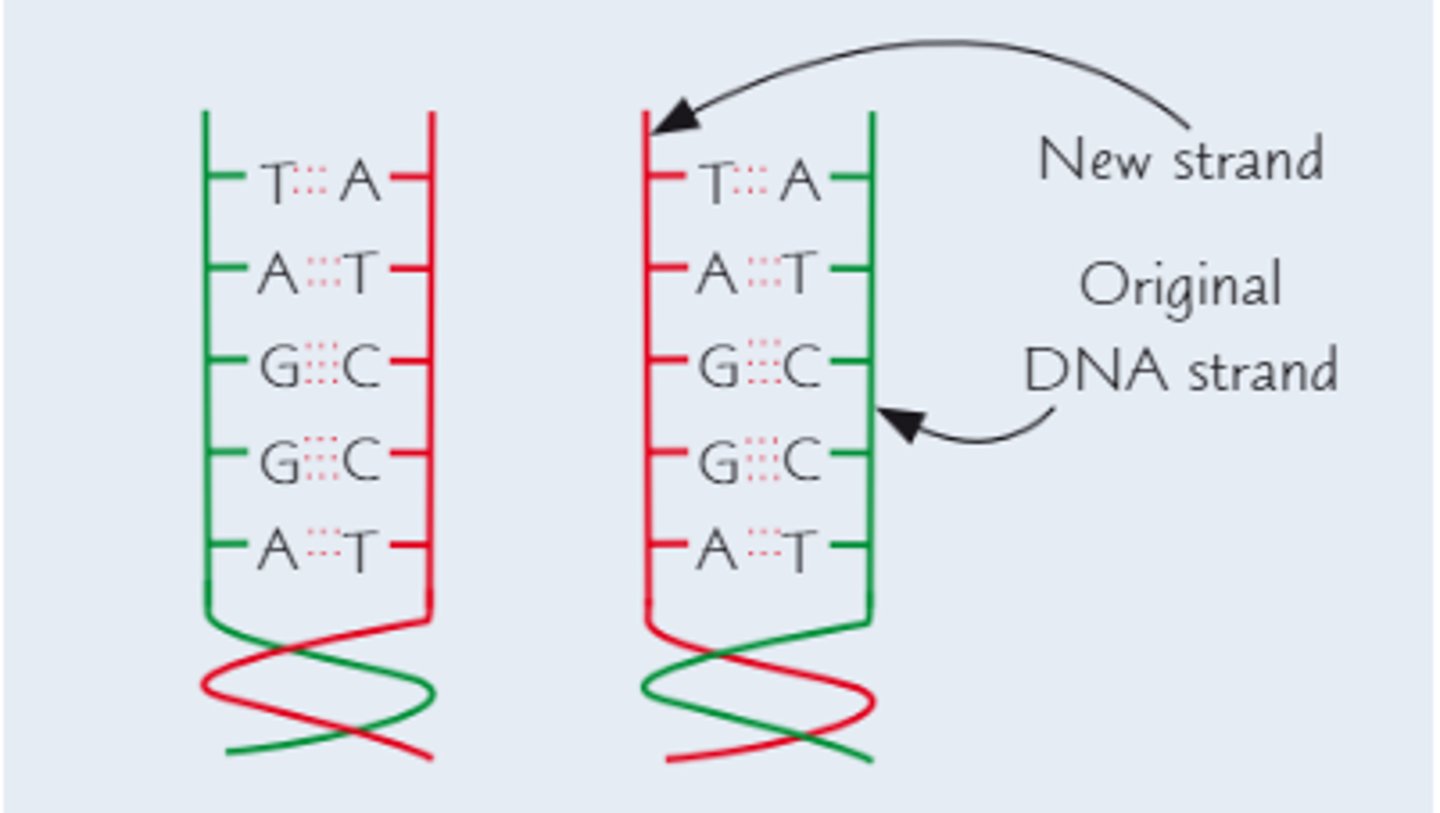
What is the product of semi-conservative replication?
A parent DNA strand is replicated, whereas a double stranded DNA splits in order to form two new DNA strands, each containing 1 strand from the parent and 1 "new" strand
Ensures that if an error is made during replication, the original strand is kept
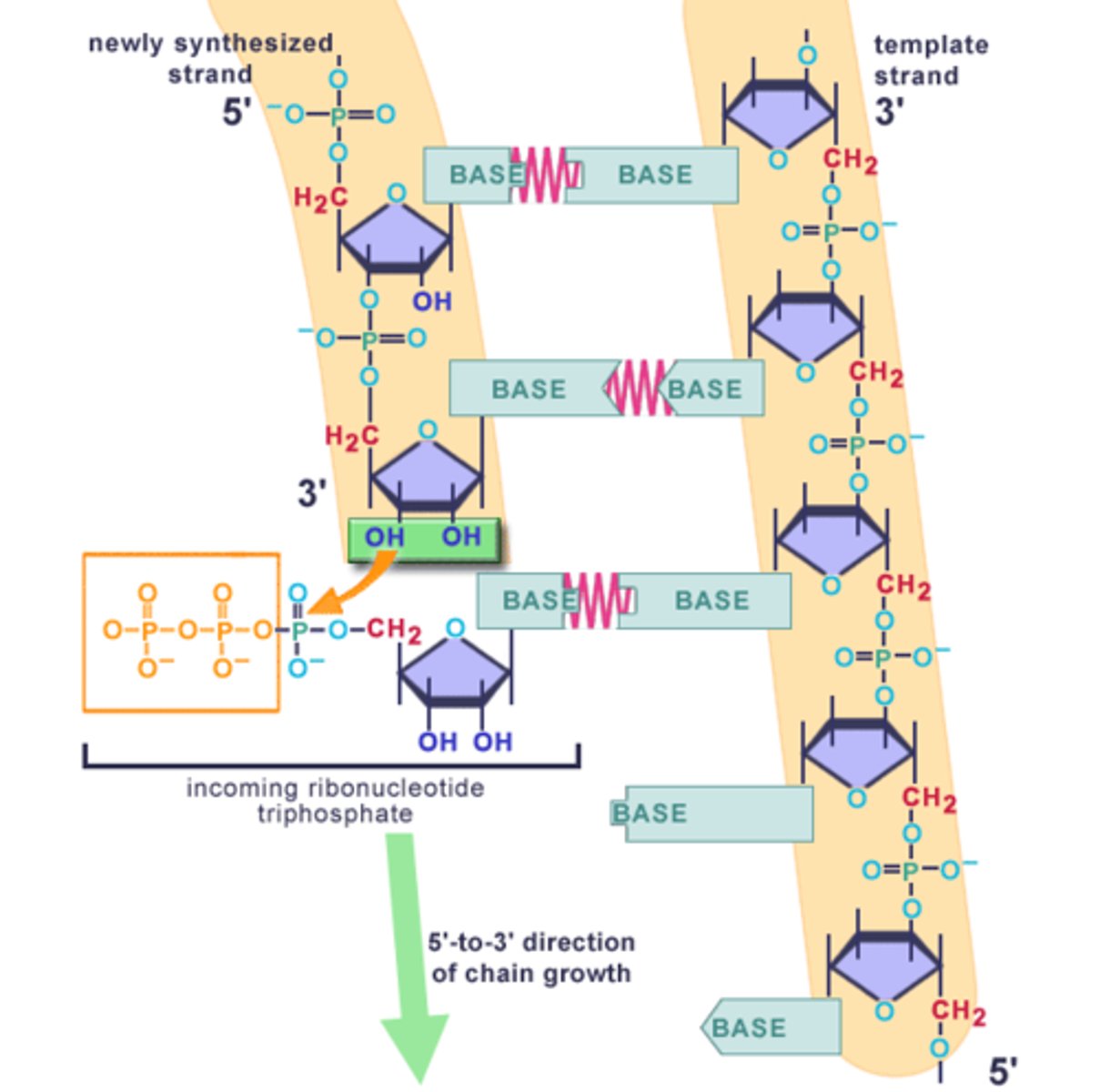
What direction is DNA synthesized?
Synthesized in the 5' to 3' direction (DNA polymerase attached the phosphate group of the new nucleotide onto the -OH on the 3' carbon of the pentose sugar)
Means that DNA polymerase "reads" the template DNA strand in the 3' to 5' direction
Which direction is each strand of DNA synthesized as opposed to each other?
Since DNA is antiparallel, DNA is synthesized in the opposite direction of each other, called bi-directional replication

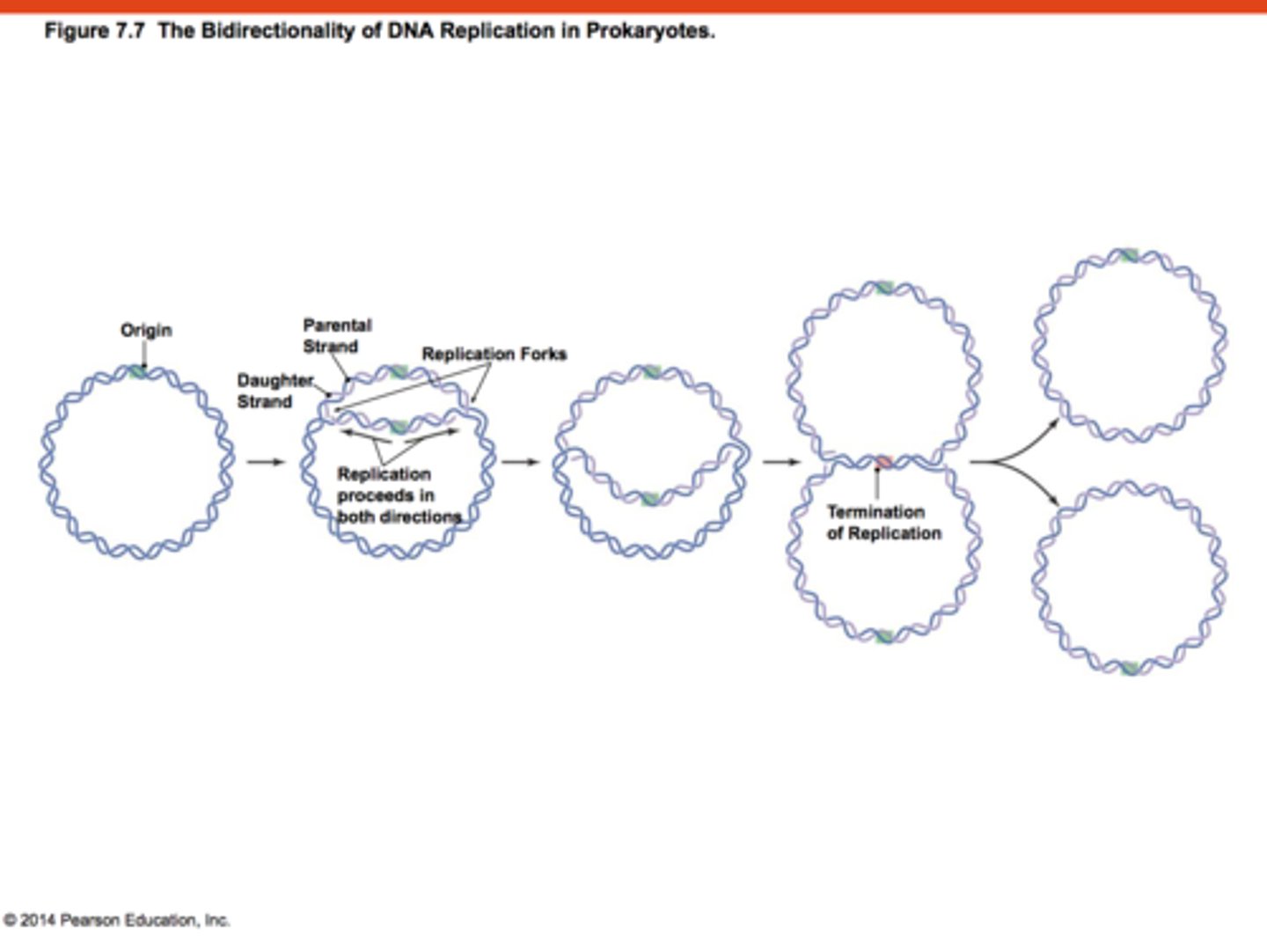
Describe DNA replication in bacteria
Bacteria contain circular DNA, with a single origin of replication.
Once replication occurs, DNA replication occurs in both directions
Bacteria is used as a model for DNA replication as it is well understood, compared to human DNA replication, which is complex and less understood
Describe the general steps of DNA replication
1. DNA helix is opened and unwound (helicase, ssb proteins, topoisomerase/gyrase
2. Original strand of DNA is used as a template to make a copy (primase, DNA poly III)
3. The RNA primers are removed and replaced by DNA (DNA Poly I, RNase H)
4. The DNA fragments on the lagging strand are attached (DNA ligase)
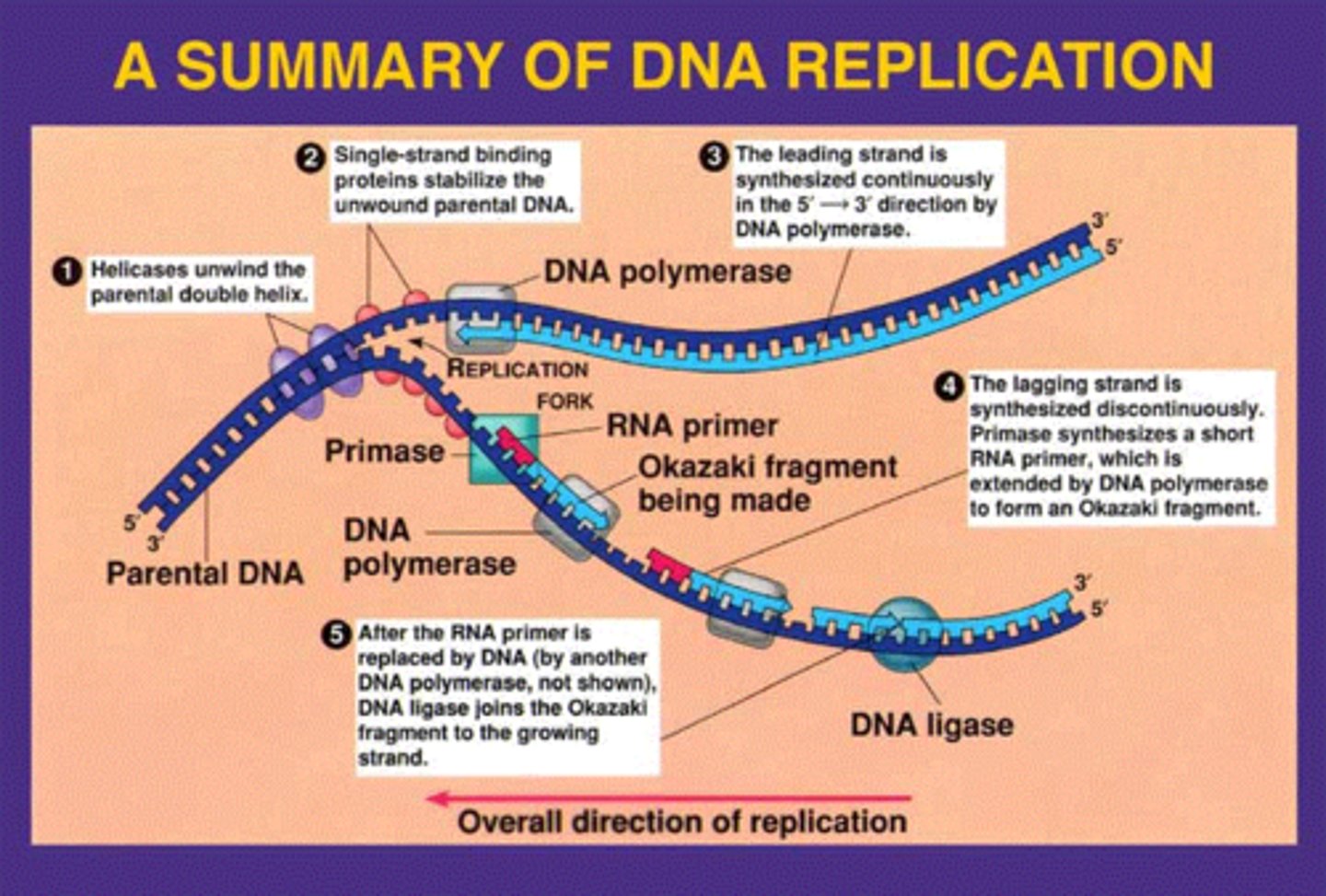
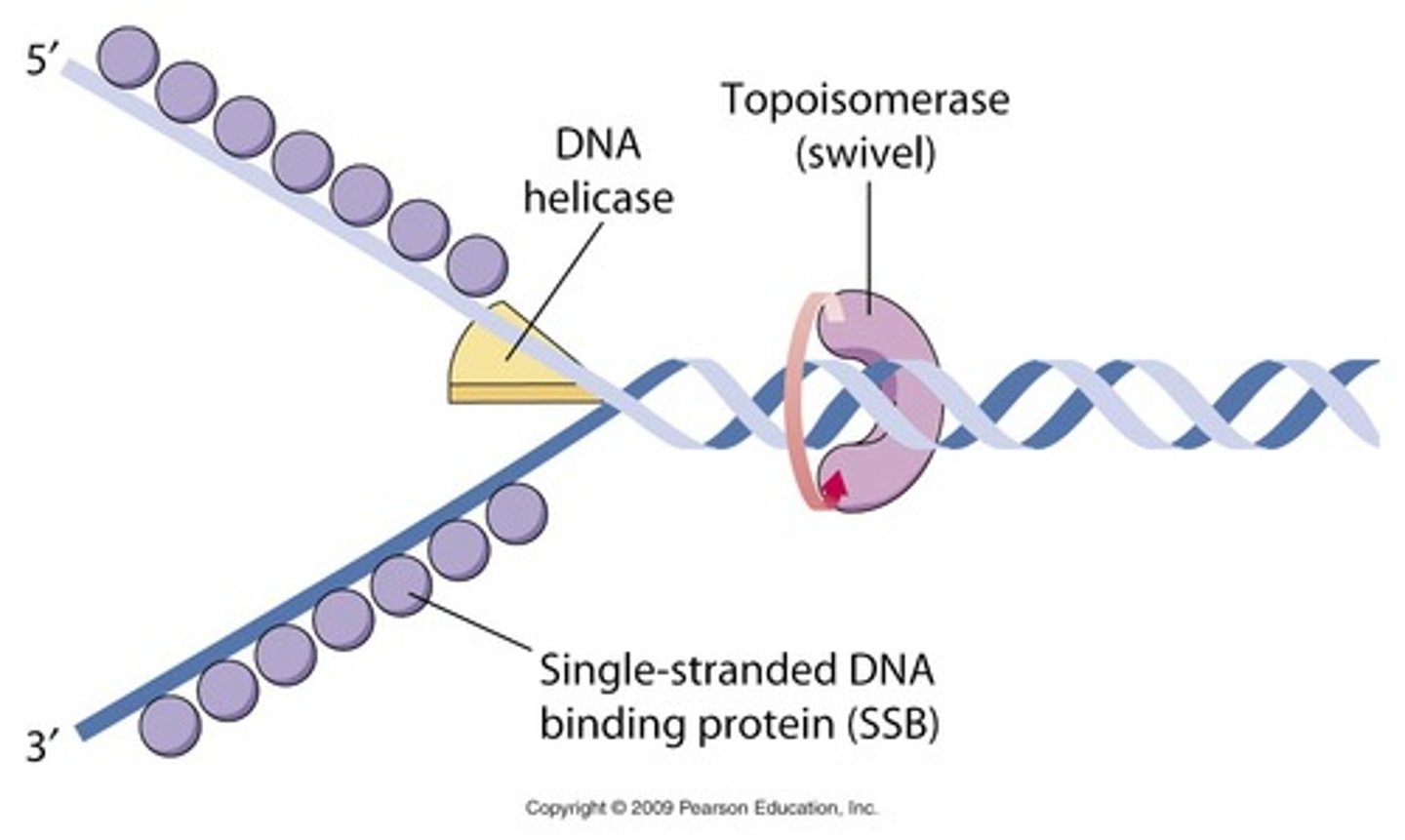
What is the role of helicase in DNA replication?
Opens the double stranded DNA so that it unwinds into two single DNA strands
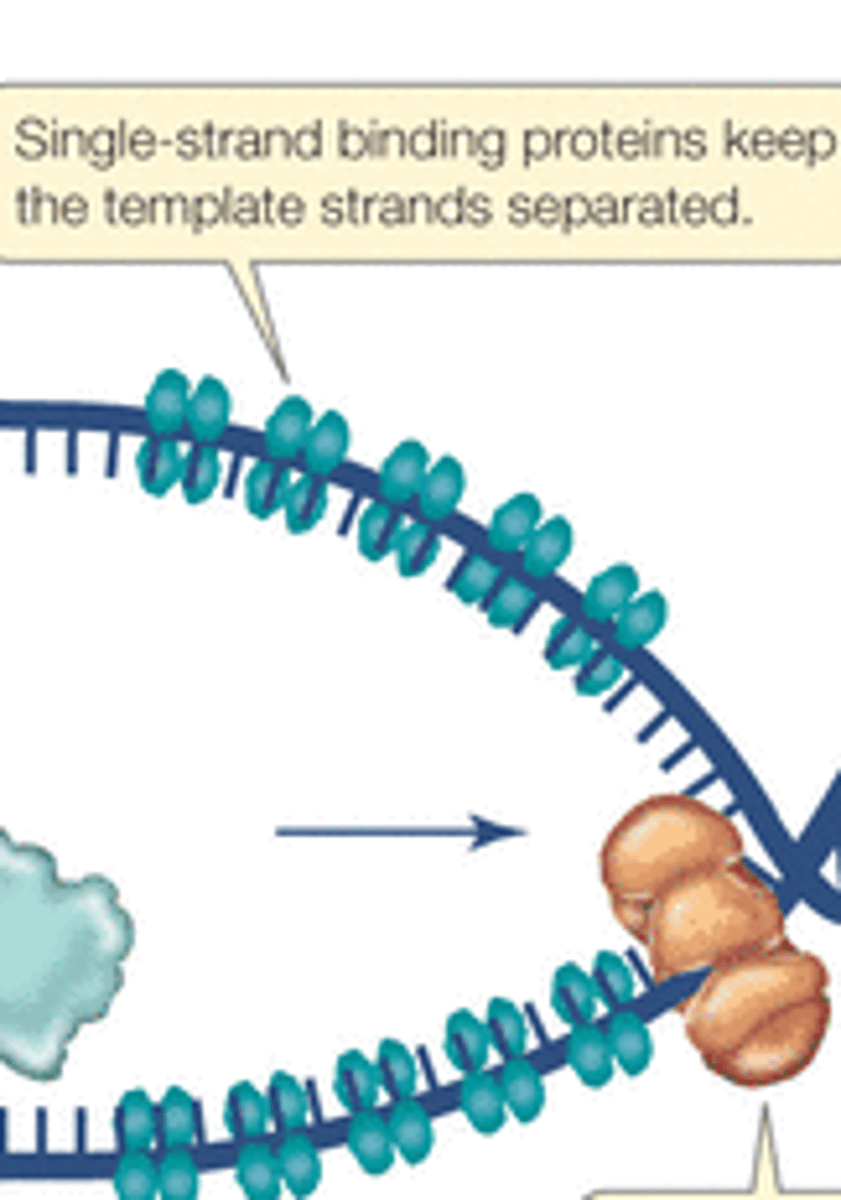
What is the role of ssb proteins in DNA replication?
Binds to the single strands of the DNA once it it unwound by helicase to prevent DNA from re-annealing (closing back up)
What is the role of topoisomerase/gyrase in DNA replication?
Relieves supercoiling/tightening of DNA while it is being unwound by breaking apart phosphodiester bonds in DNA and rejoining it
Gyrase is the bacterial version of topoisomerase

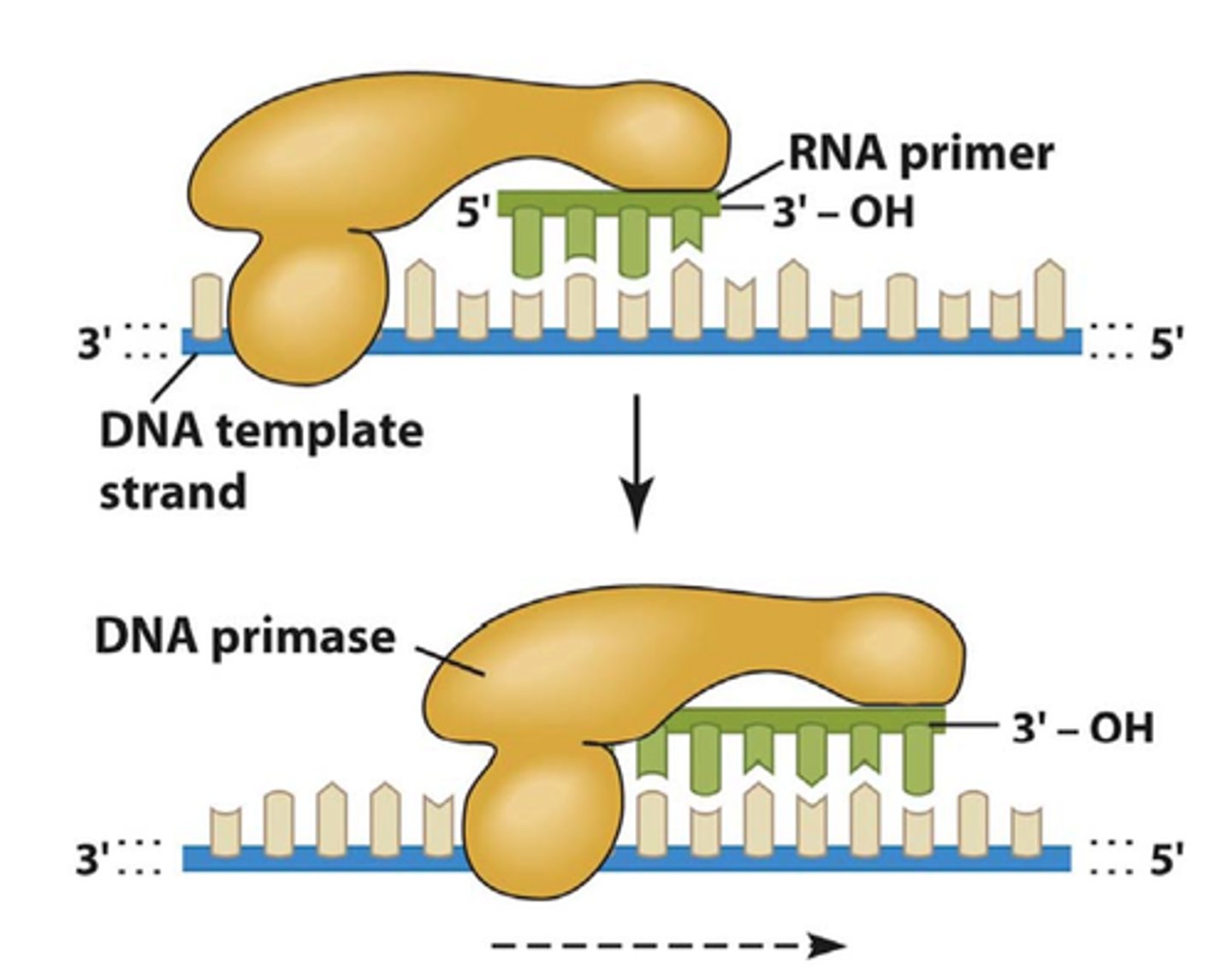
What is the role of primase in DNA replication?
Adds small RNA primers onto the strand of DNA being copied, which marks the starting point and allows DNA polymerase III to begin synthesizing new DNA (gives DNA poly III a 3' -OH carbon to work off of)
Multiple RNA primers are inserted onto the lagging strand of the DNA
Only a single RNA primer is required on the leading strand of the DNA (being synthesized in the same direction as DNA is being unwound -> reading 3' to 5')
What is the role of DNA polymerase III in DNA replication?
Adds a new complementary base pair of the DNA that it is synthesizing by catalyzing the formation of the phosphodiester bond on the 3' -OH of the previously synthesized DNA
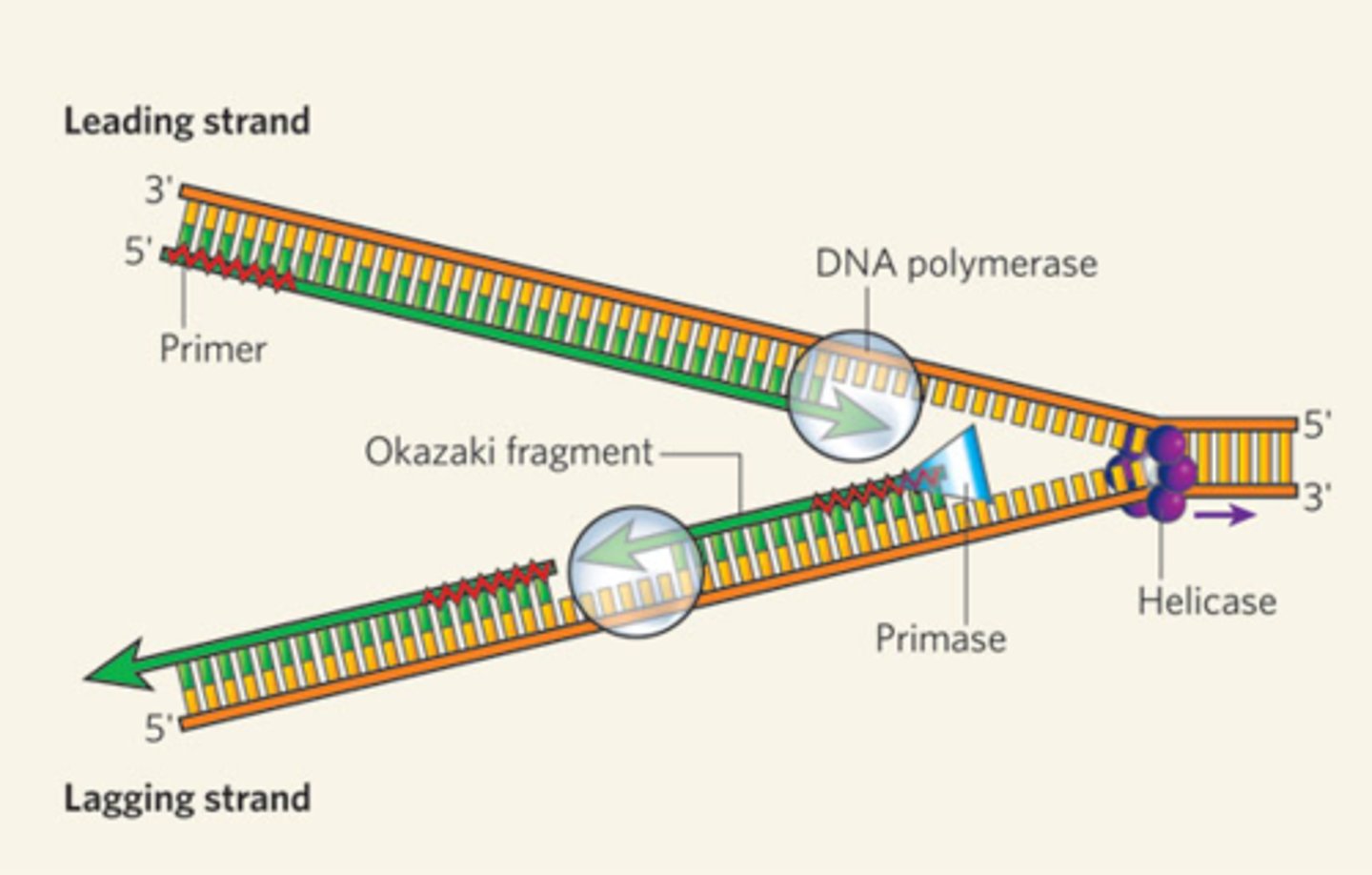
On the leading strand, DNA polymerase III creates one continuous strand as it is synthesizing DNA in the same direction as the DNA is being unwound
On the lagging strand, DNA polymerase III has to stop at RNA primers due to travelling in the opposite direction as the DNA is being unwound, causing Okazaki fragments to form
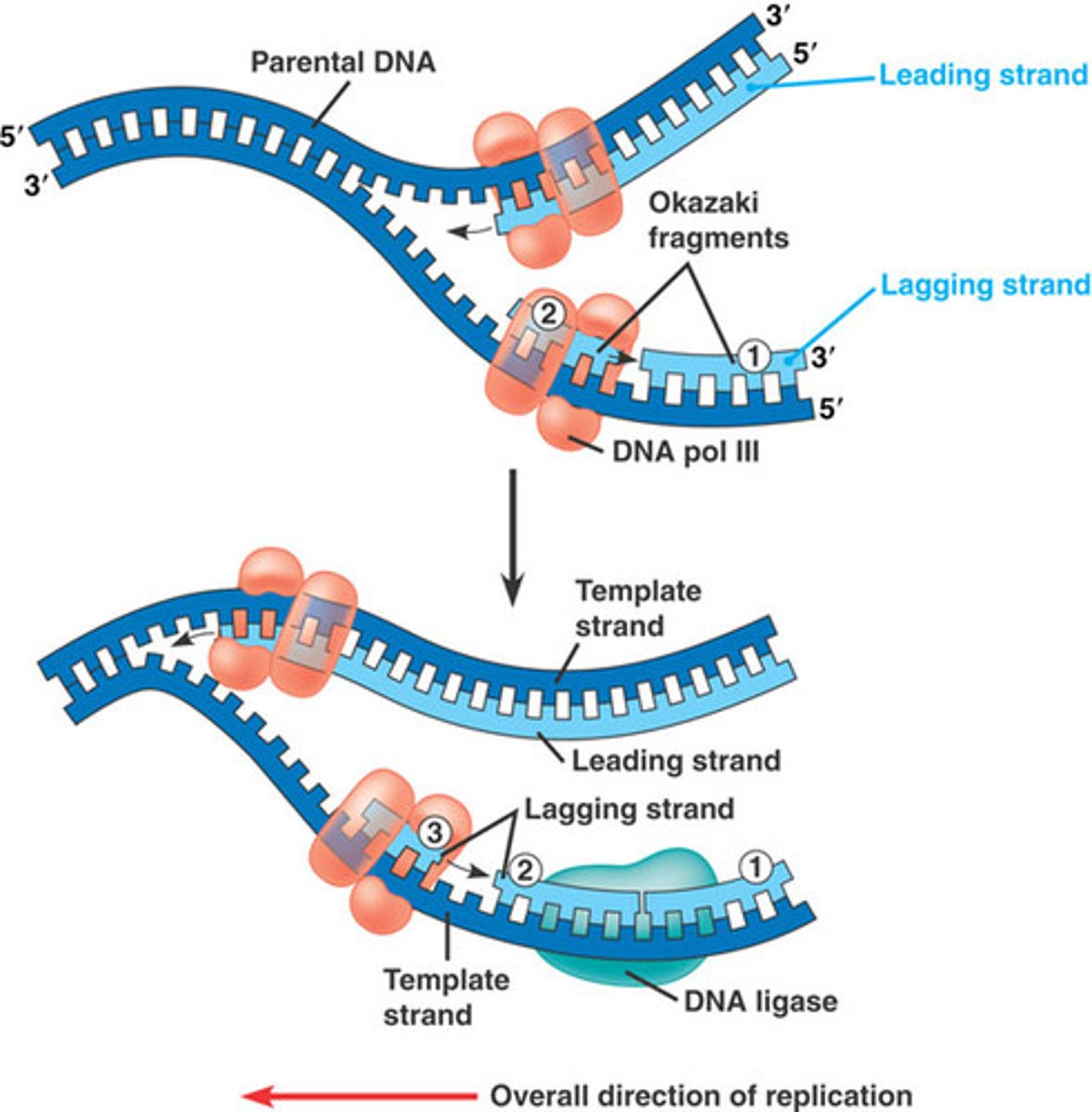
What is the role of DNA Polymerase I in DNA replication?
Works with RNase H to remove and replace RNA primers with new DNA nucleotides, however, does not join the Okazaki fragments of the lagging strand yet (DNA ligase)
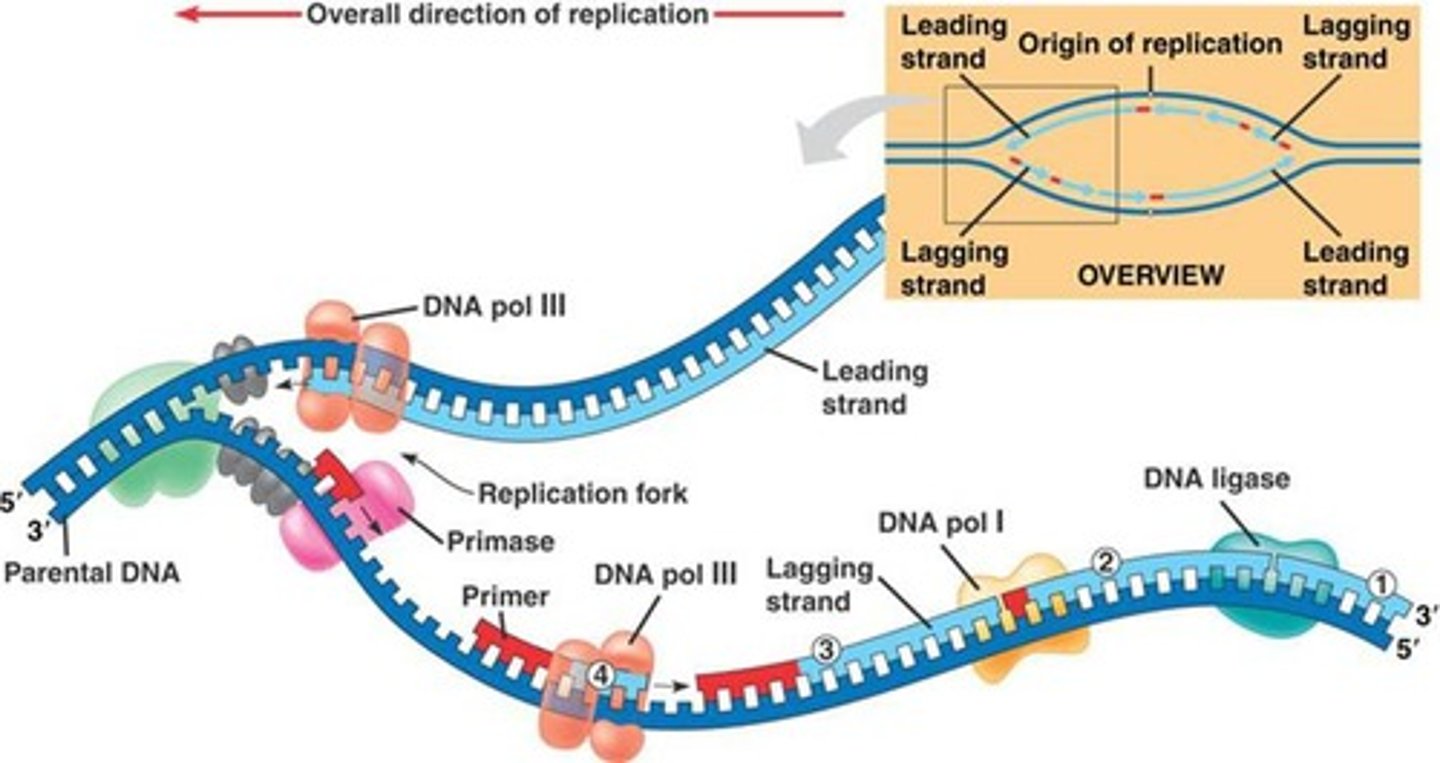
What is the role of RNase H in DNA replication?
Works of DNA poly I to remove and replace RNA primers with new DNA nucleotides, however, does not joint the Okazaki fragments of the lagging strand yet (DNA ligase)
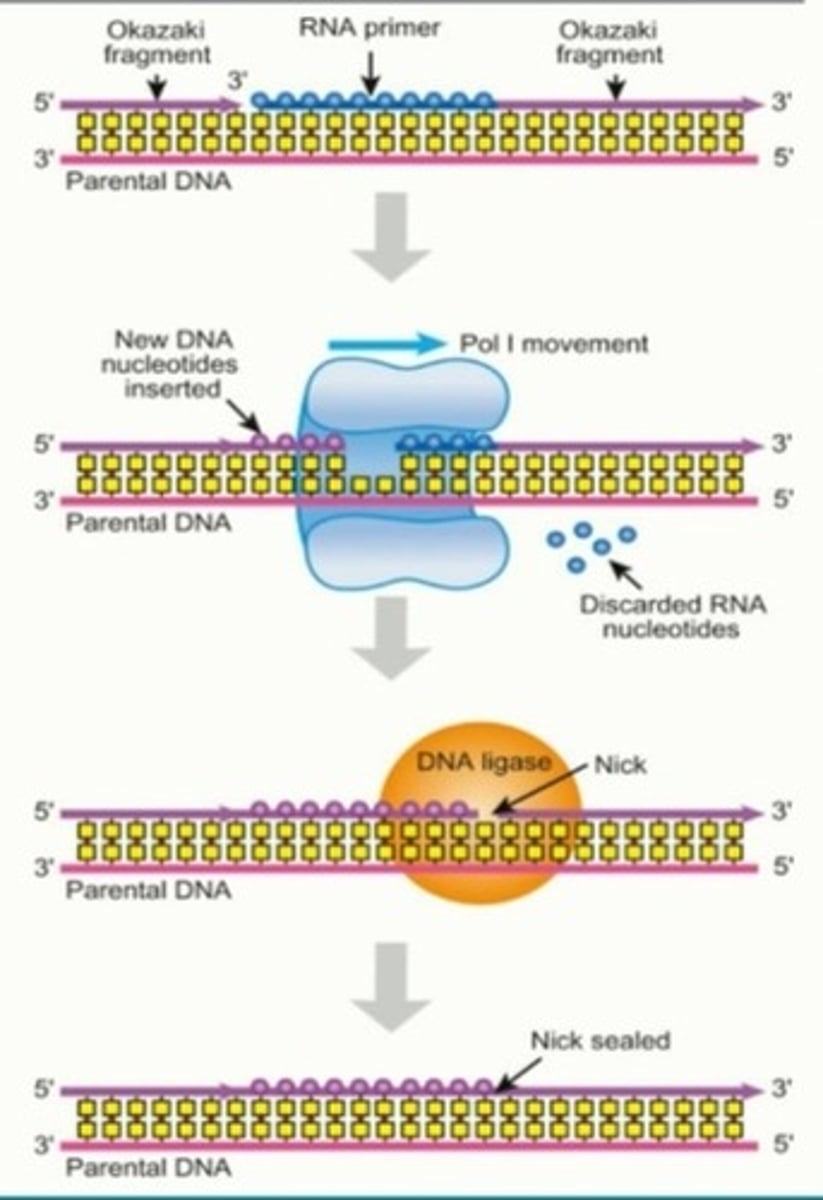
What is the role of DNA ligase in DNA replication?
Creates phosphodiester bonds between the DNA Okazaki fragments of the lagging strand, the final step of DNA replication
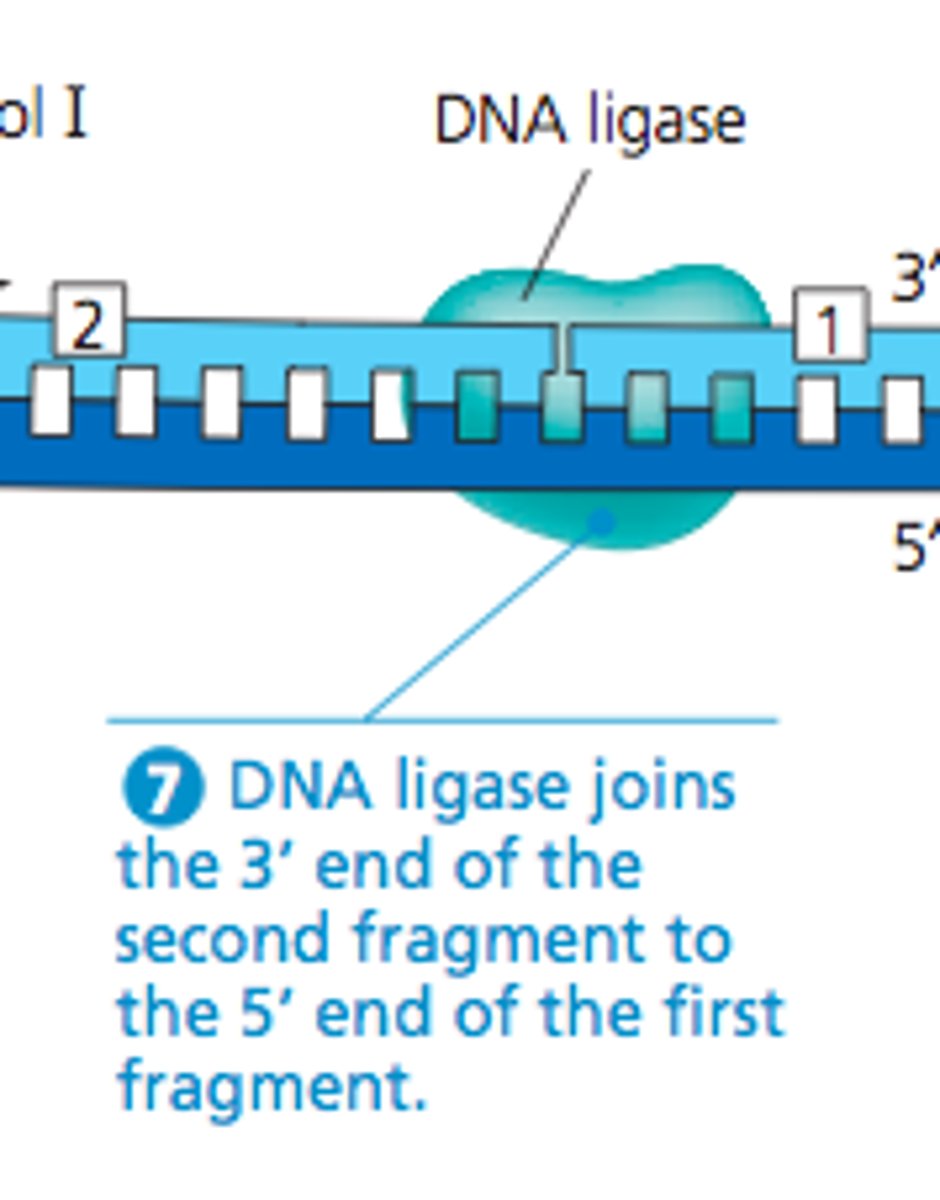
What are the differences between DNA replication on the leading strand and on the lagging strand?
On the leading strand, DNA polymerase III creates one continuous strand as it is synthesizing DNA in the same direction as the DNA is being unwound
On the lagging strand, DNA polymerase III has to stop at RNA primers due to travelling in the opposite direction as the DNA is being unwound, causing Okazaki fragments to form
Multiple RNA primers are inserted onto the lagging strand of the DNA
Only a single RNA primer is required on the leading strand of the DNA (being synthesized in the same direction as DNA is being unwound -> reading 3' to 5')
DNA Poly I, RNase H, and DNA ligase are required to remove the RNA primers and connecting the Okazaki fragments together on the lagging strand
What is the difference between cell division in bacterial cells and eukaryotic cells?
Bacterial cells can replicate very quickly and do not require a lot of resources
Eukaryotic cells require many more resources and time to replicate DNA and make a copy of the cell
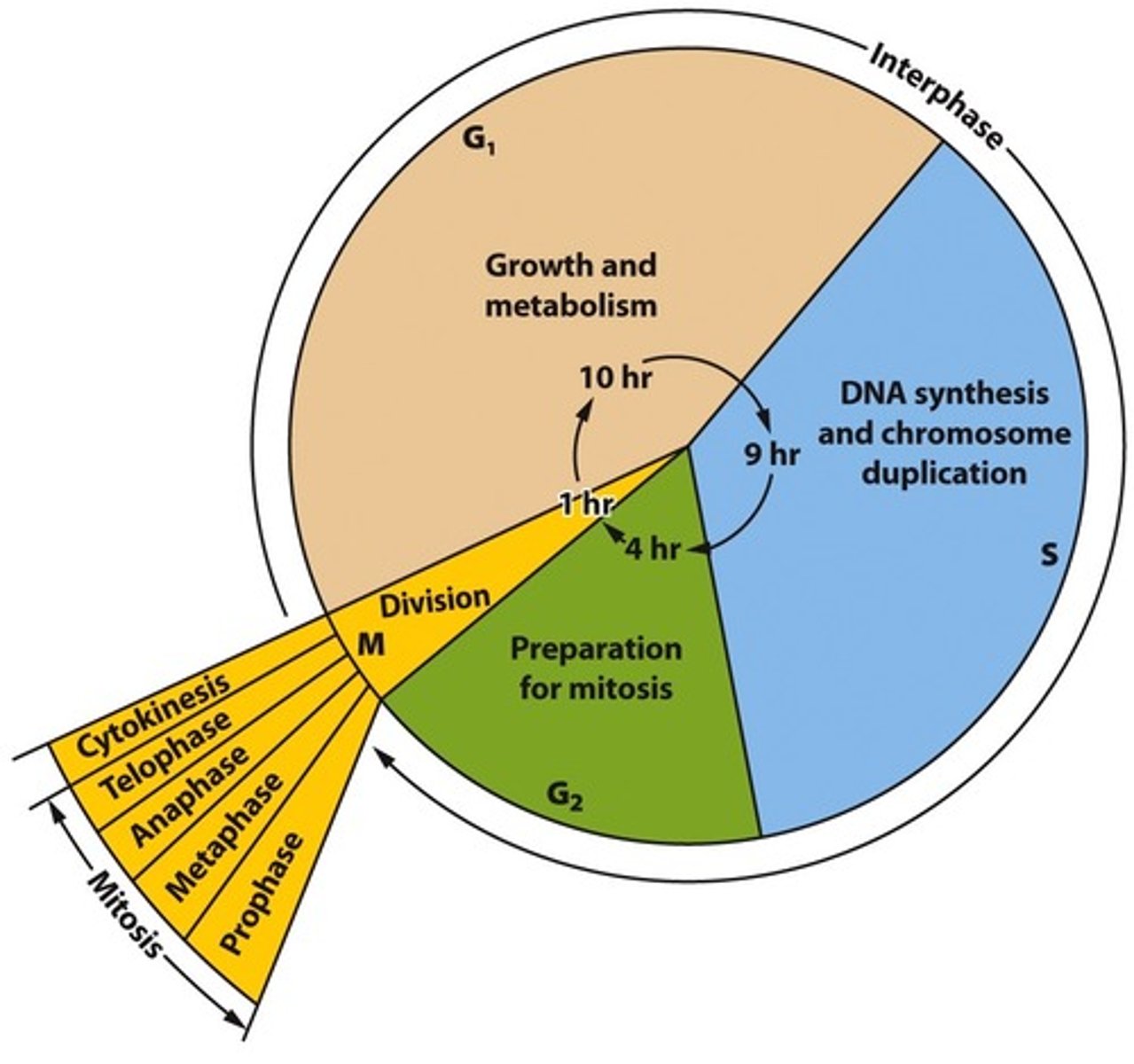
What occurs in the G0, G1,S,G2, and M phases of the cell's life cycle?
G0 phase-> Resting phase, no replication or growth occurs (some cells may skip this phase)
G1 phase-> Growth and metabolism, the cell prepares the necessary resources for division and DNA replication
S phase-> DNA replication occurs in the cell
G2 phase-> The cell prepares the necessary resources for cell division
M phase-> Mitosis, the cell divides
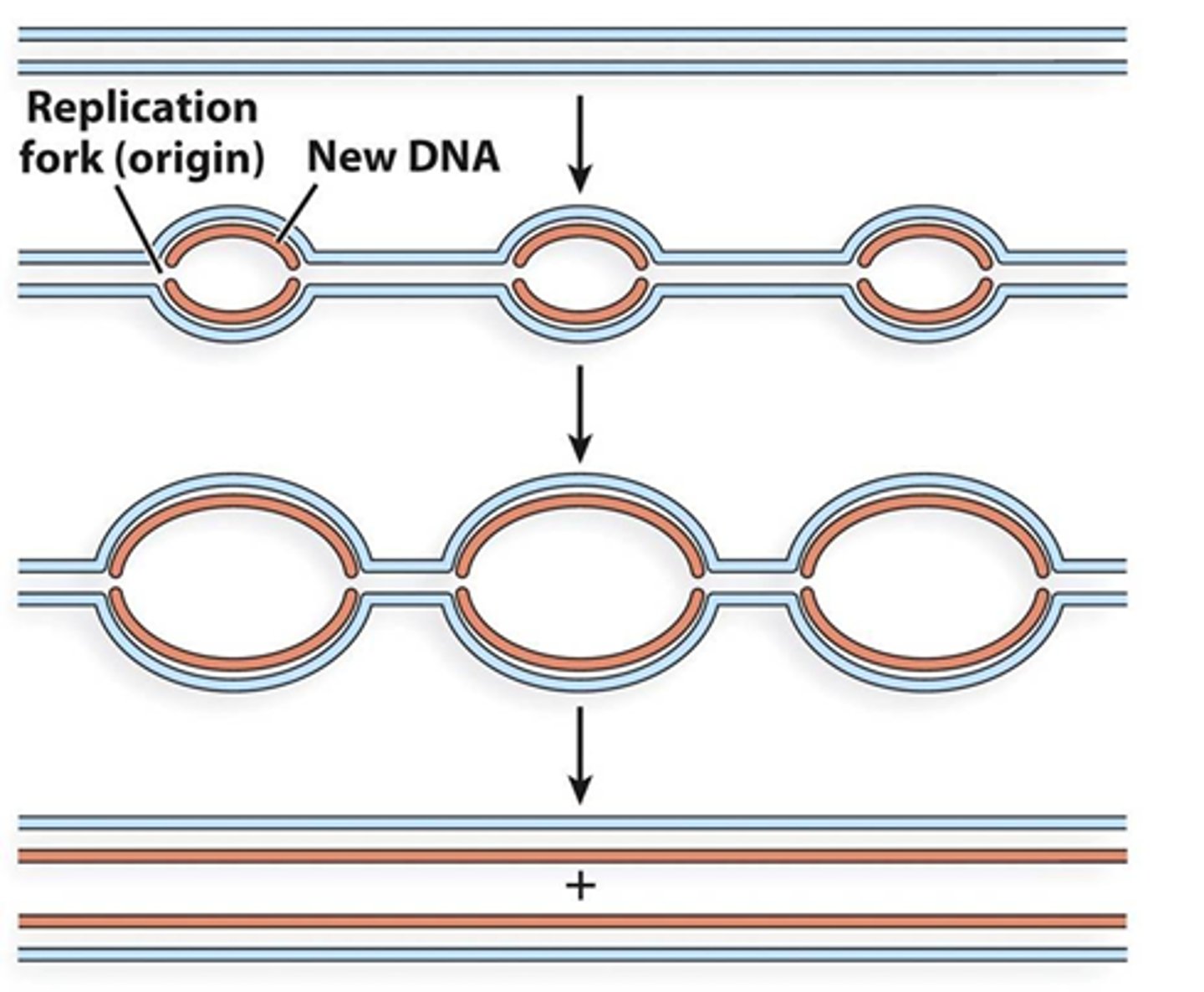
How is DNA replication sped up in eukaryotic DNA?
Due to the sheer size of the genome of eukaryotic DNA, multiple origins of replication occurs in eukaryotes via replication bubbles, which forks in each direction along the DNA, and replicates until they meet up with other replication bubbles
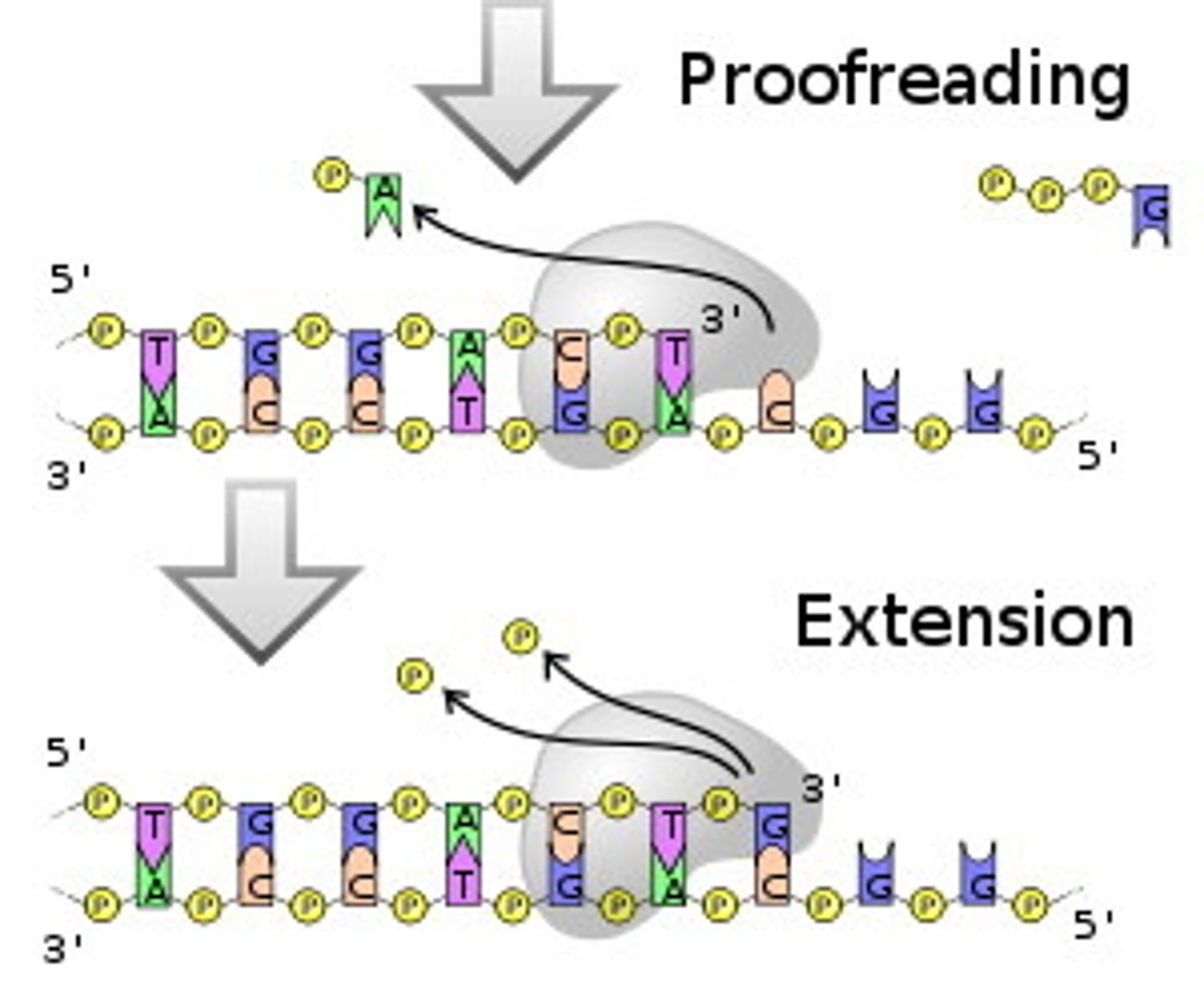
What is the role of exonucleases in DNA polymerases in eukaryotic organisms? Which DNA polymerases contain exonucleases?
Prevents mistakes from happening in DNA replication by removing wrong nucleotides (proofreading)
The follow DNA polys contain exonucleases:
DNA polymerase gamma
DNA polymerase delta
DNA polymerase epsilon
What is a Polymerase Chain Reaction (PCR) and what is it used for?
Using DNA replication enzymes to quickly replicate DNA in a lab setting, amplifying the number of copies of a piece of DNA to detect it in a small sample
A PCR can be used to screen for bacteria or viruses, or sequence DNA in order to look for mutations and ancestry

What "mistake" occurs at the end of normal DNA replication?
At the end of the lagging strand during DNA replication, there is a gap at the end of the lagging strand after the last RNA primer is removed.
Since DNA polymerase requires RNA primers to perform DNA synthesis, this gap cannot be filled.
Upon repeated replication, DNA information will be lost
What are telomeres on DNA?
Nucleotide repeats at the ends of linear chromosomes, which prevents crucial DNA information from being lost on the lagging strand.
However, these telomers become shorter each time replication occurs, and eventually, important parts of the genome get lost and the cells die
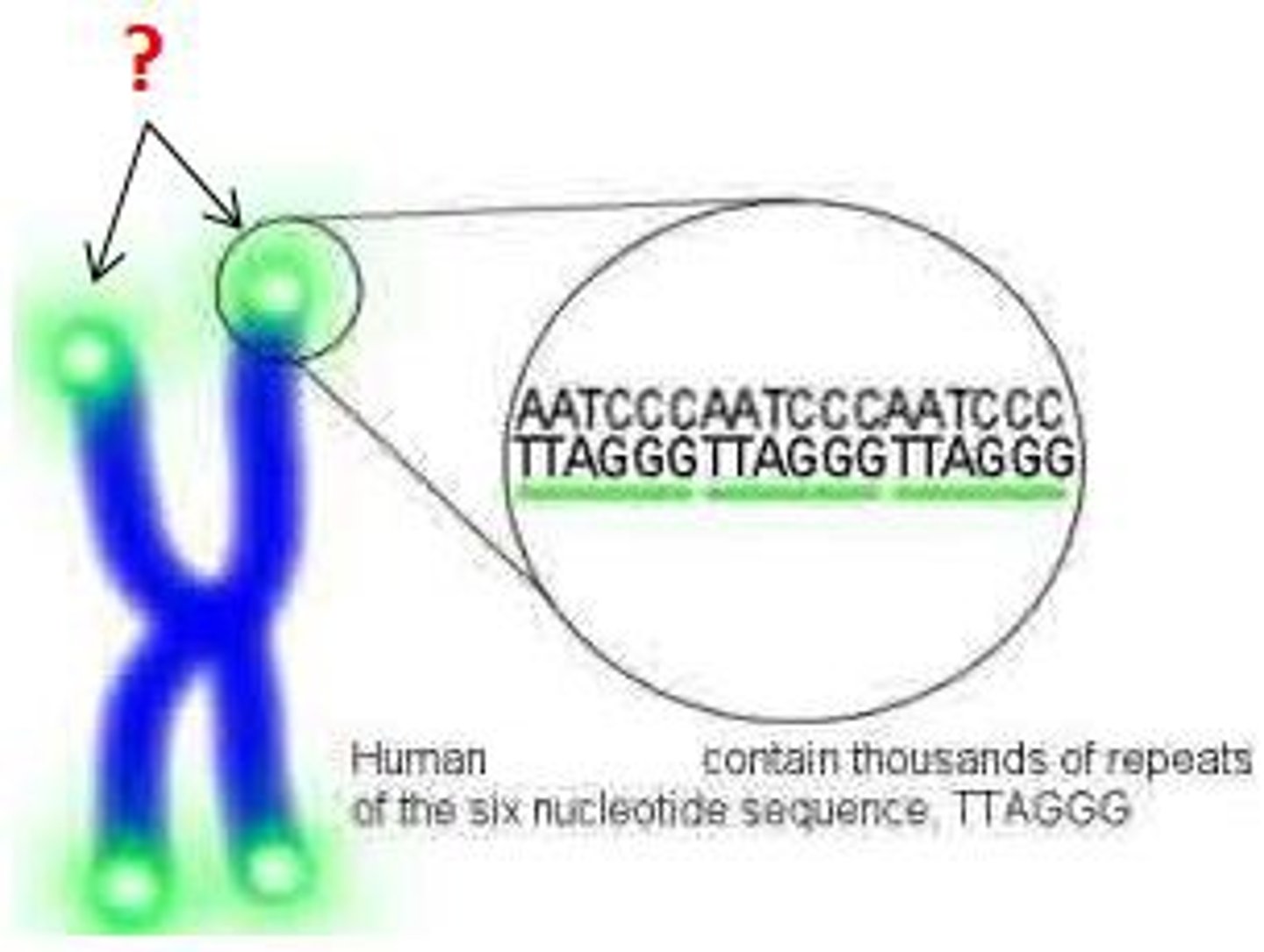
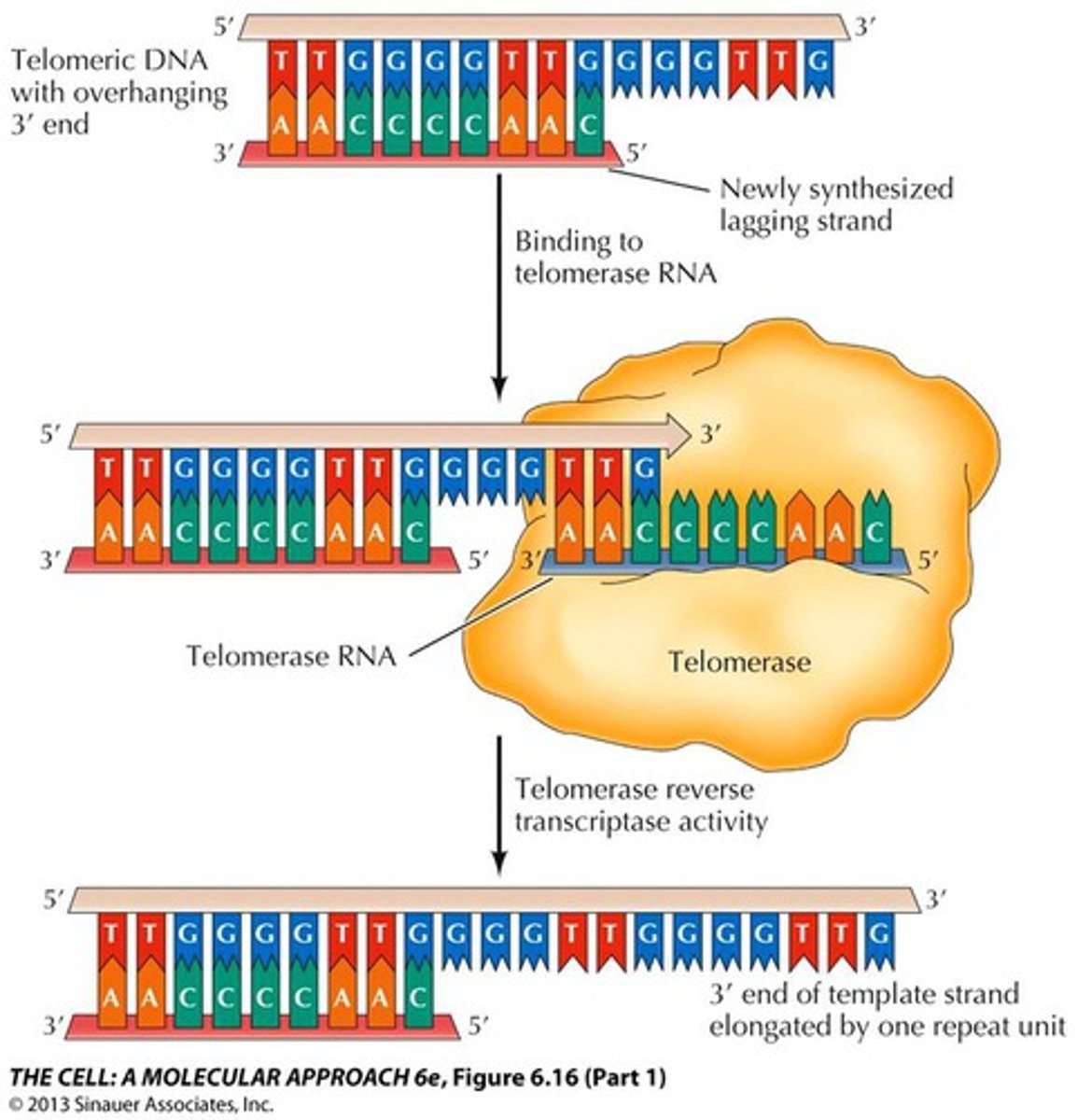
What is telomerase?
An enzyme, which contains an RNA primer specific to the telomere, which can thus lengthen the ends of telomeres, "immortalizing" cells
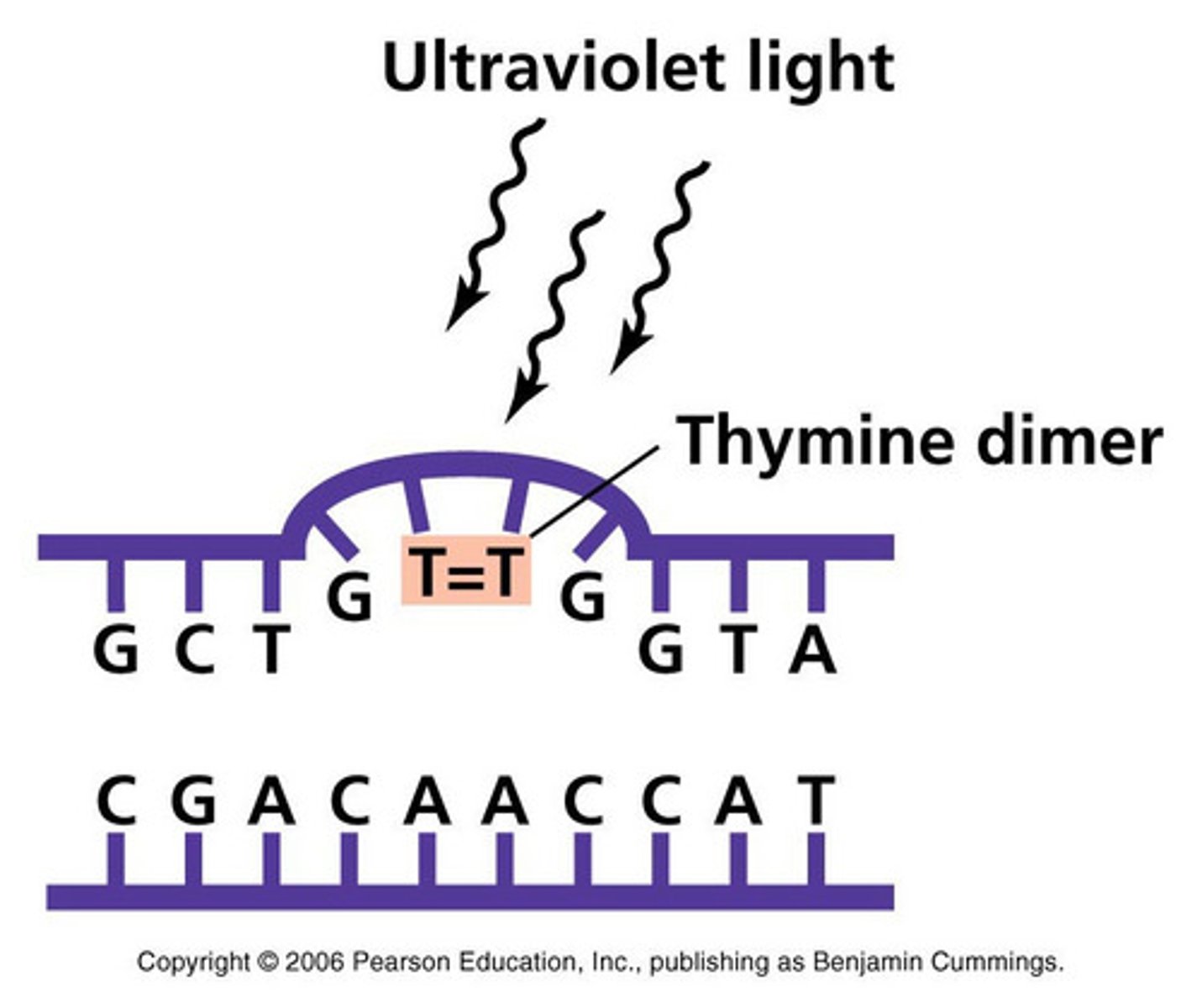
What are examples of environmental agents that can introduce mutations to DNA?
Cigarettes/smoking, UV radiation (causing thyine dimers, thymine binding to each other, stopping DNA synthesis)
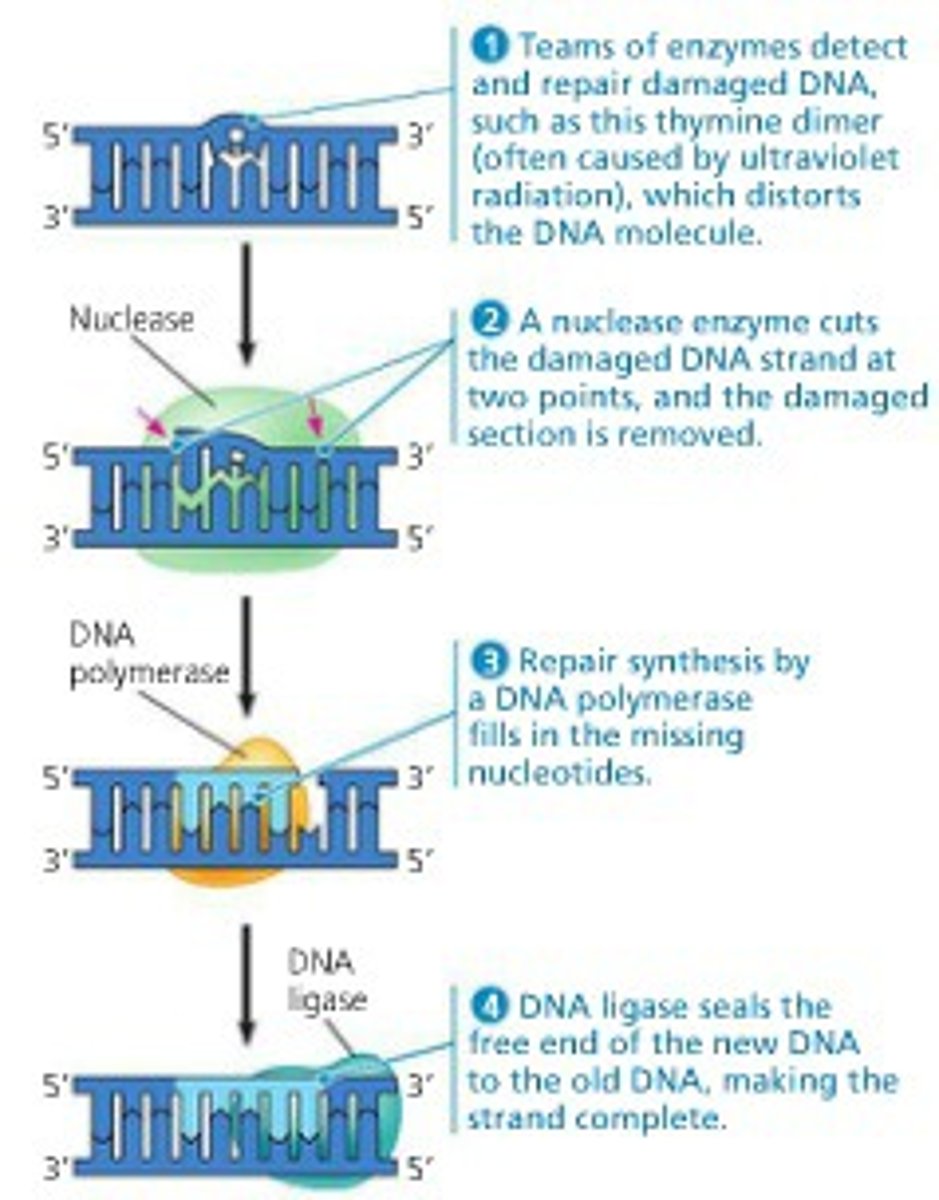
What are the general steps of DNA repair in eukaryotic cells?
1. Damaged DNA is recognized by a protein
2. Enzymes are recruited to break the phosphodiester bond and remove the damaged area of DNA
3. DNA polymerase rebuilds the damaged area
4. DNA ligase re-seals the phosphodiester bonds
How do proteins recognize the parental strand of DNA during a mismatch repair of DNA?
The original parental strand is methylated (contains CH3) - epigenetic marks, thus the nucleases are able to recognize the parental strand and instead are able to bind to the correct "new" strand
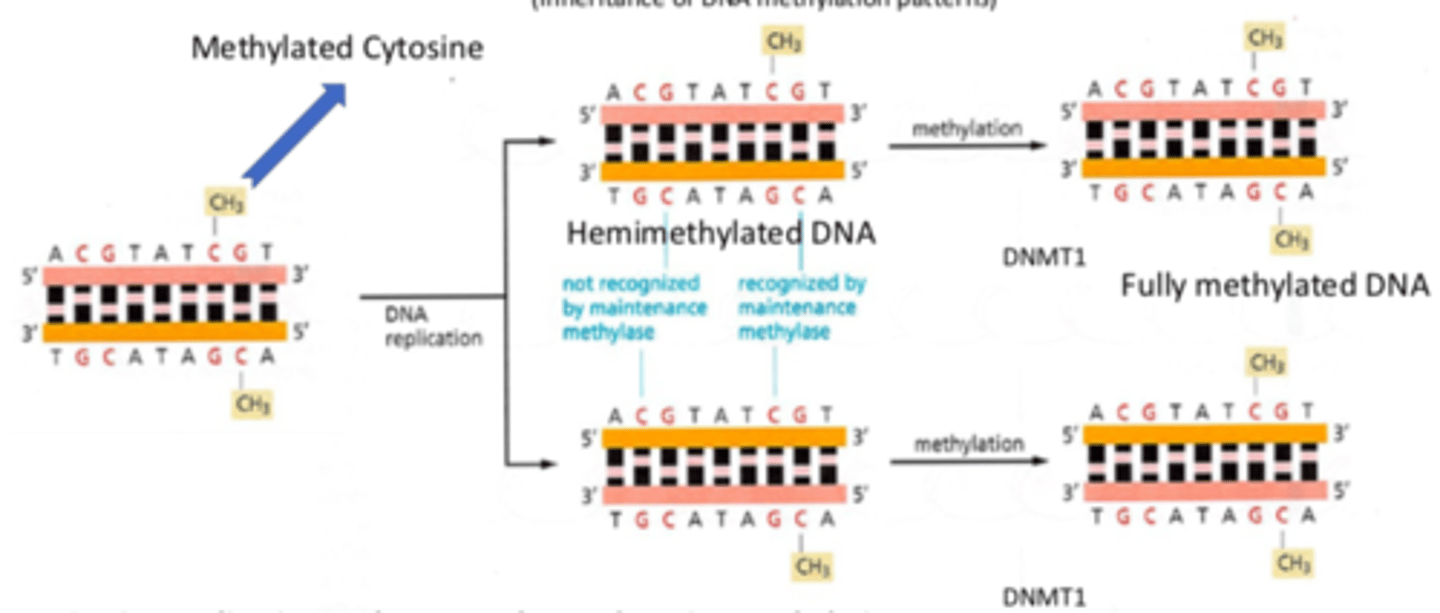
How is a repair in a DNA mismatch done in prokaryotic cells?
1. Nucleases remove bases in area around the un-methylated DNA mismatch (new strand)
2. DNA polymerase rebuilds damaged area
3. DNA ligase re-seals the DNA
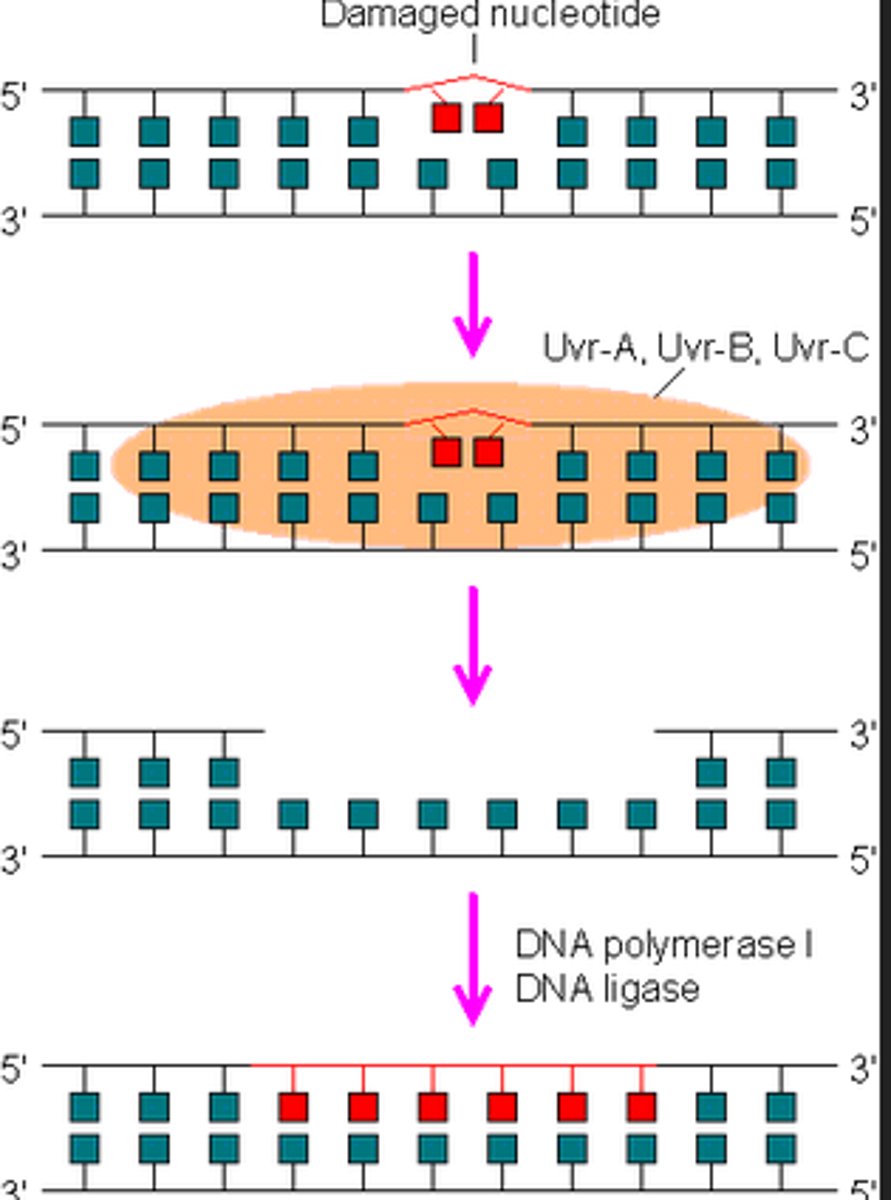
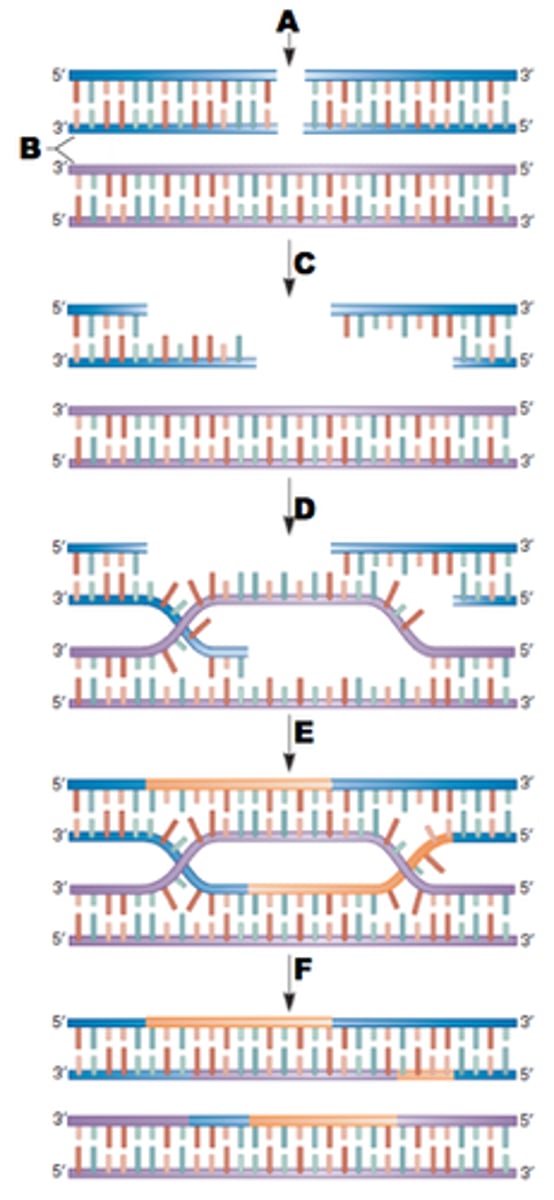
What are two methods in repairing DNA breaks in double stranded breaks? Which one is less error prone?
1. Nonhomologus end joining - joining the two ends of the breaks, basically hoping that nothing bad happens (more error prone)
2. homologous recombination -> another piece of DNA is used as a template in order to put the correct nucleotides into the break (less error prone)
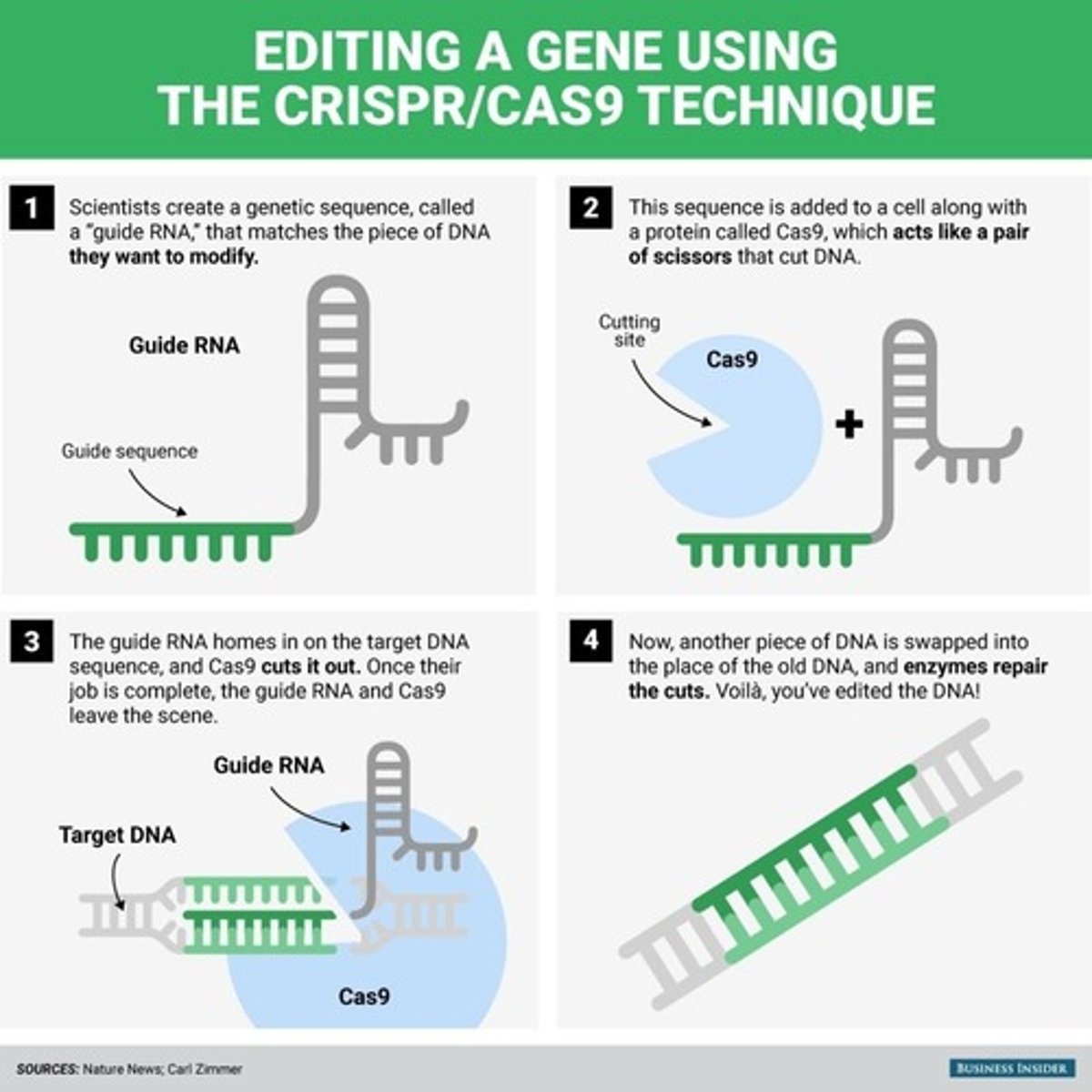
What is CRISPR? How does it work?
System found in bacteria that can be used to edit DNA
Consists of a nuclease - Cas9, which cuts the DNA at a precise location, causing a double-stranded break, which can then be repaired through homologous recombination or non-homologous end joining
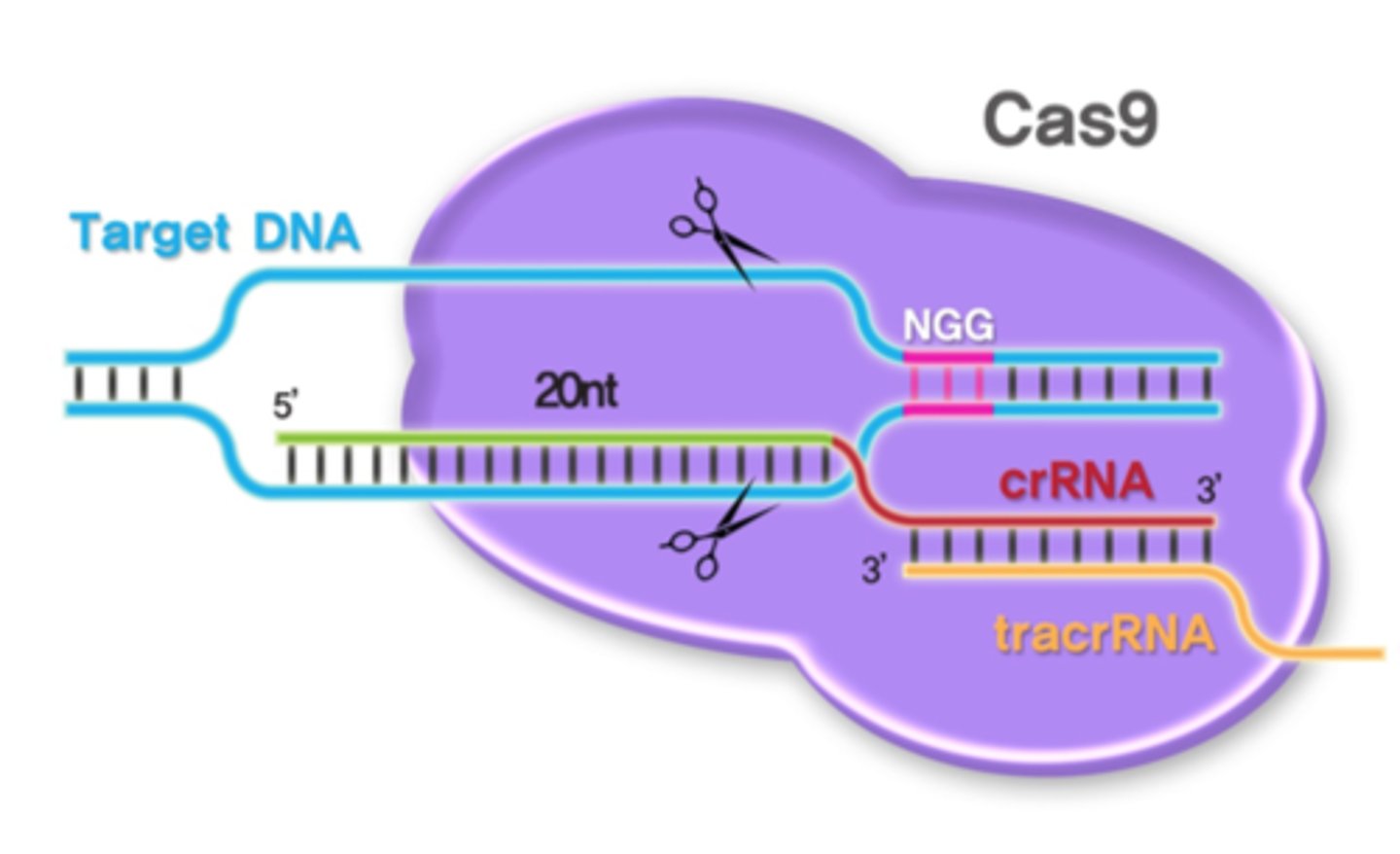
What is the basis of genome editing?
Precise changes to DNA can be made by using the excision capabilities of Cas-9 in CRISPR to make a precise cut in DNA, and then repairing it with a repair template
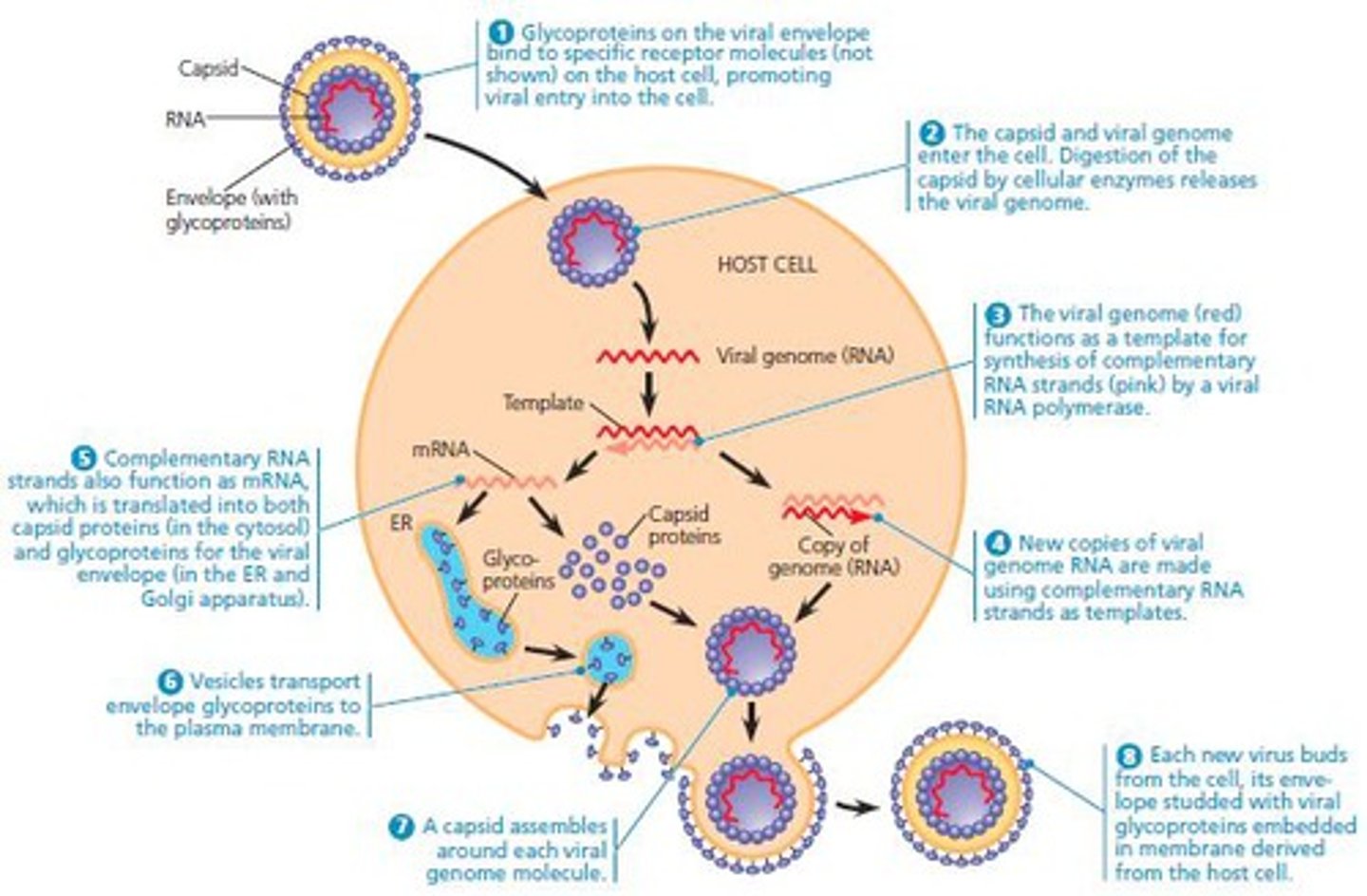
How do viruses replicate themselves?
Often "hijiack" the cell machinery from the host's cells to replicate their proteins by injecting an RNA sequence into the cell
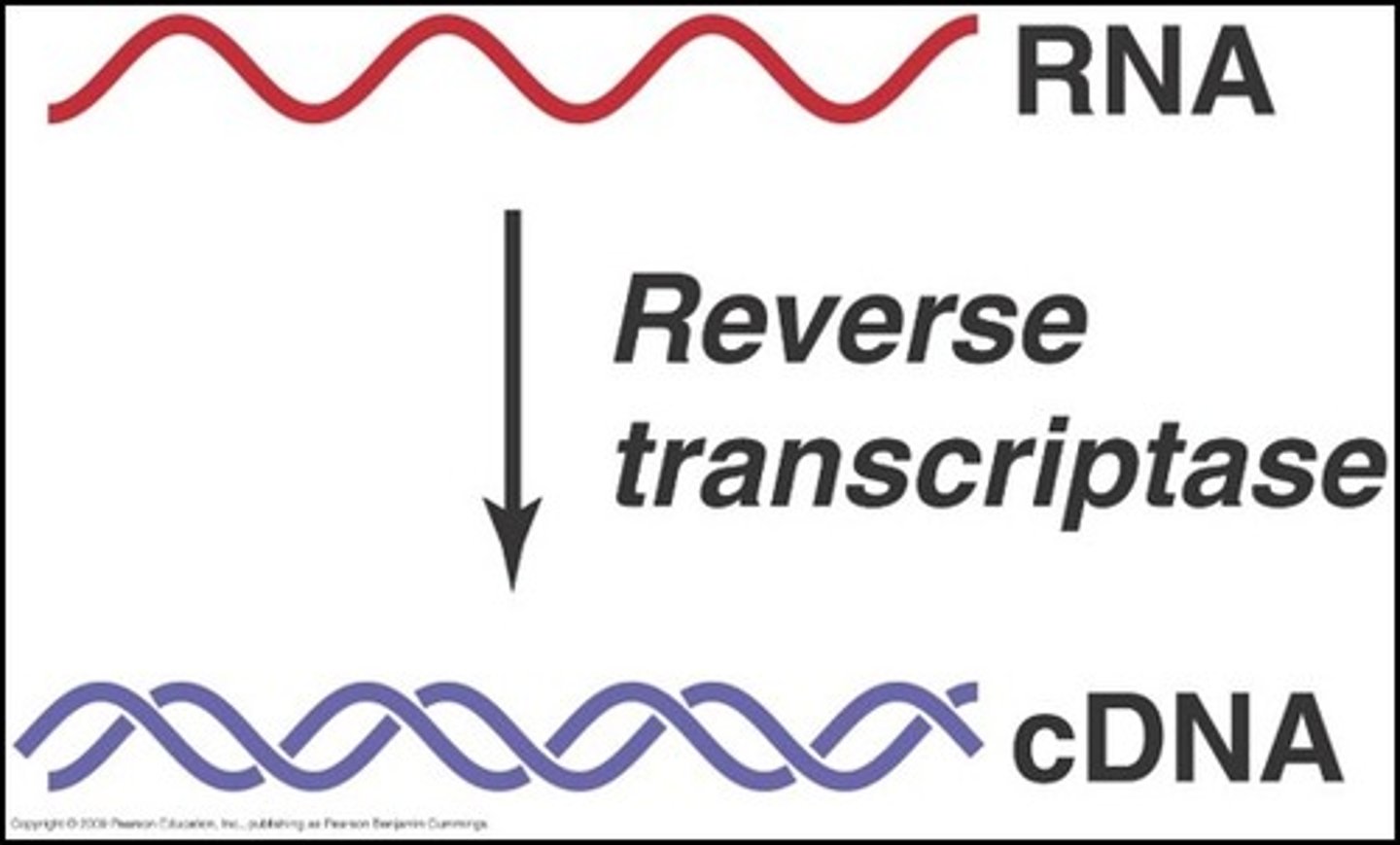
How do retroviruses replicate themselves?
Retroviruses inject an RNA template into the host's cell, which is then converted into DNA via reverse transcriptase, which creates a single-stranded, complementary DNA (cDNA)
Note that reverse transcriptase does not contain a proofreading function, therefore, many errors are made
How is norfloxacin capable of inhibiting bacteria? (CS)
Inhibits gyrase (the bacteria version of topoisomerase), which causes DNA to supercoil during replication
Norfloxacin has no effect on the host's cells