L3: Gene Regulation in Prokaryotes - how & why?
1/33
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
34 Terms
Why regulate gene expression?
Rapid adaptation to new environments
Conservation of energy - avoids unnecessary simultaneous expression of counter productive pathways being used when not needed
Correct timing of differentiation
Appropriate stress response and maintaining homeostasis within the cell.
what are examples of signals bacteria receive from the environment that affect how they respond?
Changes in pH
Temperature
Nutrients
Oxygen
Osmolarity

what are examples of responses bacteria do in relation to changes in the environment?
Stress responses i.e formation of spores
Virulence responses i.e invasion
Metabolic responses i.e change in nutrients
Transcriptional regulation essentially_____
effects how the RNA pol binds to the promoter and their ability to create a strand of mRNA
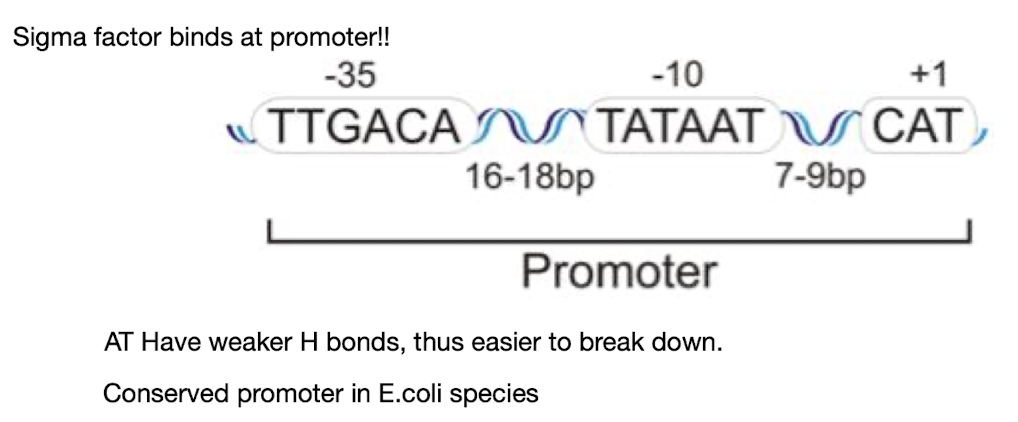
What is the general sequence of the bacterial promoter? (review)
The general sequence of the bacterial promoter is typically recognized by RNA polymerase and consists of two conserved regions,
the -10 (Pribnow box) key for unwinding DNA
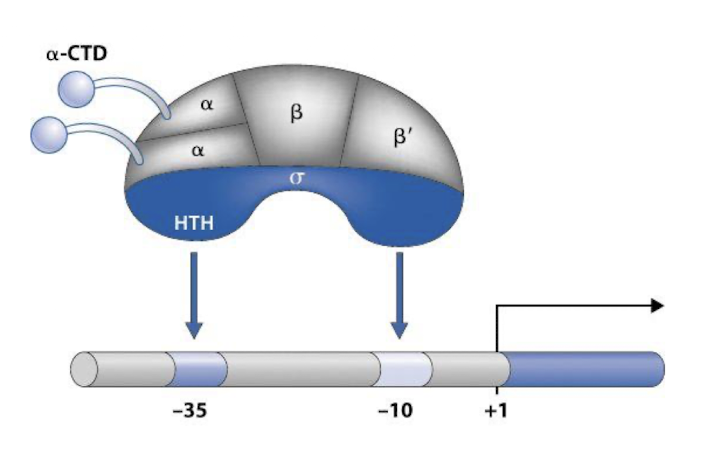
-35 elements, crucial for initiating transcription as this is the sequence that the σ factor binds to
Sequence and spacing can vary!
What is meant by promoter strength?
Basic form of regulation that dictates the efficiency of transcription initiation by RNA polymerase, influenced by the sequence and spacing of elements within the promoter region.
Differences in ________ sequence can lead to differences in amounts of _____ produced
promoter, mRNA
___________ genes have weak promoters where structural more _______ genes have stronger promoters
Tightly regulated genes
Constitutive genes eg housekeeping genes, are expressed at constant levels regardless of environmental conditions.
What are the various sigma factors and their key features?
σ 70 - Housekeeping, most common σ factor, found in healthy states
σ 54 - N limitation
σ 38 - Starvation stress
σ 28 - Flagellar sigma factor
σ 24 - Extreme heat
σ 19 - Iron cycle
Each factor allows for the selective transcription of specific sets of genes essential for survival under different environmental / stress conditions
What is the benefit of prokaryotes using different σ factors?
They can induce transcription of different genes from different promoters
What is the result of altering conserved sequences?
You can change the relative activity of the promoter and affect gene expression levels.
What is an example of a catabolic pathway that is regulated by genes that are substrate-induced?
Benzoate degradation is induced by benzoate:
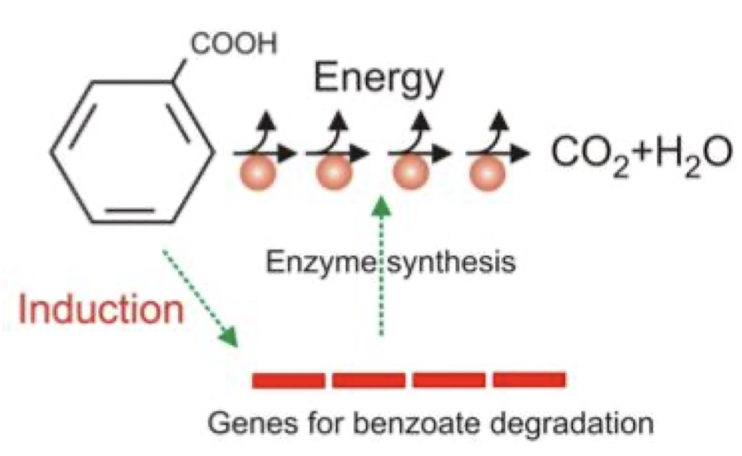
*IF NO benzoate is present, no need to “turn on” genes for when the substrate is absent
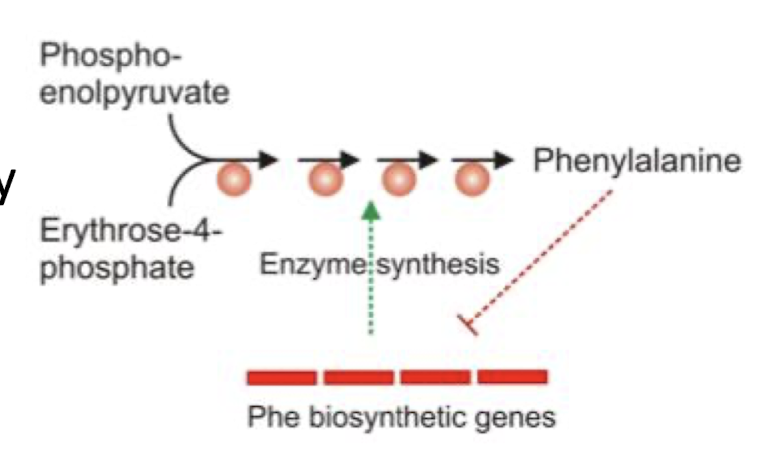
What is an example of a anabolic pathway that is regulated by genes that are substrate-induced?
Amino acid synthesis eg Phenalanine is induced by the presence of the respective amino acid precursor and REPRESSED when Phenylalanine is present , no need to have gene turned on to produce it when levels are sufficient
What are the two types of transcriptional control?
Negative control - regulatory protein is a repressor, binds to DNA inhibiting transcription
Positive control- regulatory protein is an activator stimulating transcription
In positive transcriptional control, the regulatory protein involved is an ____ which binds to DNA (typically a site other than the operator) , stimulating transcription
Activator
E.g In the lac operon, CAP binds to the promoter and increases the efficiency with which RNAP can bind to the promoter and transcribe the structural genes
__________ are those in which transcription is normally off, thus something must change to induce transcription / turn it on
Inducible operons
__________ are those in which transcription is normally on, thus something must occur to repress transcription / turn it off
Repressible operons
How do Repressors function?
In plain terms, repressors binds to the promoter so the RNA pol cannot bind, thus transcription is not initiated, which prevents gene expression.
Stop RNA Pol from binding
Prevent it from leaving the promoter sequence i.e binding to the -35 site
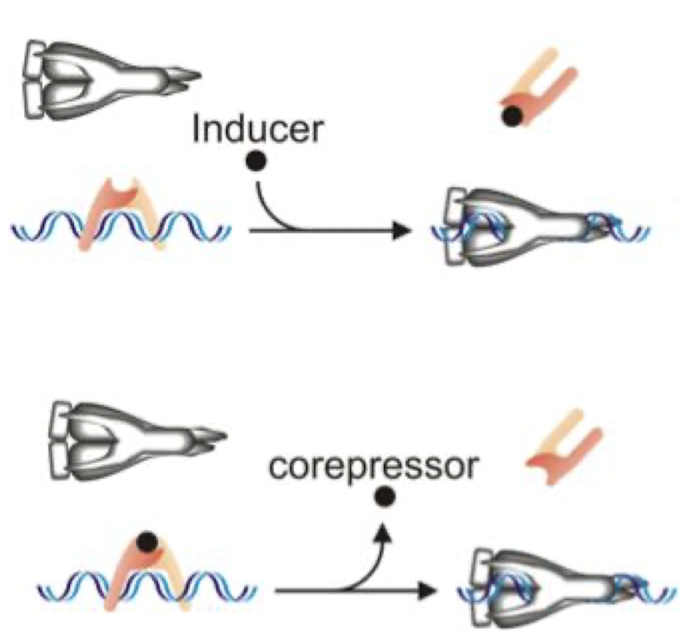
Repressors are often _____ which are controlled by ________
allosteric, controlled by small molecules
Proteins such as _______ which change shape upon binding to another molecule like ____ are called ________
repressors, DNA , allosteric proteins
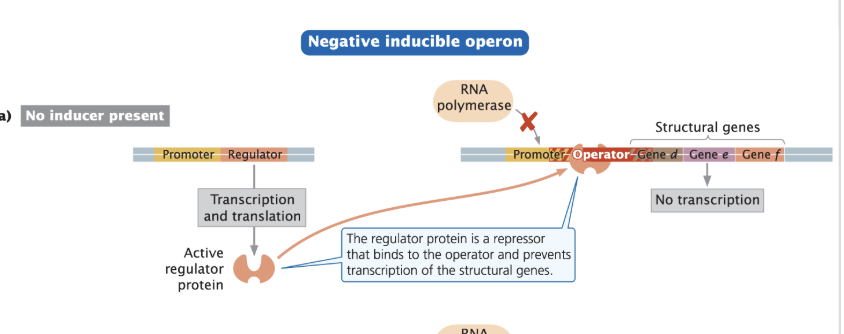
How do Activators function?
Essentially HELP the RNA Pol bind to the promoter instead of stopping it.
Either BEND DNA to make the shape of RNA Pol more attractive to bind to the DNA
Interact with CTD of ⍺ subunits of RNA Pol
Promoters that require activators have poor consensus sequences
Often allosteric (a process where a molecule binds to a protein (like an enzyme) at a site other than its active site, causing a shape change that affects the protein's function)
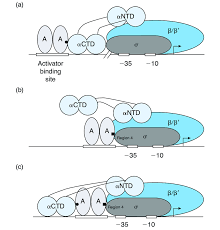
What is the function of the CTD in the alpha subunits of RNAP and how does this aid activator binding to DNA?
Domain is connected to the alpha-NTD by a flexible linker and interacts with DNA as it is highly mobile and can adopt different positions, allowing it to reach out from the core enzyme to interact with specific DNA sequences or proteins i.e. activators
activator-⍺-CTD interaction facilitates the recruitment of RNA polymerase to promoters, stabilises binding and can help induce conformational changes in RNAP to make transcription initiation more favourable
e.g. In the CRP
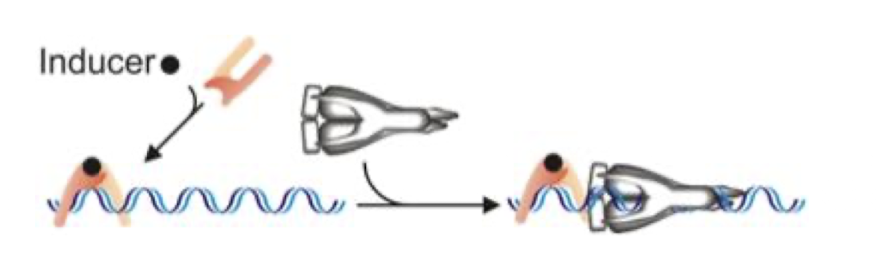
What is allosteric control?
Where the protein conformation changes when a small molecule is bound:
HTH (helix turn helix) can become exposed (protein more likely to bind) or hide it (less likely to bind)
What is an example of allosteric control?
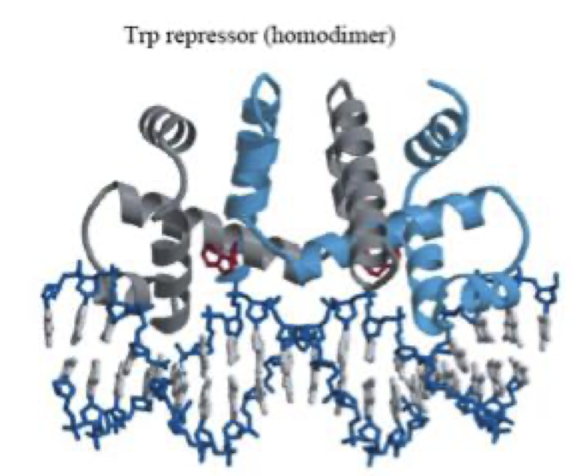
The Trp repressor cannot bind the trp operator DNA in its default state.
When Trp binds to a site on the repressor that is not the DNA-binding site (the allosteric site), it causes a conformational change in the protein.
This conformational change repositions the helix–turn–helix DNA-binding domains, enabling the repressor to bind the operator and block transcription of the trp operon
Induction can be either through ________ or ________
activation or de-repression
Repressor binding sites are generally found at ______ or ______
promoter, downstream
Activator sites generally found ________ or ______
Just before the promoter or further upstream
*harder to know exactly their location due to the fact DNA can bend
________ is a group of bacterial structural genes transcribed together into a single mRNA along with their ______ and ________ that control their transcription.
An operon, their promoter and additional sequences that control their transcription.
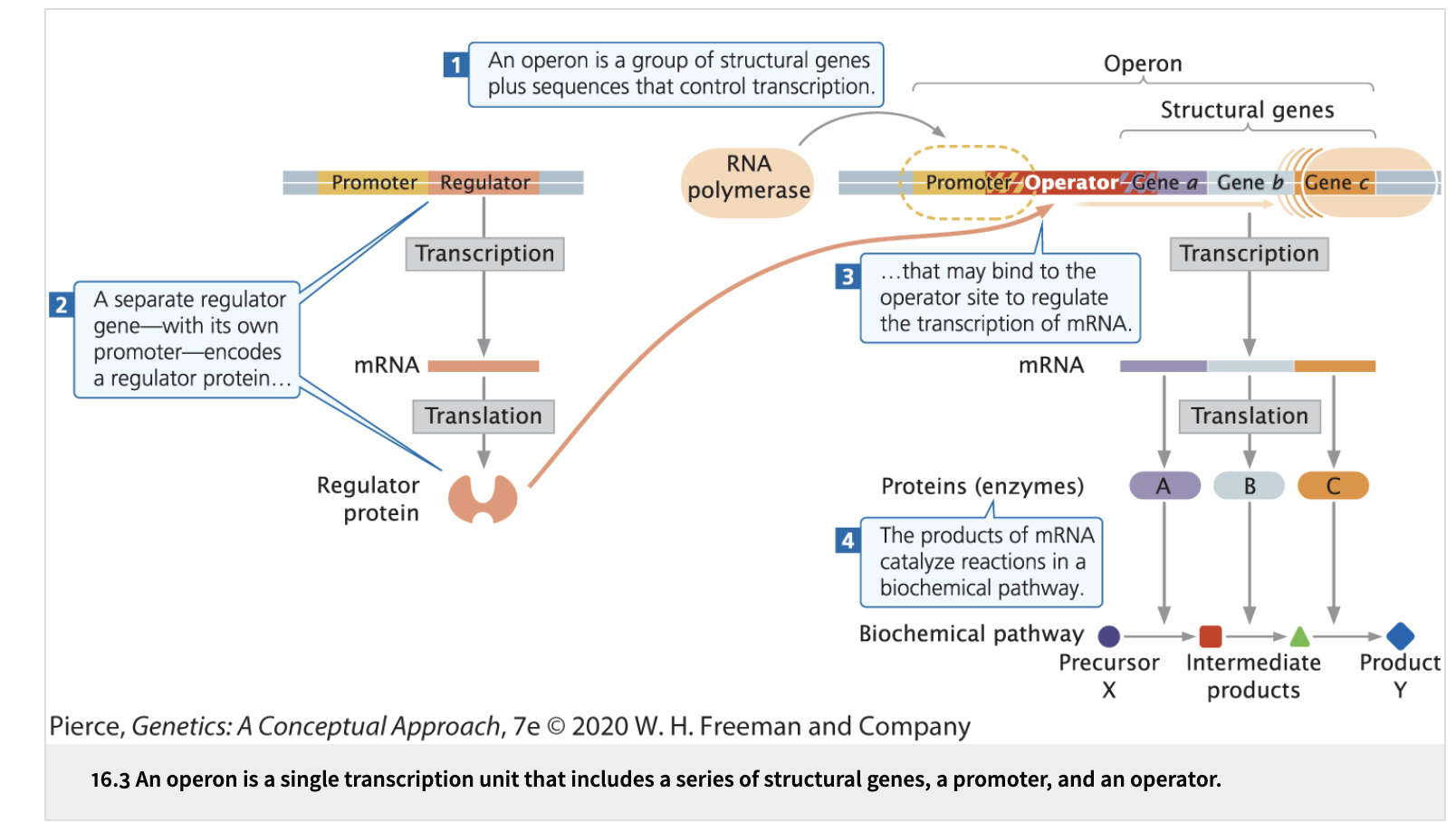
What is the importance of an operon?
Allows for the coordinated regulation and expression of a set of genes that usually work together in a related function.
Each gene is regulated at the same level of transcription
Common in bacteria for genes used in biochemical pathways
“Prokaryotic filing cabinet” - genes put under the same category
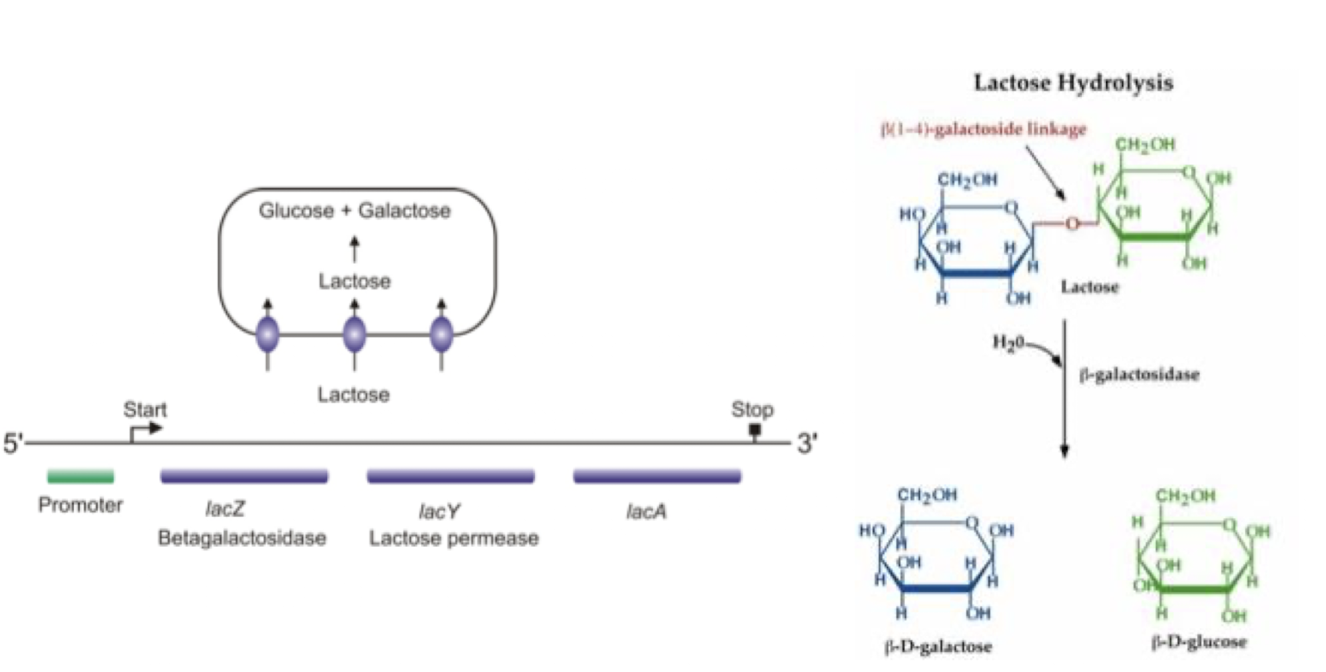
What is an example of a well-defined operon?
The lac operon, which regulates lactose metabolism in E. coli.
Polycistronic
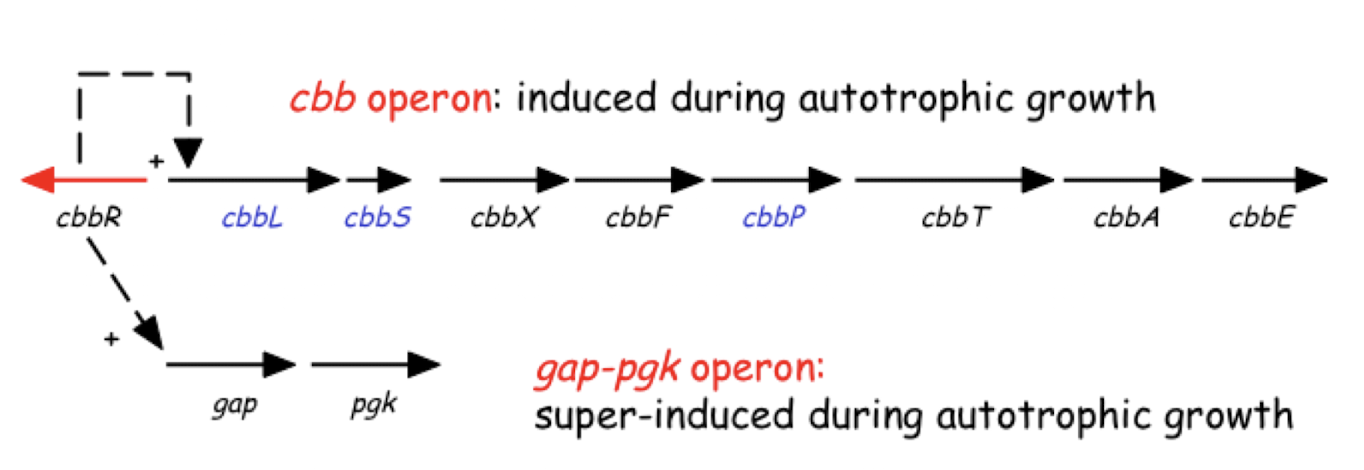
What is a regulon?
A transcriptional regulator controlling more than one operon
They can overlap to coordinate expression e.g. CbbR
How are regulons studied?
In the past individual mutants could be made and their effect on the individual genes would be studied (very time consuming)
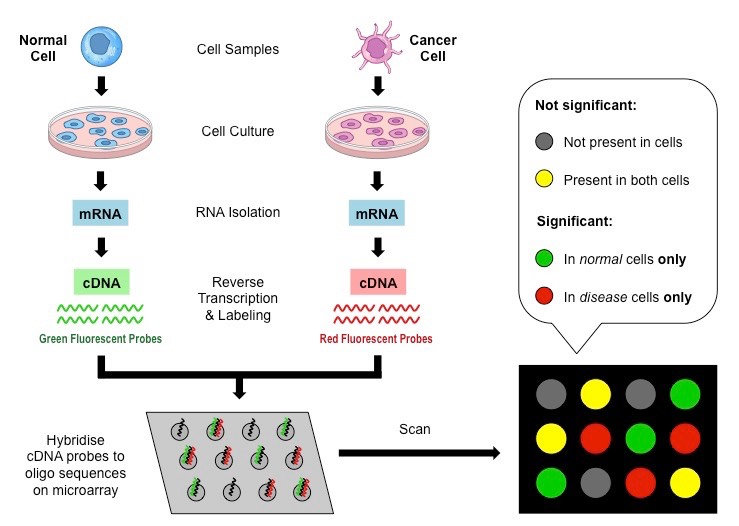
Now, microarray analysis allows for the simultaneous examination of gene expression across multiple operons and conditions, making it more efficient to study regulons and their regulatory networks.
How are microarrays generated?
Label mRNA from bacteria from 2 different conditions:
i.e Wildtype bacteria (red) and Mutant bacteria (green)
Allow to hybridise to a chip containing fragments of DNA from the entire genome
If mostly red, then gene expressed less in the mutant than the wildtype
If mostly green, then gene expressed more in the mutant than the wildtype
• Develop expression profile of entire genomes
what is a transcriptome?
The complete set of RNA transcripts produced by the genome under specific conditions, providing insights into gene expression and regulation in a cell.
Analysed using generational sequencing to identify transcriptomes