LF264 Lecture 7 - B Cell Development and Maturation
1/30
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
31 Terms
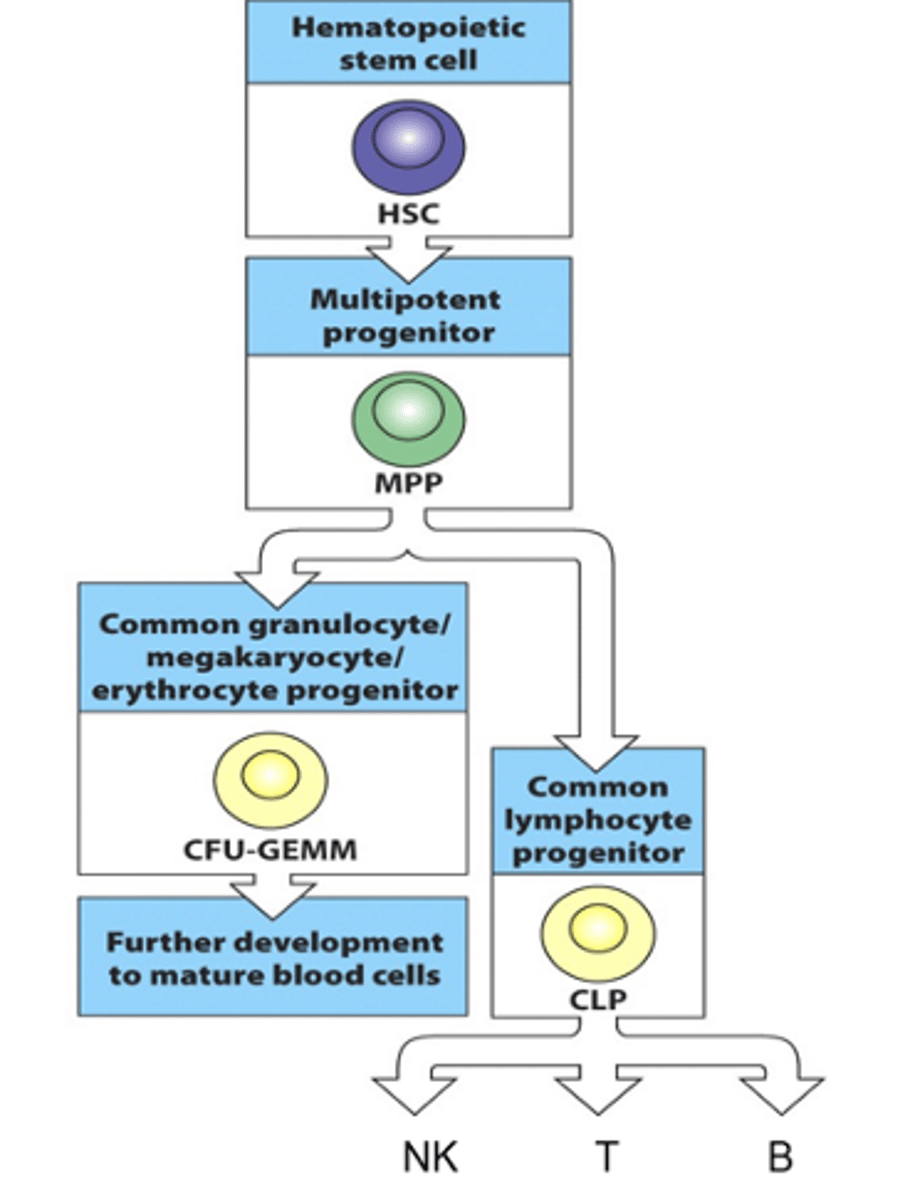
How do B cells develop? What are these stages called?
- Derived from haematopoietic stem cell in the bone marrow
- Haematopoietic stem cell goes through stages of restriction
--> As cell goes through decision trees/differentiation steps, they are restricted as they become specific and fully commited to being a certain type of cell
--> Can't go back and change their mind so are restricted to the function that was decided
- Haematopoietic cell develops to a multipotent progenitor (MPP)
- MPP then develops into a common lymphoid progenitor (CLP)
- CLPs then develop into NKs, T cells and B cells
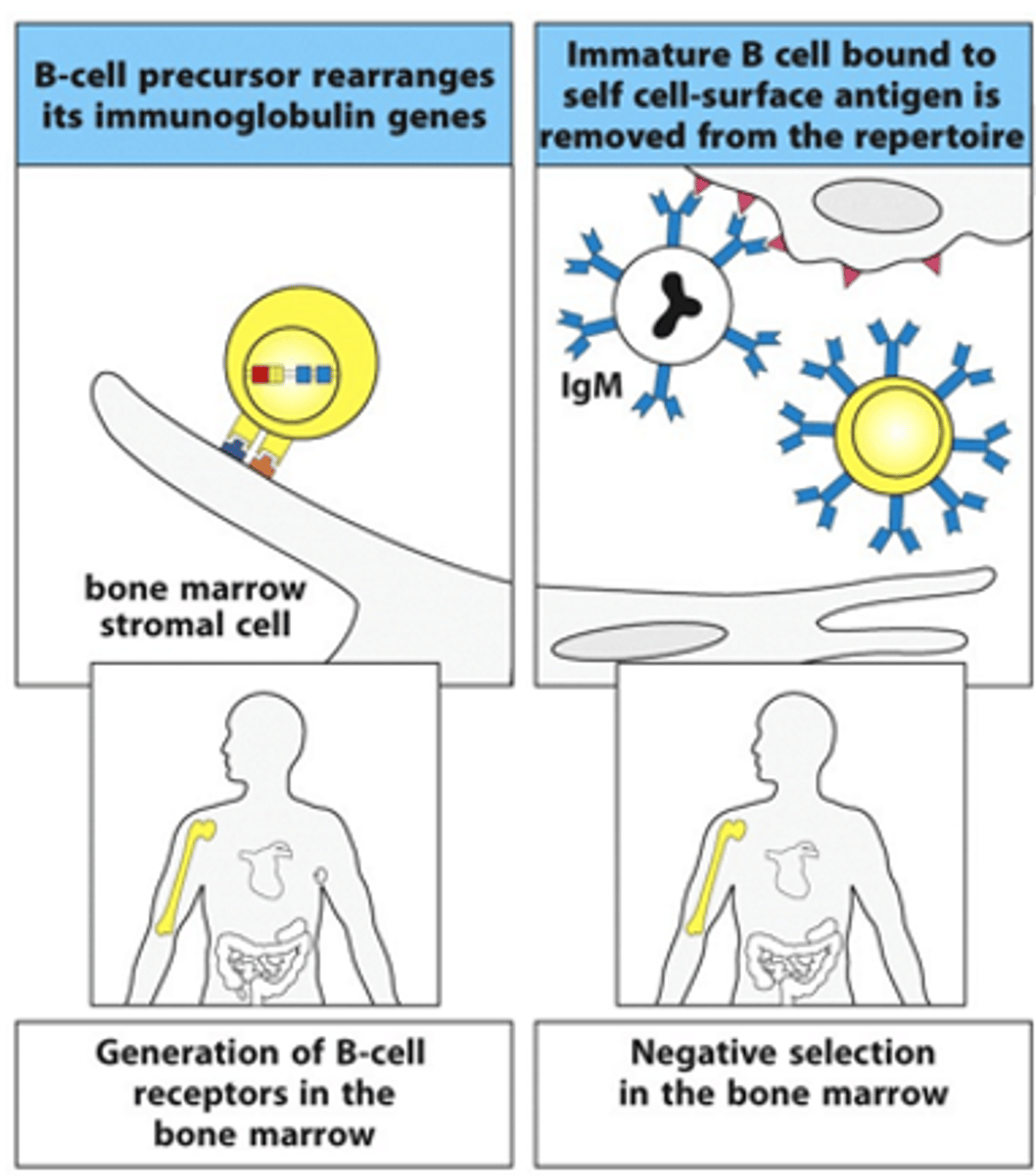
Describe the 1st phase in the B cell life cycle
- Starts in the bone marrow
- B cell precursor interacts with the bone marrow stromal cells
--> Stem cell niche in bone marrow is very important (lots of microenvironments and lots of things that help it grow, e.g growth factors)
--> Gene segment rearrangement starts to occur
--> B cell starts to generate surface receptor (1st class of this is always IgM)
- These receptors get tested on self-antigens:
--> If it recognises it, cells are given a negative selection signal and then the cells goes through apoptosis
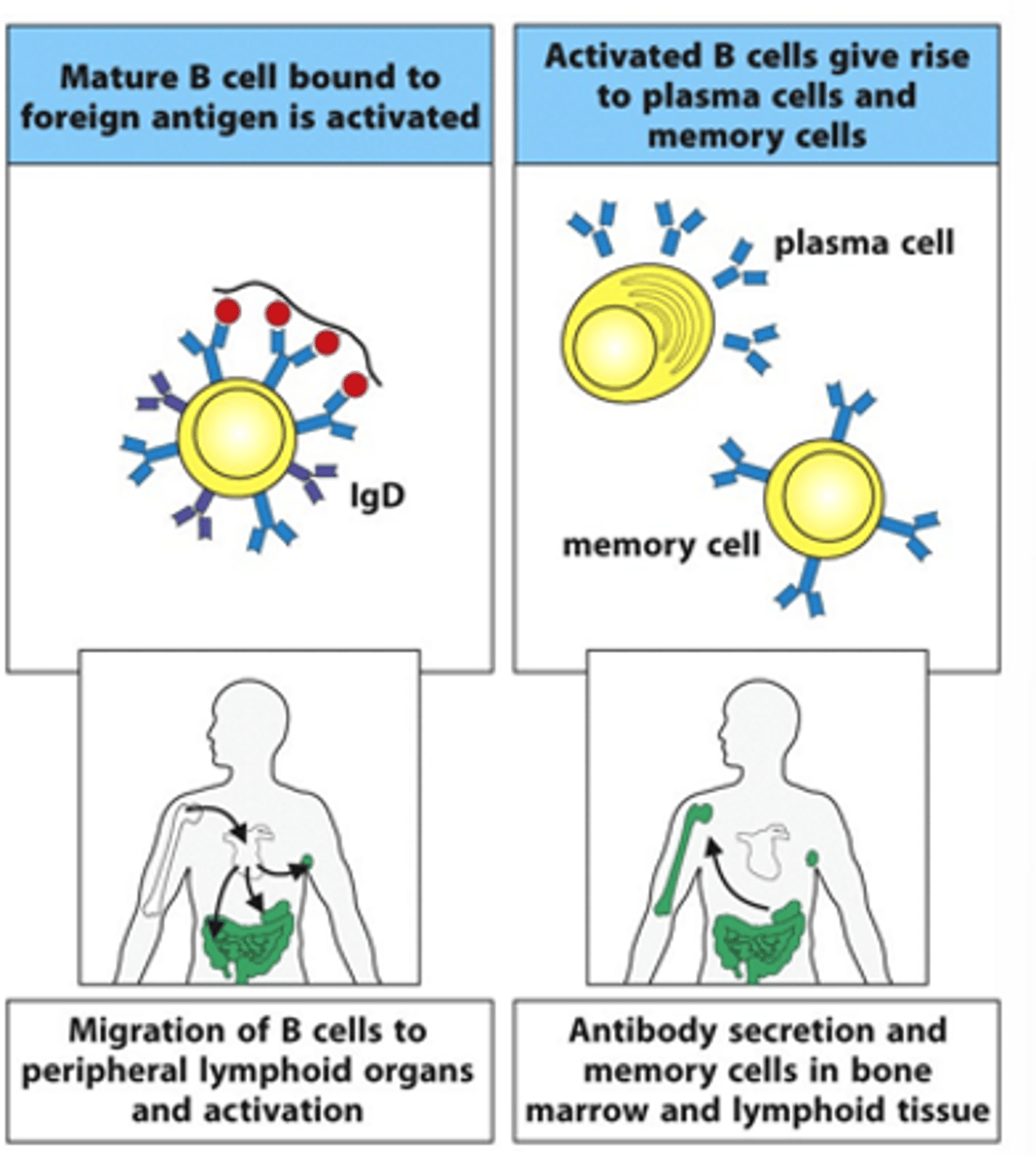
Describe the 2nd phase in the B cell life cycle
- B cell migrates from the bone marrow into the circulation and makes its way to secondary lymphoid organs, e.g lymph nodes, lymphoid follicles in gut
- If B cell then sees its cognate, 3D antigen that it recognises, it becomes activated with the help of T cells
--> They then become antibody-secreting plasma cells
--> Secrete surface receptors as antibodies
--> Also produce memory cells
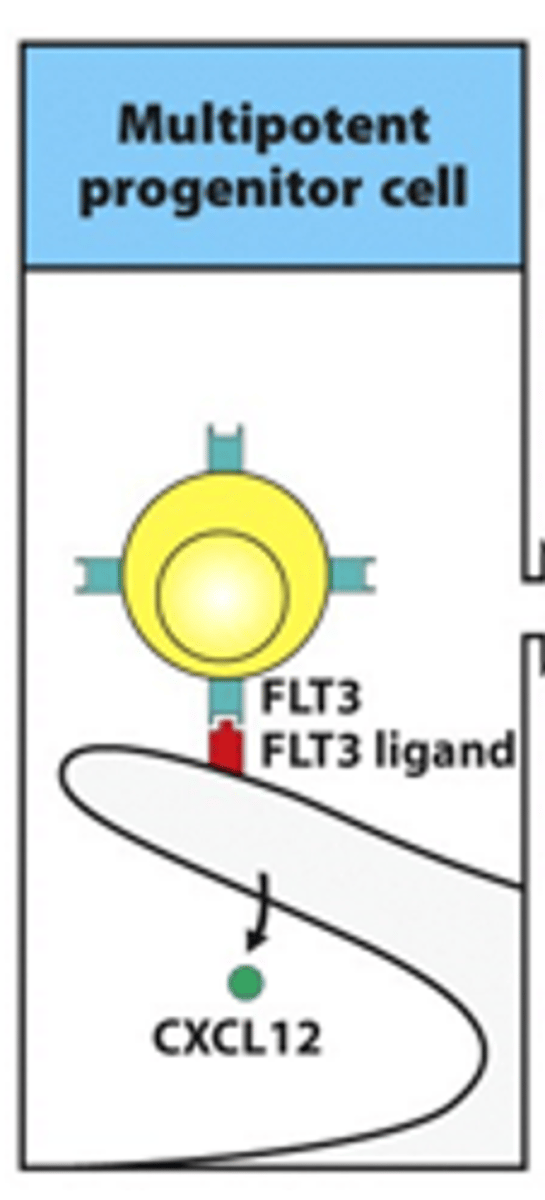
Describe the formation of common lymphoid progenitor cells from multipotent progenitor cells. Include details on the receoptors and cytokines involved
- Early phases depend on the stromal cells in the bone marrow
- Multipotent progenitor cell:
--> Interaction between FLT3 receptor and FLT3 ligand
--> Occurs on surface of stromal cells
--> Essential for progression into next phases
--> FLT = tyrosine kinase receptor
- Interaction between CXCL12 keeps interaction/attraction between the cells
--> CXCL12 = chemokine
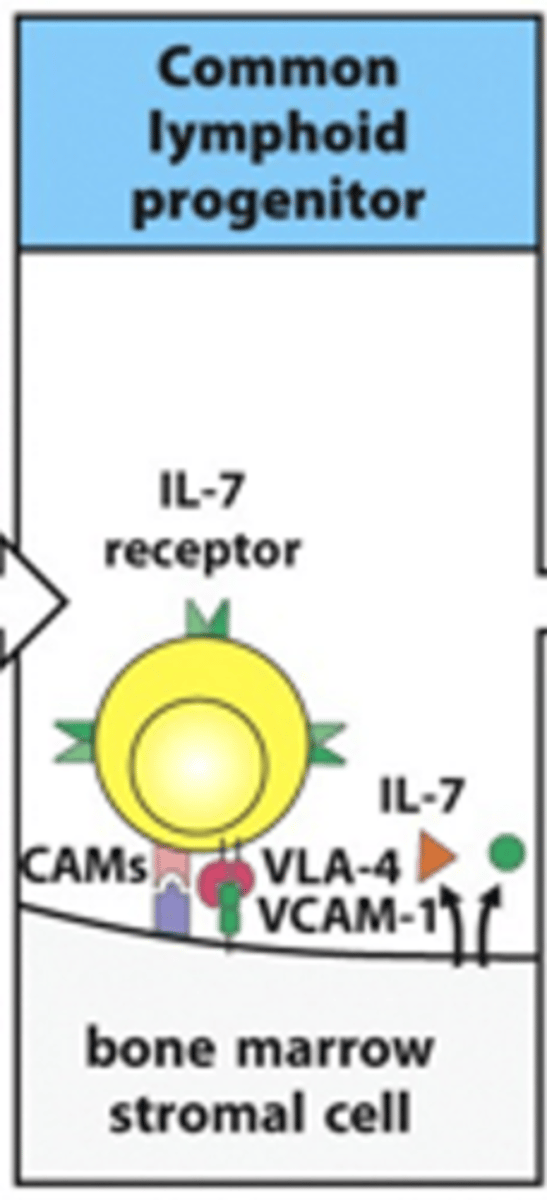
Describe the formation of early pro-B cells from common lymphoid progenitor cells. Include details on the receoptors and cytokines involved
- IL-7 receptor on B cell binds IL-7 ligand released from stromal cell
- Adhesion molecules (e.g CAMs, VLA-4 and VCAM-1) help to retain the B cell on the stromal cells
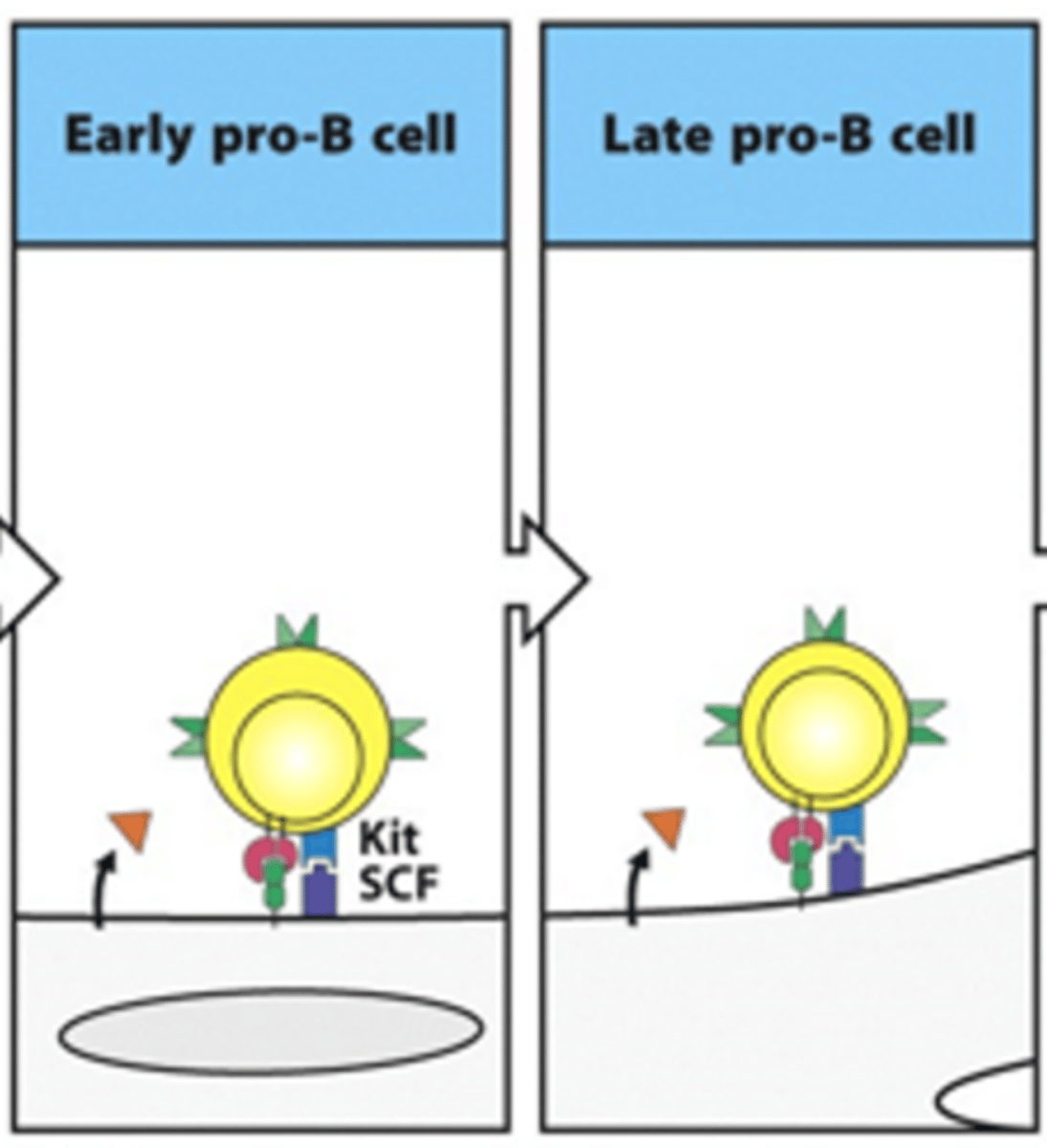
Describe the formation of late pro-B cells from early pro-B cells. Include details on the receoptors and cytokines involved
- Adhesion through adhesion molecules allows tyrosine kinase receptor (Kit) to bind its ligand stem cell factor (CSF)
- This activates the kinase and drives the cell into cell division and differentiation
--> Increased number of B cells
- During this process, IL-7 is still binding to IL-7 receptor
- In late pro-B, we start seeing gene rearrangement for BCR
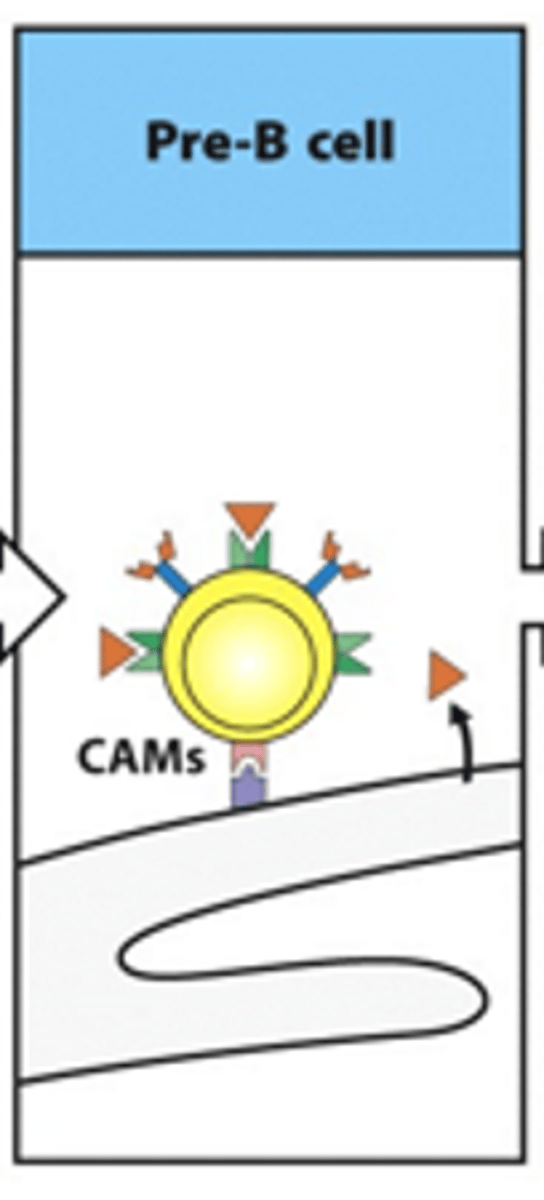
How do we describe the pre-B cell?
Pre-B cells have transient BCRs
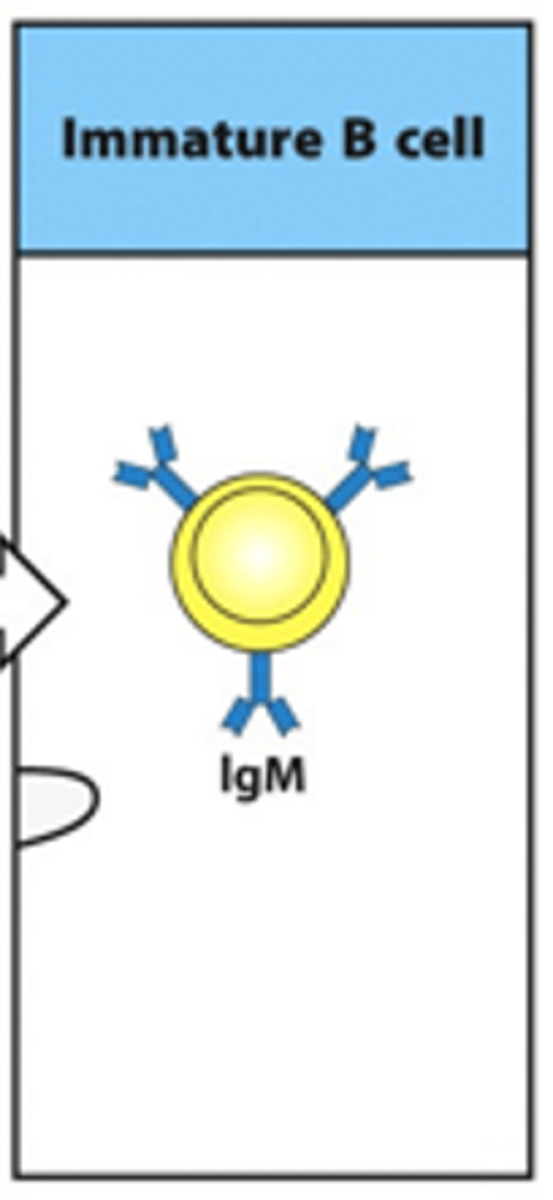
How developed is the BCR in an immature B cell?
- Immature B cells have a fully functional BCR
- 1st class of this BCR produced is IgM
What defines the stages of B cell development?
Defined by immunoglobulin gene rearrangement and expression
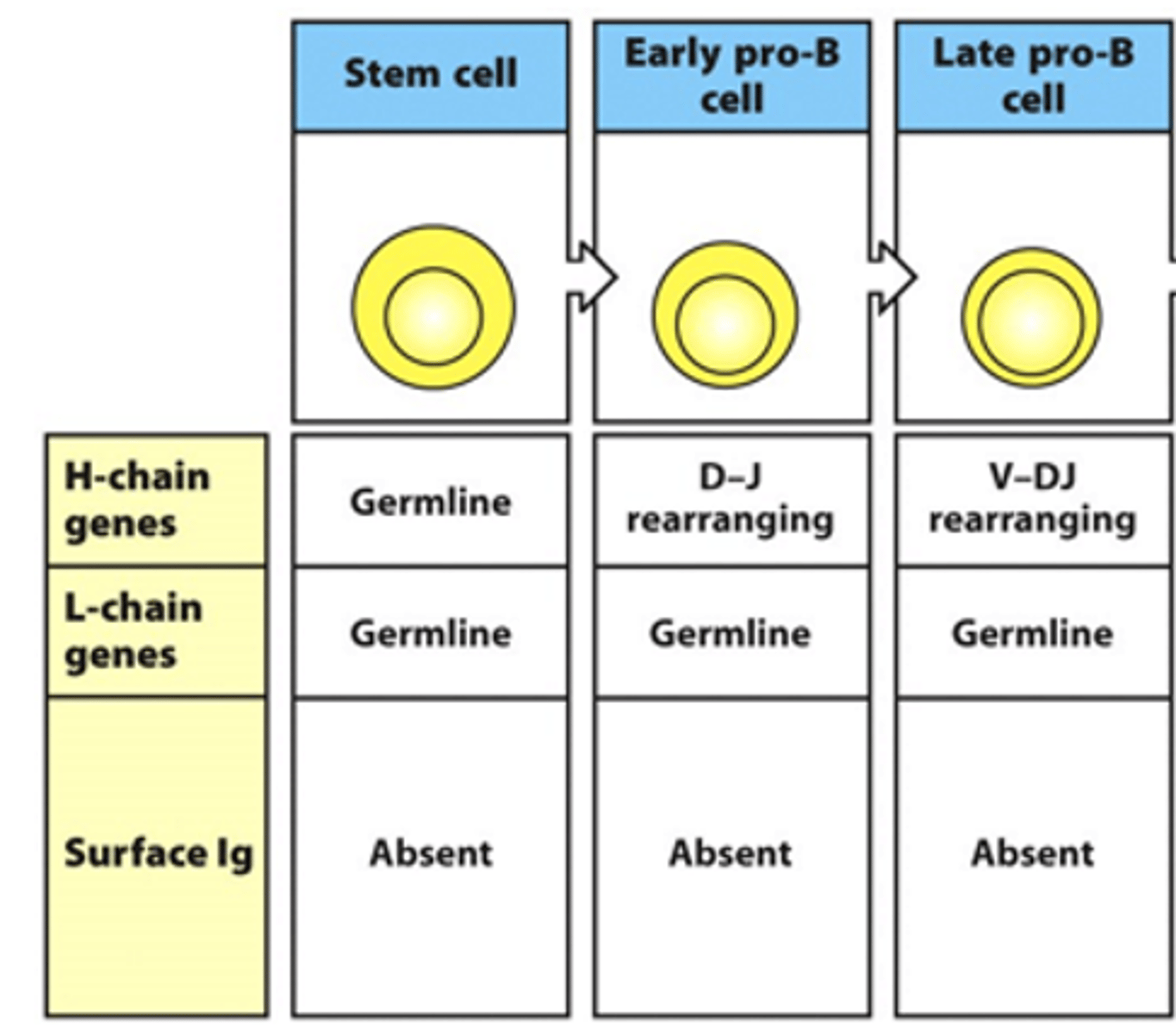
What is the expression of the 2 chains of the BCR like in the early and late pro-B cell stages? Are there any surface Ig seen?
- Heavy chain genes begin rearranging
--> D and J are rearranging in early stage
--> V and DJ rearrange in late stage
- No rearrangement of light chain genes
- No surface Ig seen
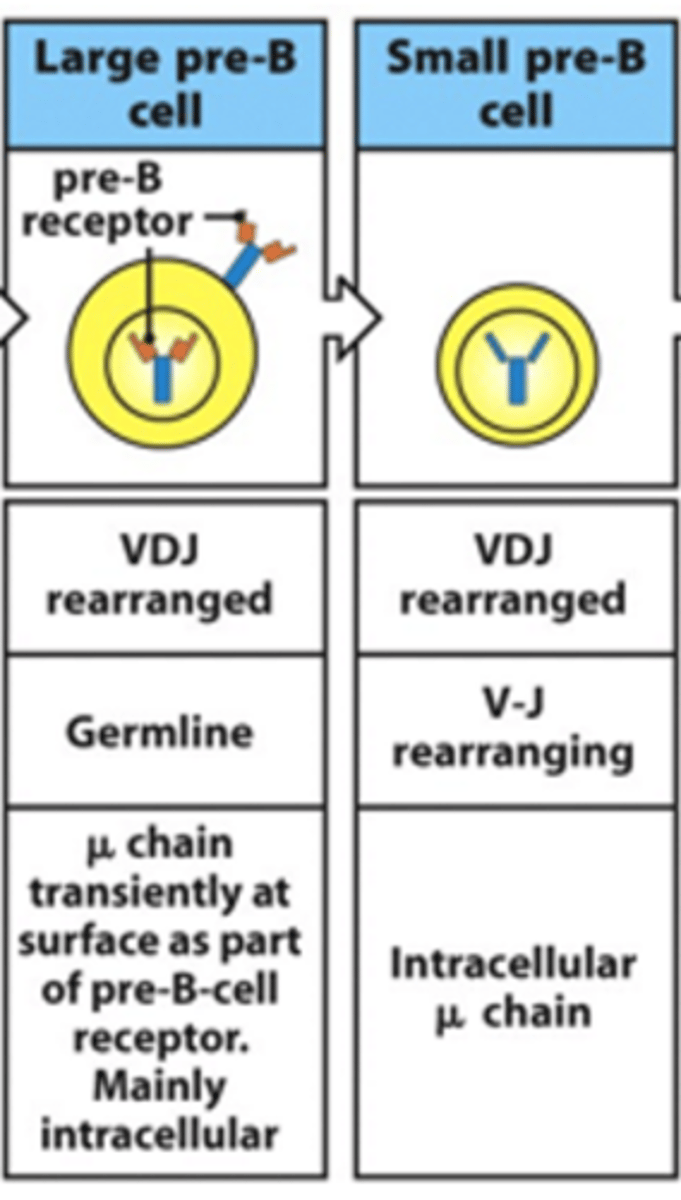
What is the expression of the 2 chains of the BCR like in the large and small pre-B cell stages? Are there any surface Ig seen?
- In large pre-B cell:
--> Heavy chain genes are fully rearranged
--> See movement of heavy chain towards surface
--> Surrogate light chain is expressed on surface with heavy chain --> allows expression of pre-B receptor on surface as heavy chain wants to be expressed immediately after being rearranged
--> Surrogate light chain signals to start light chain rearrangement as the heavy chain is confirmed to be functional
--> No rearrangement of light chain genes in large pre-B cell
--> Pre B receptor and surrogate light chain are expressed on the surface (mainly intracellular surrogate light chain)
- In small pre-B cell:
--> Heavy chain genes are fully rearranged
--> V and J start rearranging in light chain
--> Surrogate light chain is intracellular
What is the expression of the 2 chains of the BCR like in the immature B cell and the mature B cell? Are there any surface Ig seen?
- In immature B cell:
--> Heavy chain is fully rearranged
--> Light chain is now fully rearranged
--> IgM is expressed on the cell surface
- In mature B cell:
--> Heavy chain is fully rearranged
--> Light chain is fully rearranged
--> IgD and IgM are made from alternatively spliced H-chain transcipts
What are large pre-B cells?
- Dividing cells which express surrogate light chain and the pre-B receptor
- Divide into small pre-B cells
What are the genes that make up the surrogate light chain?
- VpreB
- Lambda5
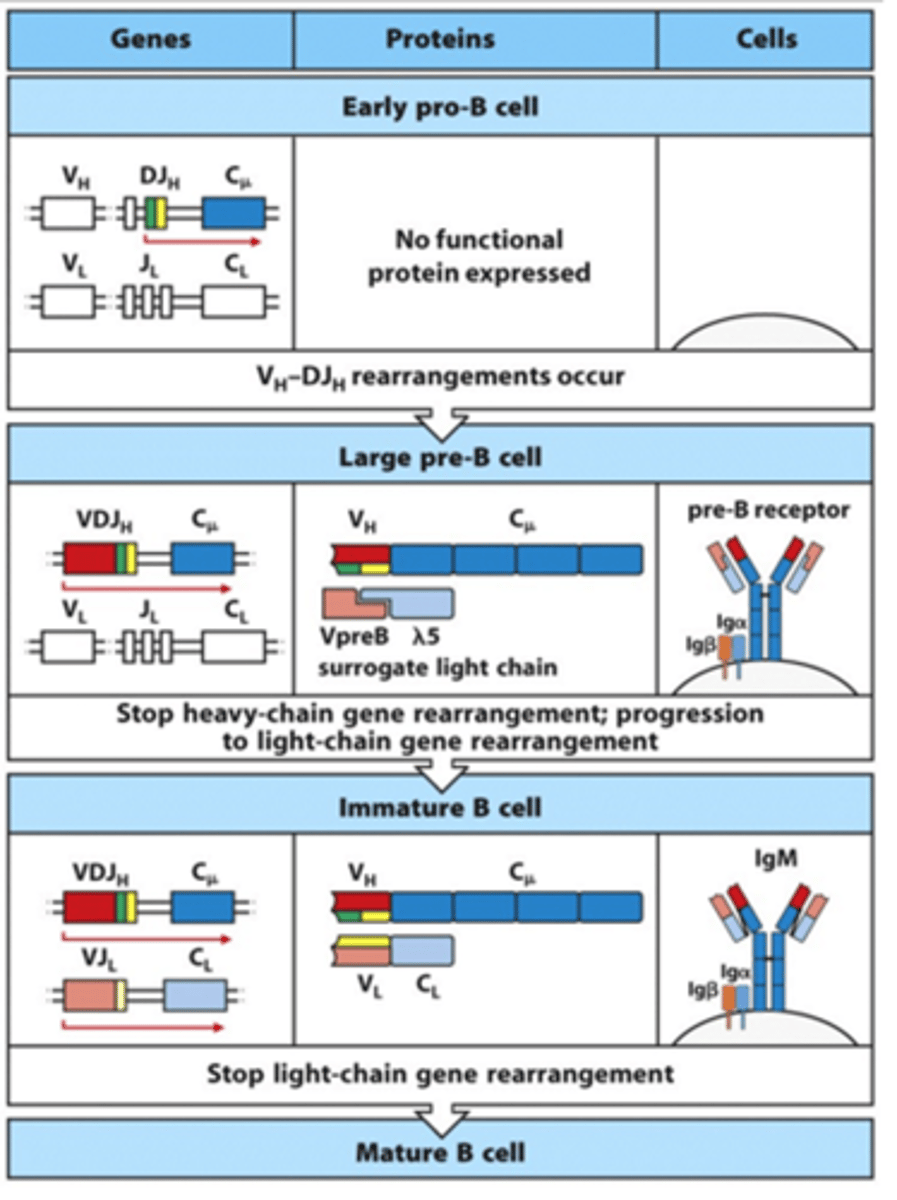
What does the BCR need for signalling?
- Need accessory molecules for signalling
--> Igbeta
--> Igalpha
--> These have little tails that indicate they are better at signalling
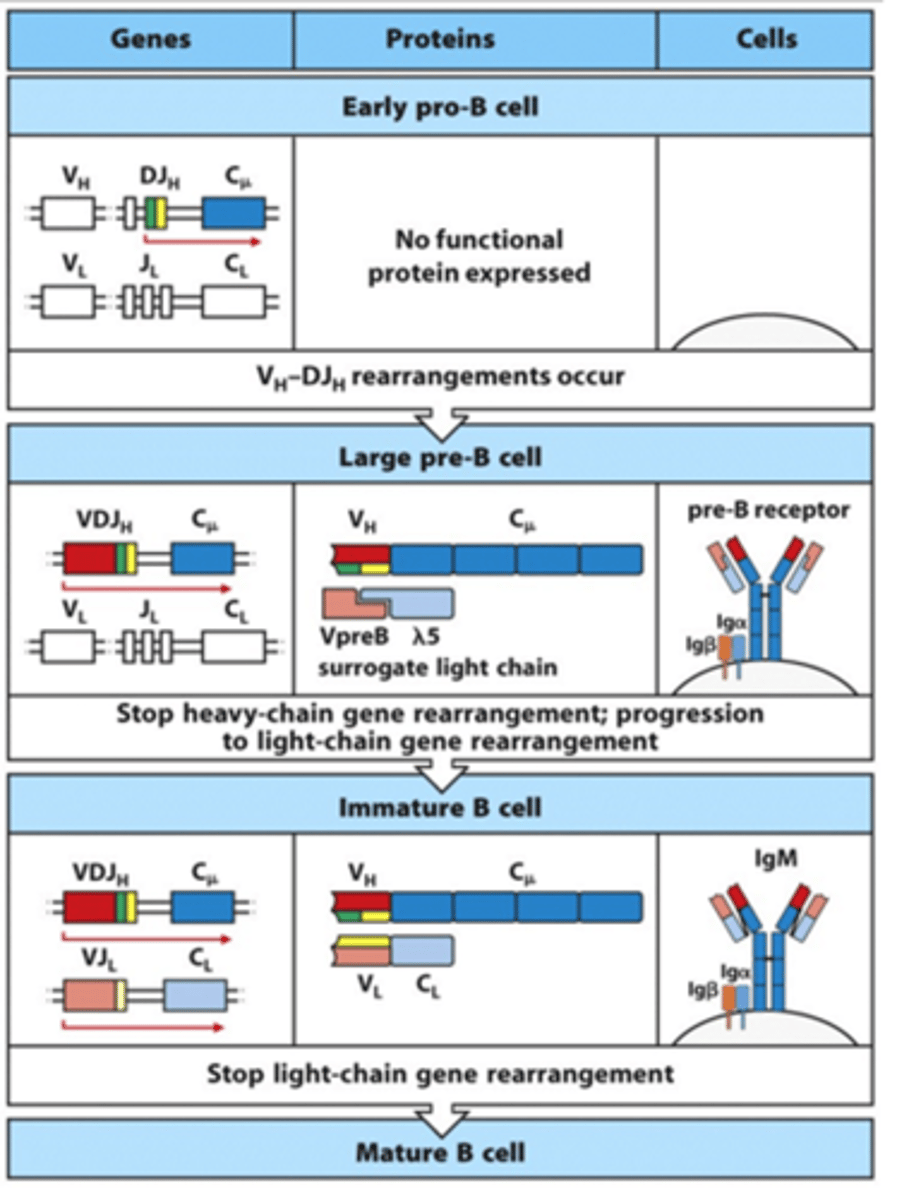
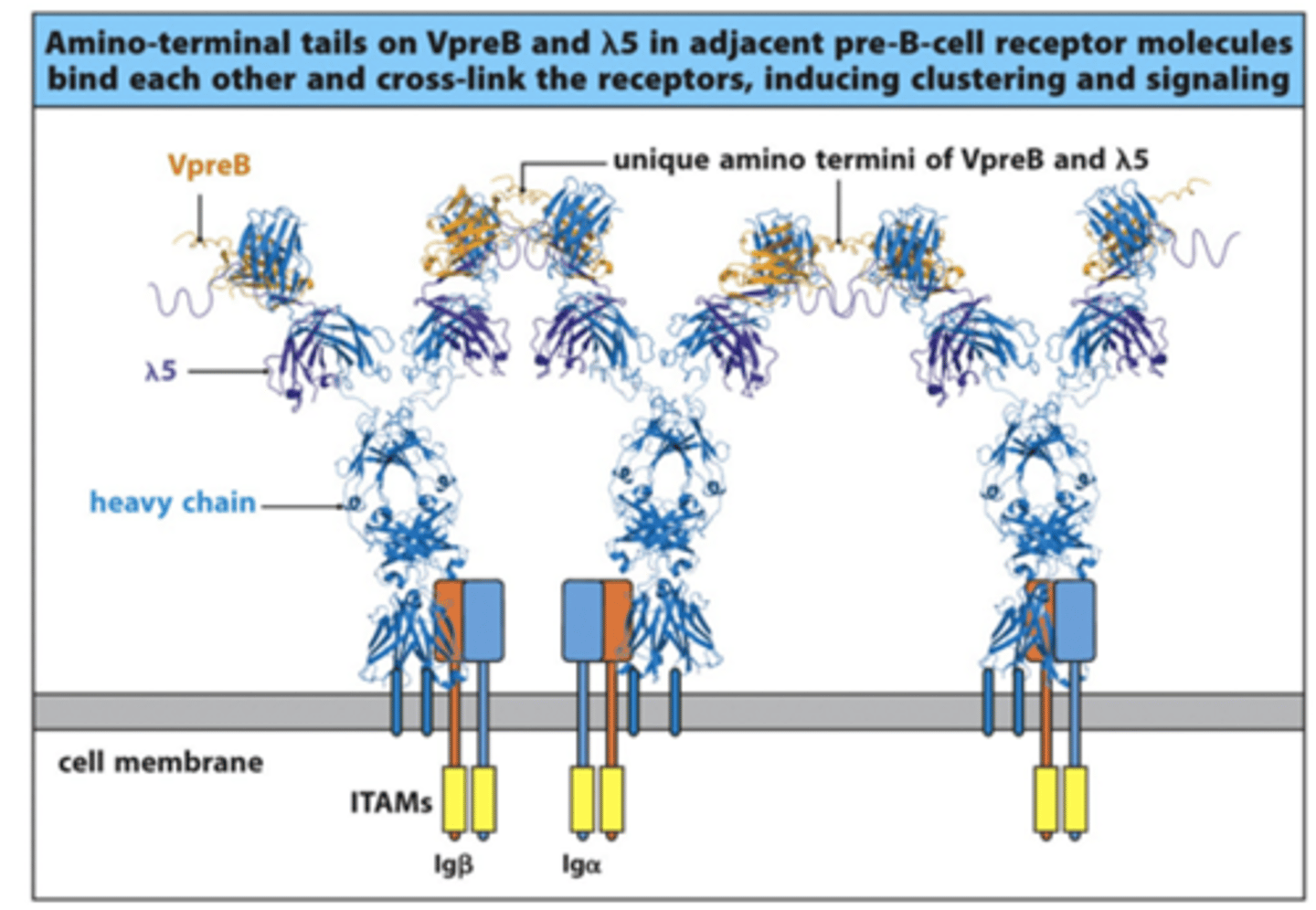
What do VpreB and lambda 5 do?
- Form surrogate light chain and bind to the heavy chain of the BCR
- VpreB and lambda5 of one surrogate light chain will bind to the adjacent surrogate ligth chain when multiple BCR heavy chains are being expressed on the surface
--> This forms cross-links between the different BCRs and forms a complex
--> This drives a signal down through the Igbeta and Igalpha through ITAMs (Immunoreceptor Tyrosine-based Activation Motifs) to stop heavy chain rearrangement and start light chain rearrangement
Through which process does heavy chain rearrangement stop?
Allelic exclusion
Why do we need to stop heavy chain gene rearrangement?
- B cell will want to secrete antibodies once it is fully matured
- If we have different variations of BCRs on the surface, the B cells will pump out different types of antibodies and they won't know their function/identity
- Want to make monoclonal antibodies so need only 1 type of BCR and only 1 type of heavy chain to be made
How many times can the BCR light chain be rearranged? What does this allow?
- Have 2 different versions of the light chain gene: kappa and lambda
- On 1 chromosome, have a lambda and a kappa gene
- Therefore, have 4 chances for light chain rearrangement
--> Rearrange kappa gene on 1st and 2nd chromosomes
--> Rearrange lambda gene on 1st and 2nd chromosomes
- Can rescue any non-productive light chain rearrangements
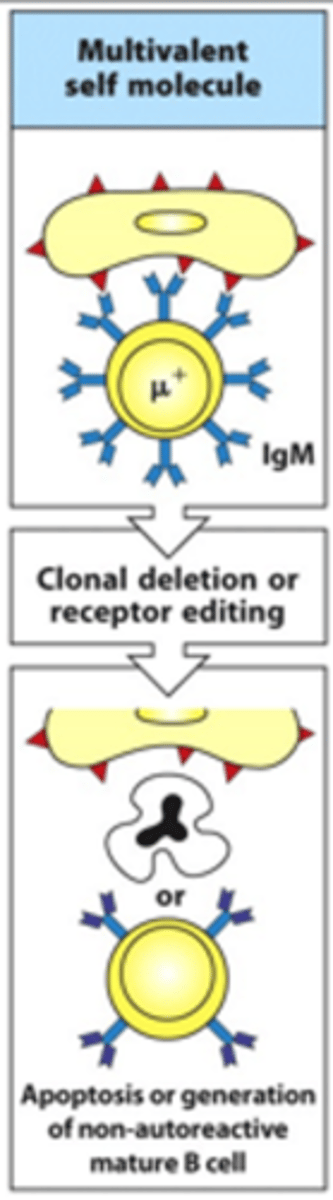
What happens if immature B cells bind quite strongly to multiple different positions on a cell that expresses self-antigens?
- This cell is multivalent
- Produces quite a strong signal which is bad so clonal deletion occurs
--> Can undergo receptor editing where you give the BCR a 'last chance' to change and adapt to stop binding self-antigens
--> If this doesn't work, B cell is given apoptotic signals and dies
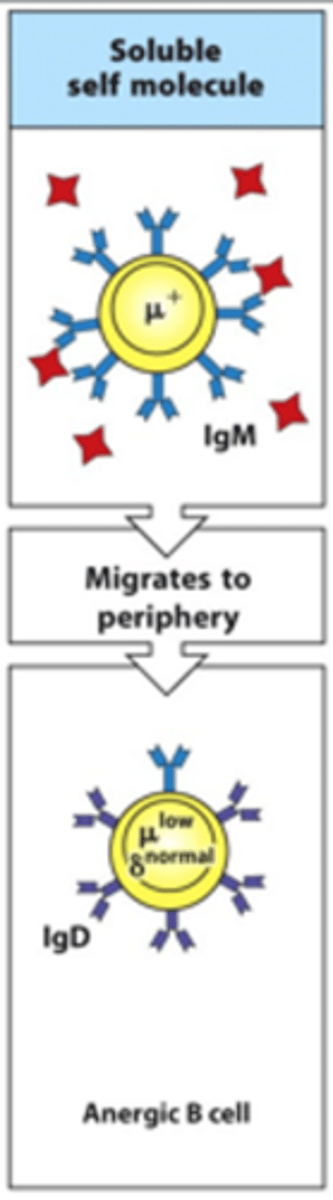
What happens if B cells bind soluble self-antigens?
- These migrate to the periphery and are anergic so are unable to react to the self-antigen binding
- Essentially they are rapidly lost through neglect
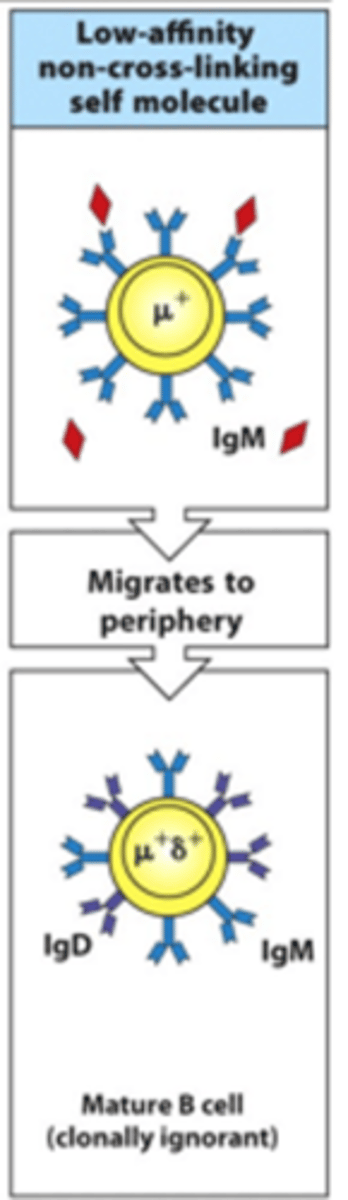
What happens if B cells bind a low-affinity non-cross-linking self-molecule?
- These migrate to the periphery and mature
- They produce very low activation signals so are not very reactive
- These are clonally ignorant --> not activated by binding to antigen
- Can maybe be self-reactive
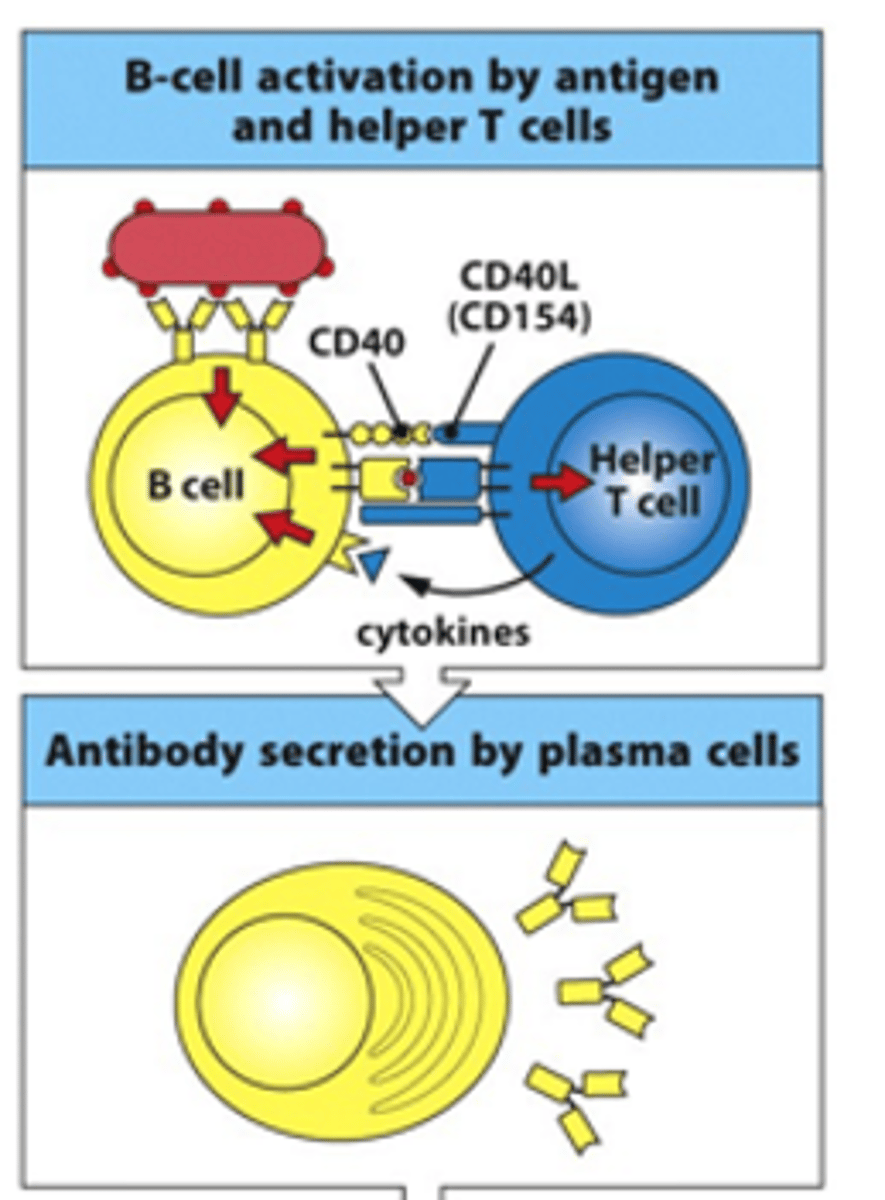
How are B cells activated by antigens and helper T cells?
- B cell binds to T helper cell via MHCII:peptide complex binding to TCR
- This binding is stabilised by interaction of CD40R on the B cell and CD40L on the T cell
- T helper cell helps activate B cell by releasing cytokines
- B cell becomes activated and becomes an antibody-secreting plasma cell
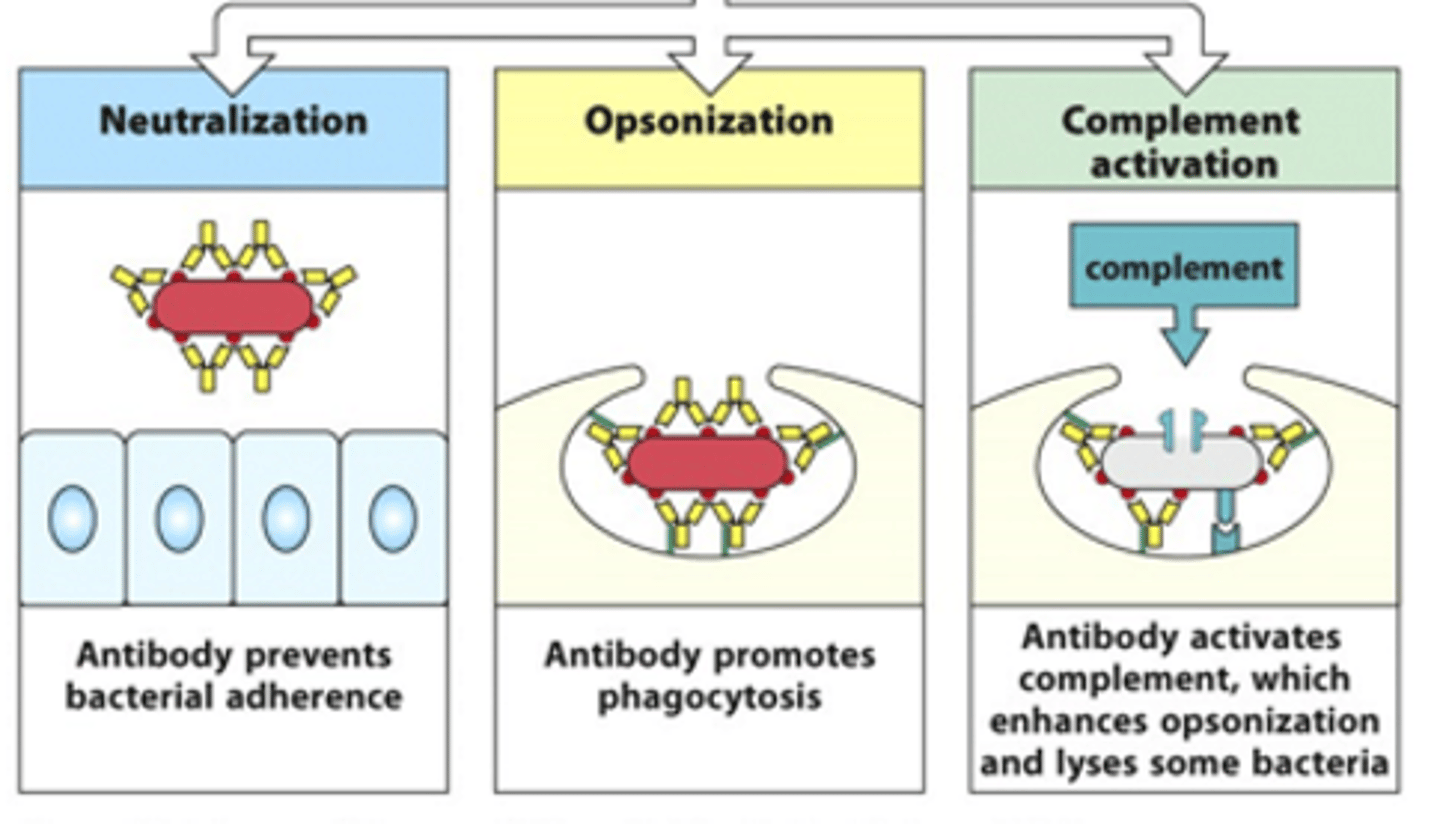
What can activated B cells go on and do?
- Can neutralise toxins/bacteria
- Can opsonise pathogens --> coat it in molecules that signal for phagocytosis
- Can activate the complement system which enhances opsonisation and phagocytosis
How many signals are required to activate B cells in thymus-dependent? What are these signals?
- 2 signals needed
- 1st signal = binding of BCR to antigen
- 2nd signal = interaction/binding of T helper cell to peptide on B cell surface (through MHCII) and release of cytokines from T cell
What happens during thymus-independent activation of B cells?
- Large antigens can cross-link several BCRs and produces large enough signal to not need activation from T cells
- LPS from pathogen can also bind TLRs on B cell surface which helps enhance the activation signal
What is proliferation? What does it produce?
- Proliferation = clonal expansion of B cells
- Produces monoclonal antibodies (one type of antibody) and memory cells

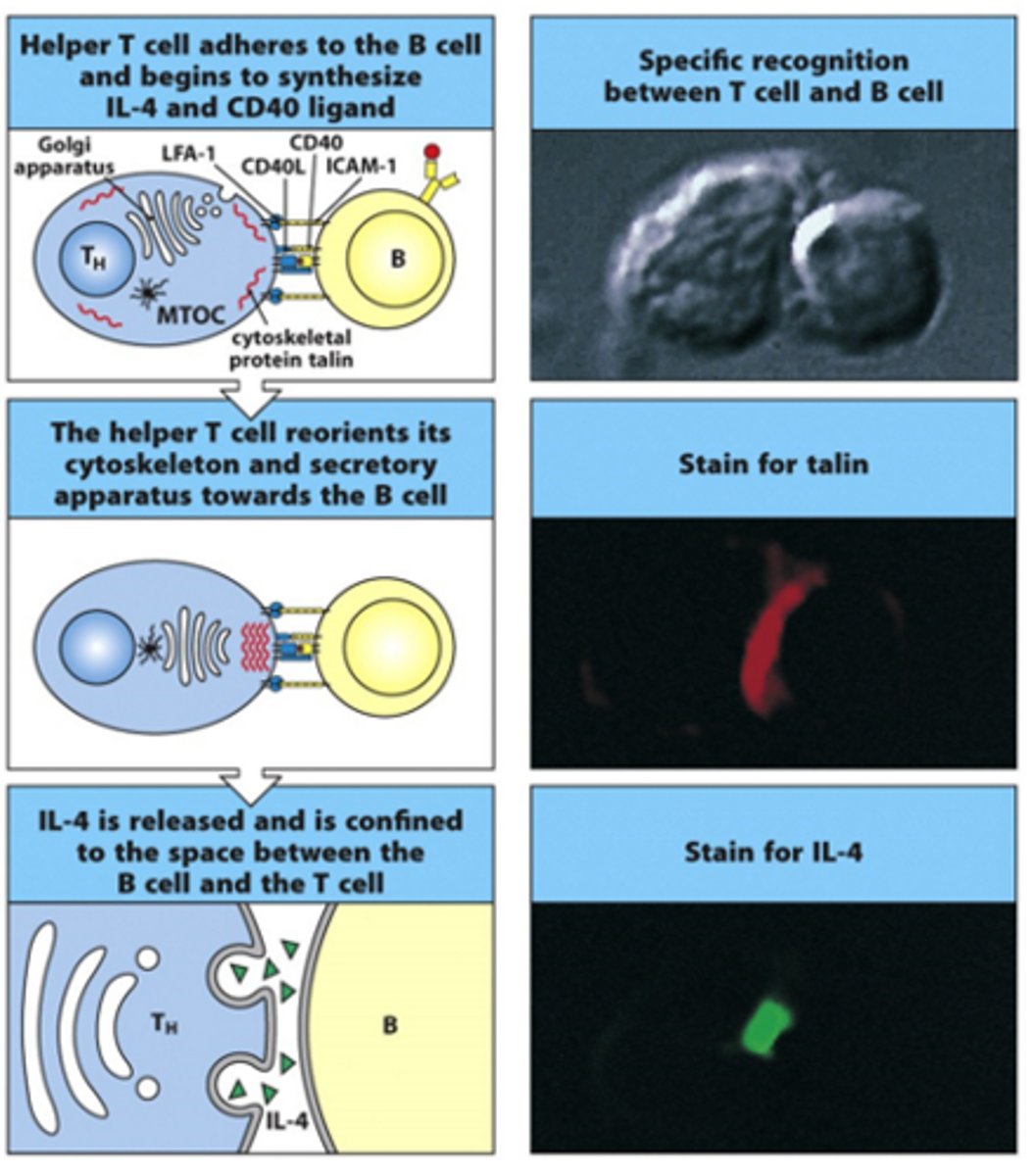
How does release of signals/cytokines from T cells occur?
- Release occurs in a polarised manner
- Have apparatus in T cell that needs to align itself to ensure signals are passed in right direction to the B cell
- Can stain T cells and see exactly where talin and IL-4 (apparatus) are being expressed
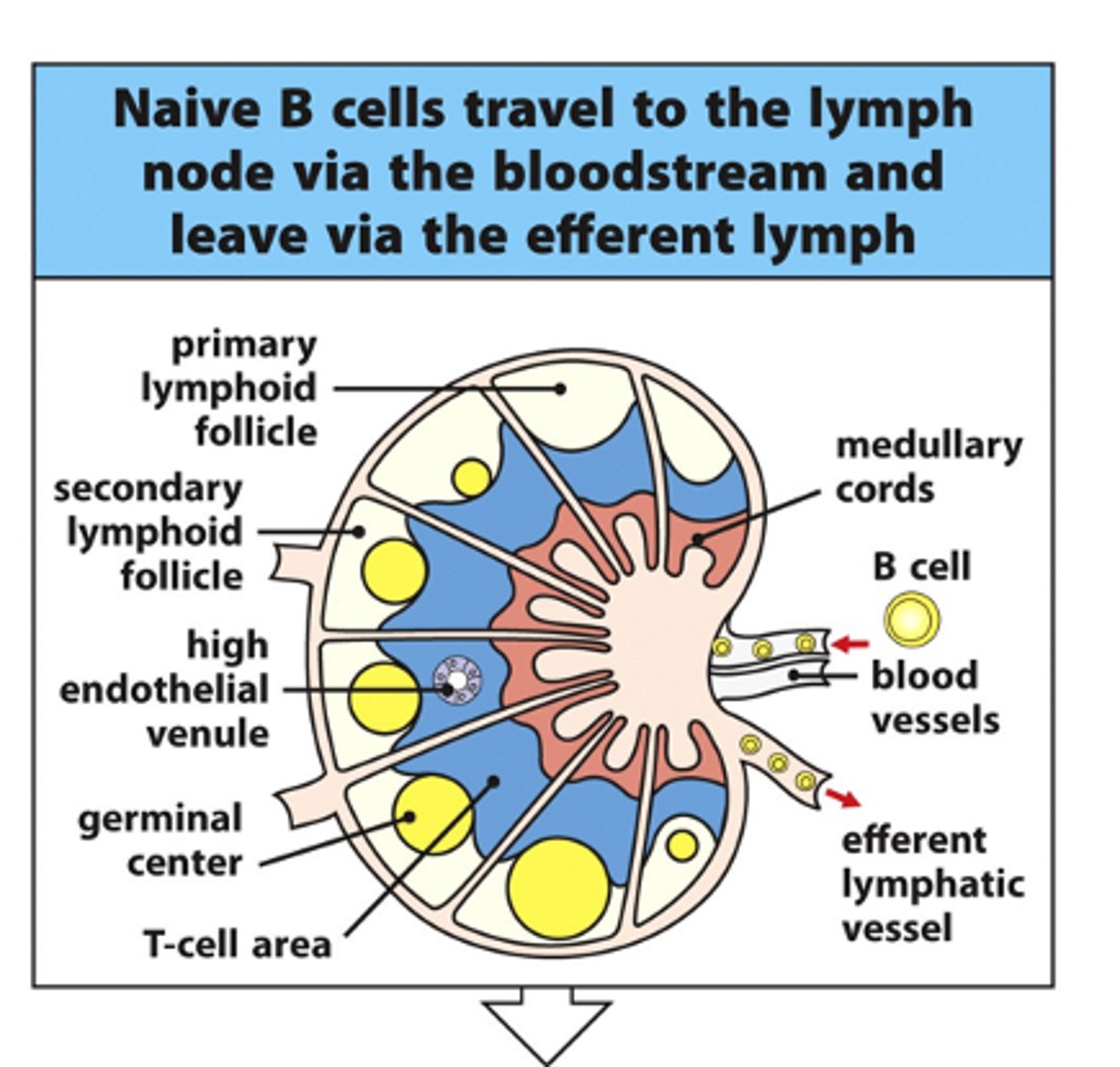
Describe what happens to B cells in lymph nodes
- For B cells to be activated, they must be in the same place as the T cells --> e.g lymph nodes
- B cells are forced through the T cell region (blue region) and interact with the T cells
--> If the T cell recognises the antigen on the B cell when they interact, B cell can become activated and B cell undergoes clonal expansion in the germinal centres
- In high endothelial venules (HEVs), B cells form a little group of cells called a primary focus
Why do lymph nodes swell during infection?
Swell due to B cell proliferation in germinal centres
What information can we get from analysing serum antibody levels?
- If there is loads of IgM present and not a lot of the other Ig subclasses, can see that this is the first response to this pathogen
- If there is loads of IgG and other antibodies, can see this is a secondary response as these antibodies are being produced very quickly from memory cells
--> IgM doesn't recognise antigens very well/not very specific but the strength of binding is high due to 10 binding sites (pentameric structure of 5 IgM molecules)