BIO1A03 T3M3: Euk Transcriptional Regulation
1/24
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
25 Terms
embryonic stem cells
cells from the early-blastocyst stage, pluripotent (can diff. into any cell type in body — but not placenta and baby germ layers)⤷ all of stem cells = identical
⤷ from the final divisions in early developmental cascades, cells communicate to form various genetic interactions that regulate continual development + cellular differentiation
⤷ diff. Embryonic cells will have diff fates depending on the signals that are exchanged and which genes are turned on/off
gene regulation
process of controlling which genes are turned on or off, allowing cells to produce specific products only when needed
⤷ responsible for creating various cell types in multicellular organisms →
⇒ all cells developed from the same embryonic stem cells, but they eventually specialize into their final cell functions after receiving diff. cues or signals during embryonic development
transcription factors
proteins that bind to specific DNA sequences
⤷ contain certain structures that allow them to interact w the DNA double helix and then control transcription of DNA and RNA and contribute to gene regulation
⤷ important in determining the pathway of a specific cell type (ʙ help determine what the mature cell will differentiate into)
⤷ gene expression triggers within dividing cells (which genes are active along chromosomes) + extracellular cues = differentiate
∴ certain proteins can only be found in certain cell types
chromatin
highly compacted DNA into fibers, to allow all the genetic information to fit inside the nucleus and be moved around during cell division.
nucleosome
small clump structure after the chromatin wraps around histone proteins
⤷ each contains an octamer of 8 histone proteins (wrapped around ~150 DNA bp of chromatin)
⤷ in the form of nucleosome = DNA is not accessible(must unravel for transcription to occur)
⤷ in euk
how is DNA tightly wrapped around histone proteins
Due to the interactions of the ⊕ charged tails of histones and the ⊖ charged phosphates in DNA ⇒ opposites attract, keeping DNA tightly wrapped around histones
chromatin remodelling
process of unravelling nucleosome/chromatin
Begins when an activator protein or transcription factor is able to bind to an accessible enhancer site
process of chromatin remodelling (acetylation)
- Activator proteins bind to specific DNA regions (enhancers)
- Activator recruit the coactivator enzyme histone, acetyltransferase (HAT)
- HAT adds acetyl groups to the lysine AA on the ⊕ charged tails of nucleosome histone ⇒ becoming acetylated
- Acetylation neutralizes the ⊕ charge on histones ⇒ weakens the grip on DNA and makes it looser and more accessible for transcription (allow transcription factors to bind onto)
** Note: histones have tails (flexible ends), made of AA like lysine, arginine, serine, and threonine –
⤷ they stick out from nucleosome and can be chemically modified after the protein is made
⤷ modifications act like switches/signals that tell cell to open up DNA for transcription or keep it tightly pack
types of post-translational modifications
Acetylation and methylation of lysine and arginine, and phosphorylation of serine and threonine ⇒ regulate protein function, stability, and interactions, particularly in processes like gene transcription
⤷ may alter charges of these tails, and ∴ alter binding to DNA thats wrapped around them (allowing space for transcriptional proteins to work)
⇒ results in modification of strings of AA that protrude from histone proteins
⇒ whether transcription is activated or repressed affects the degree of modifications to the histone tail
Eg. acetylation and methylation w single methyl group ⇒ transcriptional activation (seen as + symbol)
Eg. methylation with 3 methyl groups ⇒ repression of transcription
classifying TF
Transcription factors are classified based on structures of distinct DNA binding motifs like:
Basic helix-loop-helix
Helix-turn-helix
Zinc finger
Leucine zipper regions
⇒ most transcription factors have alpha-helical domains that fit in the grooves of DNA
⤷ molecular interactions (eg. H-bonds) occur btw AA in alpha helix and functional groups of the bases along the grooves
⤷ when strong interaction → TF assumes conformation for the control of transcription (eg. recruitment of other TF, RNA poly, or the activation of transcription at the target gene)
cis-sequence
specific DNA sequences that are required to initiate transcription ⇒ the “specific promoter regions”
core promoters
the binding site thats required for binding RNA polymerase and associated TF
⤷ composed of TATA box + transcriptional start sites
⇒ TATA box and BRE regions = close proximity to the transcription start site → the binding of TF = allow for protein to protein interactions… assisting w the assembly of transcription initiation complex
TATA box
consensus sequence TATAAA, located in the promoter region of many genes, about 25-35 base pairs upstream of the transcription start site
⤷ recognized by the TATA-binding protein (TBP) subunit of the TF “TFIID”
B recognition element (BRE region)
recognized and bound by the TFIIB general TF
enhancer regions
a DNA sequence that increases the likelihood of gene transcription
⤷ found in the regulatory promoter regions (upstream of GOI)
⤷ able to bind cells or region-specific TF’s
When FT bind to enhancer, they help gather machinery needed for transcription
⇒ helps RNA poly to attach to promoter and start transcription
silencer regions
DNA sequences that can bind transcriptional repressors, halting gene expression by interfering general transcriptional factor assembly and mediator activity → which is necessary for RNA poly to bind and start transcription
⤷ ū upstream of GOI
transcriptional repressors
proteins that inhibit gene expression by binding to DNA and blocking the transcription process
hemoglobin
protein that binds to oxygen and carries it to all cells in the body
progenitors
stem cells
blood cell progenitors
can differentiate into RBC (w hemoglobin)
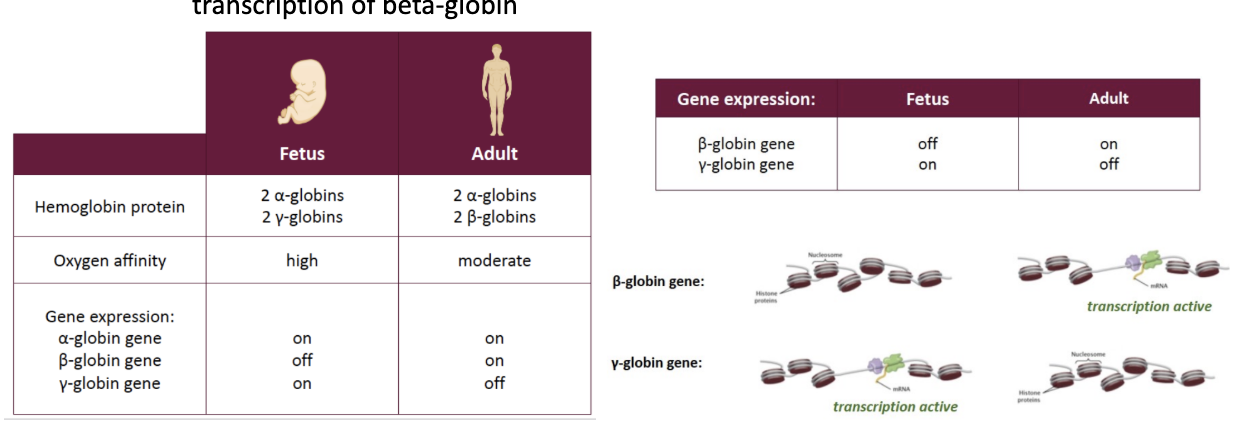
in order for hemoglobin to be a functional protein (from stem cell)
progenitor cell must activate transcription of globin proteins that are appropriate for the fetus or adult
**In humans… hemoglobin = tetrameric protein (4 subunits)**
In fetus – ½: 2 alpha-globin proteins + ½: 2 gamma-globin proteins
→ gamma-globin can bind oxygen stronger than beta-globin ⇒ allowing the fetus to be able to have more O2 while growing in womb
⤷ chromatin wrapped around gamma-globin = open ⇒ yes transcription
HAS beta-globin genes, but chromatin is wrapped around to inhibit transcription
In adult – ½: 2 alpha-globin proteins + ½: beta-globin proteins
→ no need for gamma-globin anymore ⇒ so transcription changes
HAS gamma-globin… but chromatin inhibits transcription of gamma, while allows transcription of beta-globin
Developmental switch from gamma to beta-globin proteins:
- regulated by TFs that are able to silence/repress (OR activate beta-globin) gamma-globin gene transcriptions
repression (stop) of transcription by methylation
recall: histone modifications allow DNA to unwind from nucleosome —> allow DNA-binding TF’s to associate w enhancers + promoter sequences and start transcription
… ^^ but DNA modifications can still inhibit transcription
In euk – transcription is affected by chemical modifications of cytosine in DNA sequence
Modification = adding a methyl group to cytosine (NOT in every cytosine, but on CpG island)
when cytosine IS NOT methylated… DNA binding proteins (incl. RNA poly) will recognize the promoter and bind ⇒ transcription
When cytosine IS HEAVILY methylated… shape of DNA binding site for proteins has changed, and proteins cannot bind → heavily methylated promoters are not transcriptionally active bc RNA cant bind to methylated sequences
⇒ the methylation state = changes in response to environamnetal and developmental cues
DNA methylation = example of epigenetic mechanism that controls gene expression
⤷ leaves to functionally relevant changes to genome ⇒ altering gene expression levels like transcription, without changing the specific nucleotide sequences of DNA
⇒ methylation = heritable from mother cell to daughter cell (able to maintain same state of transcriptional activation w next cell division)
**RNA poly & associated TFs cant bind to methylated DNA…
CpG island
string of cytosine and guanin bases short, GC-rich DNA regions, ū unmethylated
⤷ uu u located in/near promoter sequences of mammalian genes
⤷ “p” = phosphate in the DNA backbone btw 2 bases
histone deacetylase (HDAC)
bind to methylated DNA and promote the removal of acetyl groups from the neighboring histones
⤷ deacetylation of histones allow nucleosomes to reassemble ⇒ leads to masking of enhancer and promoter sequences of DNA, ∴ repress transcription
what is the default state of transcription in prok and euk
In prok
Chromatin = wound up in default “on” conformation
In euk
Chromatin = wound up in a default “off” conformation –
Genes are only transcription when chromatin is remodelled to be exposed to the promoter sequence