D103 ER Protein Sorting (ALS 10, Videos 17 and 18)
1/31
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
32 Terms
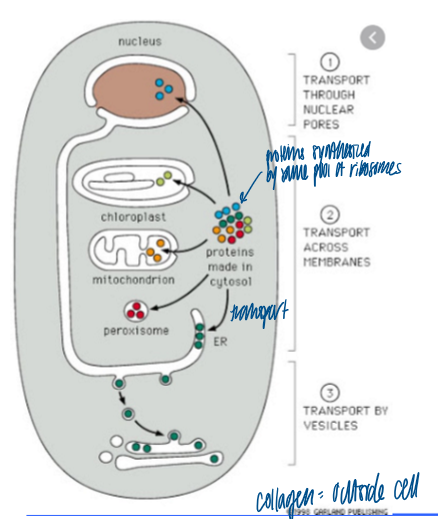
what is the entry point into the secretory pathway
the ER
ER signal sequence and receptor
SRP (signal recognition particle as sig sequence) = binds its substrate via a hydrophobic pocket, which can accomodate N-terminal hydrophobic sorting signals and hydrophobic domains inside a protein
hydrophobic region at N term
SRP receptor
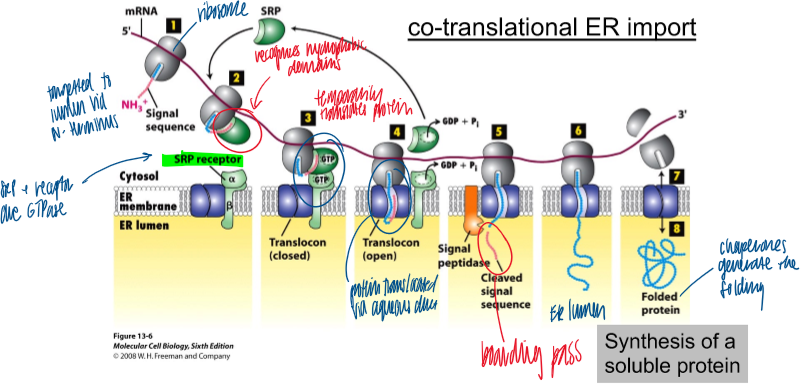
how are ribosomes directed to the ER membrane
via co-translational ER import; no chaperones
hydrophobic signal sequence on N term associates with SRP on the ribosome; moving along the mRNA
SRP receptor GTPase on the ER membrane associates with SRP bound to ribosome
when bound, translocon opens to allow protein thru ER membrane; GTP hydrolysis to inactivate srp receptor
signal peptidase cleaves signal sequence to release protein into er
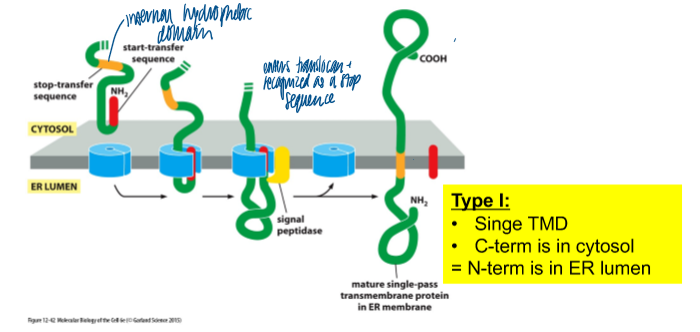
synthesis of type I membrane protein
type I: single TMD; C-term is in cytosol = N-term in ER lumen
N term signal sequence: directs the protein for import into the ER
subsequent hydrophobic domain: TMD
type ii protein
single tmd
c-term is in ER lumen = N term in cytosol
if + charged AA precede the domain, N-term is in the cytosol
what determines membrane protein topology
its insertion into the ER
if no N term signal sequence is present, hydrophobic domains can serve as internal signal sequence
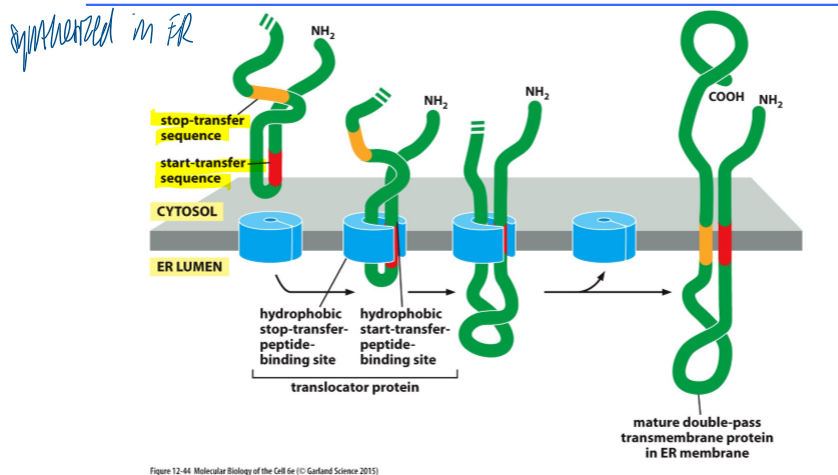
synthesis of a multi-spanning membrane protein
synthesized in ER
start transfer sequence associates with binding site
continues moving thru the binding site until the stop transfer sequence is reaches
mature double-pass TM protein in ER membrane
challenges for the translocon
the translocon has to:
form a pore to translocate proteins across the mem
recognize hydrophobic domains
release TMDs into the membrane
be impermeable to ions
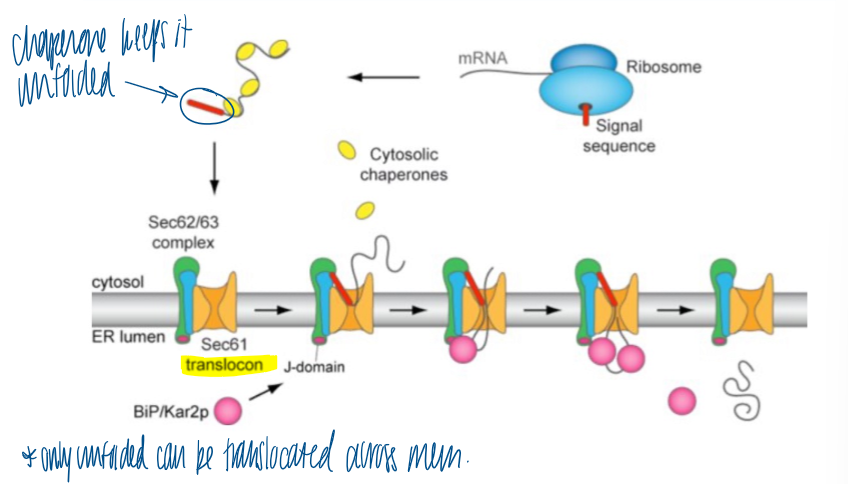
post translational ER membrane insertion
ribosome synthesizes the protein with signal sequence
chaperones keep it unfolded in the cytosol
binds to the Sec translocons → releases the chaperones
BiP in the ER lumen binds to translocon to release protein into lumen and cleave sig seq
ER import overview
sorting sig: hydrophobic at N-term; SRP
receptor: SRP receptor
translocation: ER translocon
energy: N/A; driven by polypeptide creation
folding state: unfolded bc entering as being translated
cleavage: yes; at membrane
proteasome and ER
1/3 of the proteome is imported into the ER, then trafficked to organelles of the secretory pathway or secreted out
ER functions
surrounded by lipid bilayer
determines site of mito fission
extends throughout the cell and this spread-out orgnization depends on kinesin-mediated transport
ER stores Ca2+ (SR) that stimulated muscle contraction
ER domains
rough ER: sheet-like
major site of protein import into ER
entry point of the secretory pathway
protien topology is determiend
smooth ER: tubular
synthesis of cholesterol and hormones
phospholipid synthesis
Ca2+ storage
vesicle formation
contact sites with other organelles (mitos, endosomes)
similarities between smooth and rough ER
protein folding
quality control
stress response
er contact sites
lipid synthesis and exchange via ER-mitochondria interaction
endosome fission via ER-mito interaction
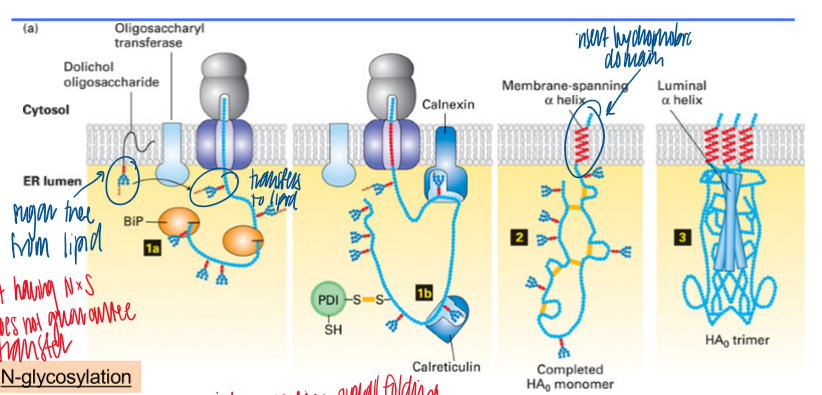
ER as a folding environment
sugar tree on lipid gets trasnfered to the growing lipid via N-glycosylation; BiP and lectin are chaperones for folding
form disulfide bonds with PDI
arrange the peptide bond between proline and other AAs by peptide prolyl isomerase (PPI)
membrane insertion as a membrane-spanning alpha helix
protein oligomerization (form luminal alpha helix)
N-glycosylation
initiated in the ER
Asn-X-Ser/Thr (consensus sequence)
initiated in the ER
continued in the Golgi
motif is necessary but not sufficient
O-linked glycosylation
Ser/Thr
occurs only in the Golgi
role of glycosylation
protective layer at cell surface
quality control
lysosomal sorting (M6P)
ER and disulfide bond formation
ER as site for disulfide bond formation (oxidative environment → promotes disulfide bond formation)
formation of disulfide bond: reduced substrate protein binds to oxidized PDI; PDI gets reduced and the oxidized substrate protein conencts
rearragement of disulfide bonds: protein with incorrect disulfide bonds reattach to reduced PDI, then pdi releases once correctly organized
chaperones and er
evaluate and modify the quality of the proteins
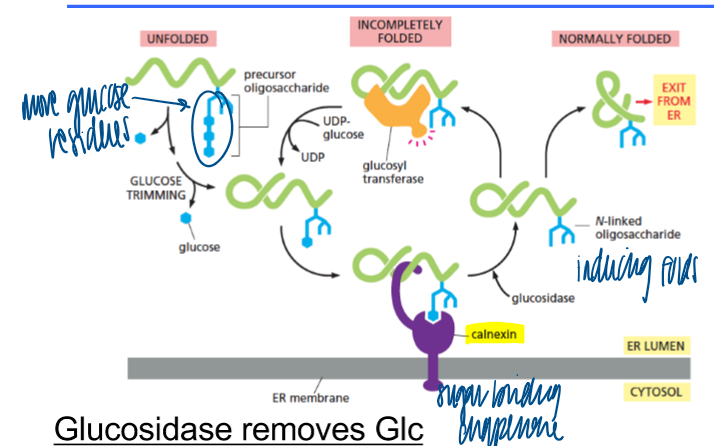
calnexin
membrane-bound lectin (=sugar-binding proteins) with chaperone activity
calreticulin
soluble “relative” of calnexin with the same function
chaperones when glucosidase removes glucose
membrane-bound chaperone (calnexin) binds to sugar tree with 1 Glc
chaperones when glucosidase removes another glucose
if folded correctly: exit from ER
if not: binds to soluble chaperone, Glc residue is added again
purpose of new chaperone cycle
“buys” time for correct folding
protein folding problems
trapped in misfolded conform
mutation that leads to misfolding
unassembled multimer subunits
what happens if a protein cannot be folded properly
glucosidases (trim glucose residues from N-linked glycans)
glucosyltransferases (folding sensor for newly synthesized N-linked glycoproteins)
misfolded proteins bind and sequester BiP
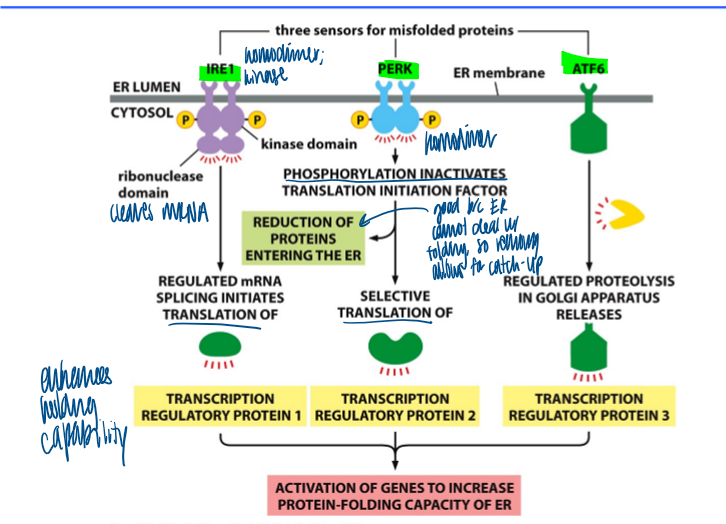
unfolded protein response pathway
unfolding response
first line of defense against er stress
IRE1: first sensor (homodimer, kinase)
ribonuclease domain cleaves mRNA and iniates translation of transcription reg protein 1
PERK
phosphorylation inactivates translation initiation factor → reduction of proteins entering the er
selective translation of transcription reg porotein 2
ATF6
regulated proteolysis in golgy releases transcription reg protein 3
results in activation of genes to increase protein-folding cap of er
ER-associated degradation (ERAD)
a cell’s repsonse to misfolded proteins in the ER
recog of malfolded protein
export from ER (pulling by ATPase)
poly-ubiquination (E3 ligase)
deglycosylation (N-glycanase)
degradation (proteasome)
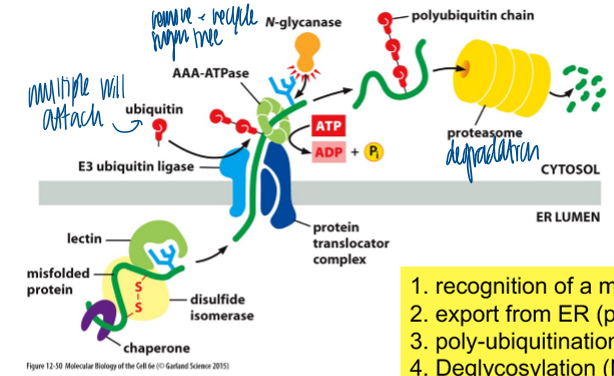
ubiquitin
conserved small protein (76 amino acids)
attached to lysines on target protein (no consensus) → poly-ubiquitination
polyubiquitination leads to proteasome-dependent degradation
ER folding overview
er chaps help the protein fold (BiP), form disulfide bridges (disulfide isomerase), and peptidul prolyl bonds (peptidylprolylisomerase)