GIO: L4 02-05 FROM DNA TO PROTEIN
1/36
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
37 Terms
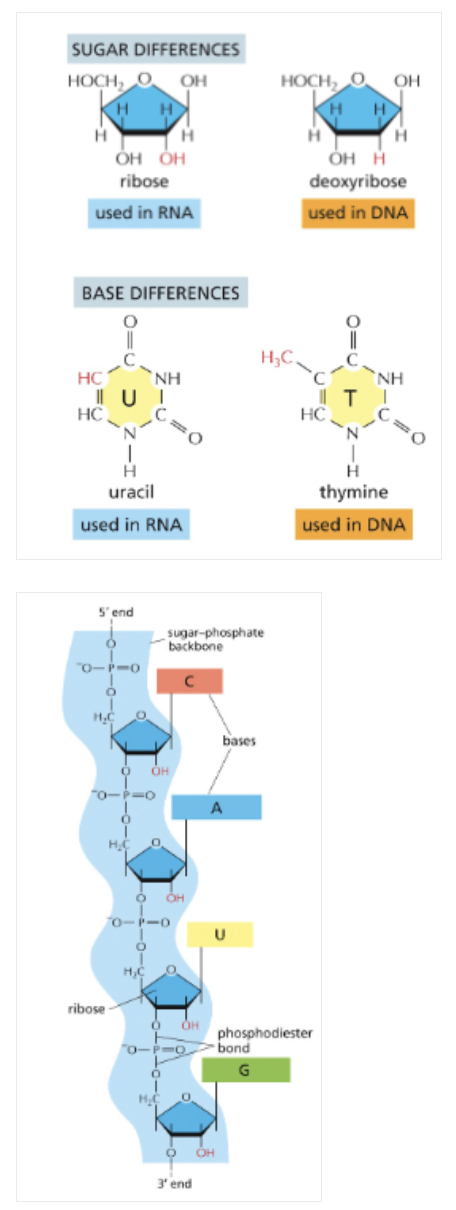
WHAT IS THE DIFFERENCE BETWEEN RNA VS. DNA?
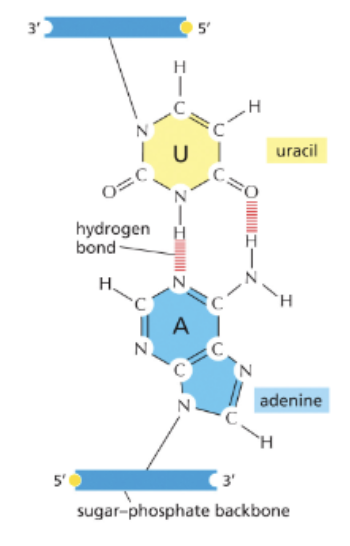
RNA CONTAINS A HYDROXYL (-OH) GROUP ON THE 2’ CARBON OF THE SUGAR
RNA CONTAINS URACIL INSTEAD OF THYMINE
U PAIRS WITH A
RNA IS SINGLE-STRANDED
RNA CAN…(4 FLASHCARDS)
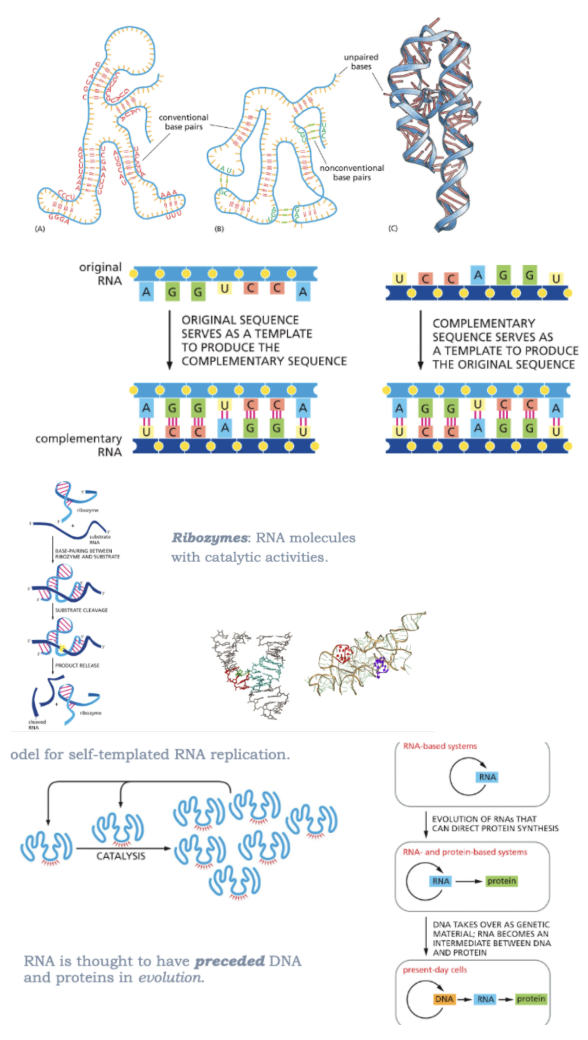
CAN FOLD INTO STABLE 3D STRUCTURES
HYDROGEN BONDS STABILIZE RNA STRUCTURES
CAN STORE INFORMATION AND COPY BY ITSELF
RNA STRANDS=ANTI-PARALLEL
FOR EXAMPLE: RNA VIRUSES STORE THEIR GENETIC INFORMATION AS RNA
MODEL FOR SELF-TEMPLATED RNA REPLICATION
RNA IS THOUGHT TO HAVE PRECEDED DNA AND PROTEINS IN EVOLUTION
CAN HAVE CATALYTIC ACTIVITY
RIBOZYMES: RNA MOLECULES WITH CATALYTIC ACTIVITIES
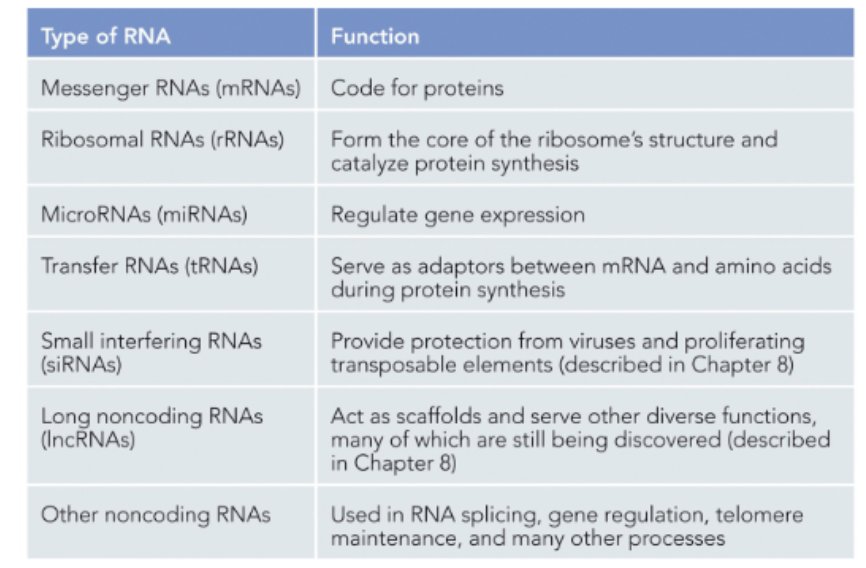
WHAT ARE THE TYPES OF RNA PRODUCED IN CELLS?
IMAGE!
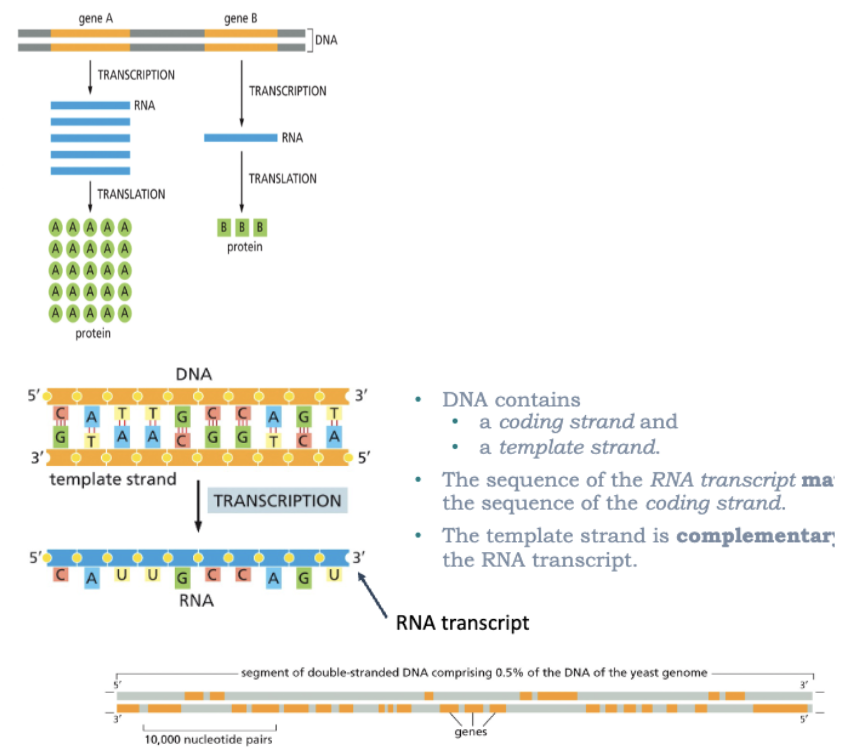
WHAT IS TRANSCRIPTION?
DNA—>RNA
THE RATES OF BOTH TRANSCRIPTION AND TRANSLATION CAN DIFFER BETWEEN GENES
ALL RNA MOLECULES (EXCEPT IN SOME VIRUSES) ARE DERIVED FROM INFORMATION PERMANENTLY STORED IN DNA
DURING REPLICATION, THE ENTIRE GENOME IS USUALLY COPIED
DURING TRANSCRIPTION, ONLY PARTICULAR GENES OR GROUPS OF GENES ARE TRANSCRIBED AT ANY ONE TIME (SOME PORTIONS OF THE DNA GENOME ARE NEVER TRANSCRIBED)
ONLY ONE DNA STRAND SERVES AS A TEMPLATE FOR TRANSCRIPTION
DNA CONTAINS
A CODING STAND
A TEMPLATE STRAND
THE SEQUENCE OF THE RNA TRANSCRIPT MATCHES THE SEQUENCE OF THE CODING STRAND
THE TEMPLATE STRAND IS COMPLEMENTARY TO THE RNA TRANSCRIPT
*REMEMBER-GENES CAN BE ORIENTED IN EITHER DIRECTION ON THE DNA
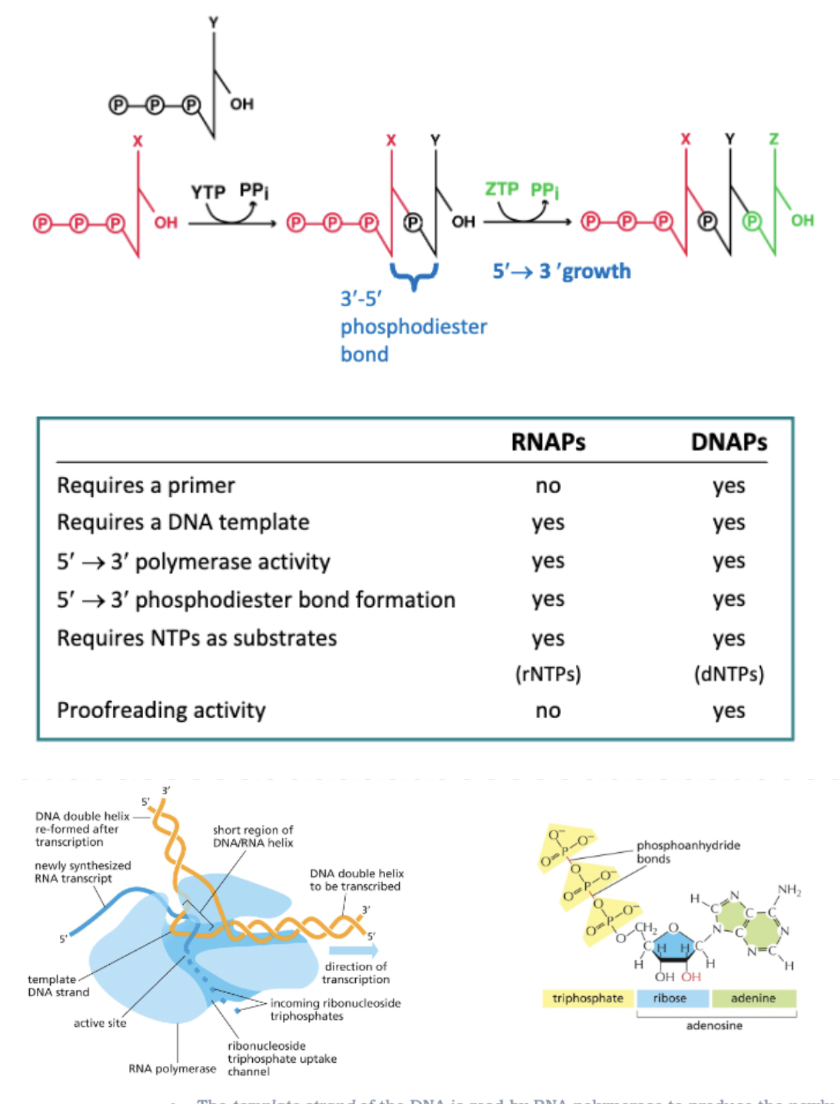
REPLICATION IS CARRIED OUT BY DNA POLYMERASES
TRANSCRIPTION IS CARRIED OUT BY RNA POLYMERASES
DNA IS TRANSCRIBED INTO…
RNA BY ENZYMES CALLED RNA POLYMERASES
SYNTHESIS IS 5’—>3’
SYNTHESIS IS DE NOVO
NO PRIMER IS REQUIRED
THE TEMPLATE STRAND OF THE DNA IS READ BY RNA POLYMERASE TO PRODUCE THE NEWLY SYNTHESIZED RNA TRANCRIPT
RNA POLYMERASE CATALYZES THE FORMATION OF PHOSPHODIESTER BONDS, ELONGATING THE TRANSCRIPT ONE NUCLEOTIDE AT A TIME
THE ENERGY FOR PHOSPHODIESTER BOND FORMATION COMES FROM THE PHOSPHOANHYDRIDE BOND IN THE ATP, GTP, CTP, OR UTP BEING INCORPORATED
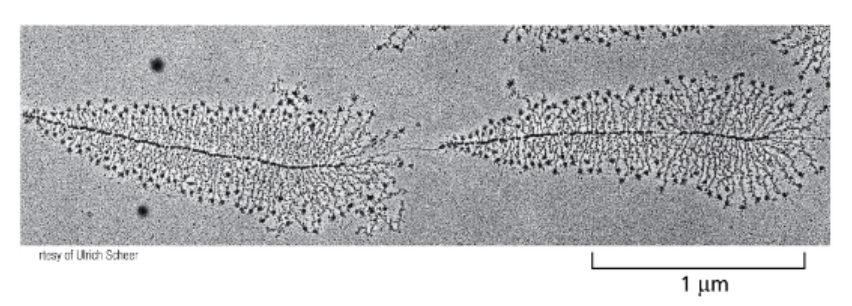
MANY MOLECULES OF RNA POLYMERASE….
CAN SIMULTANEOUSLY TRANSCRIBE THE SAME GENE
DIRECTION?
>1000 TRANSCRIPT COPIES CAN BE SYNTHESIZED IN 1 HOUR. HOWEVER, MOST GENES ARE NOT THAT HIGHLY TRANSCRIBED
WHAT ARE THE 3 MAJOR STAGES OF TRANSCRIPTION?
1) INITIATION
2) ELONGATION
3) TERMINATION
RNA SYNTHESIS IS DE NOVO
NO PRIMER. IS REQUIRED!
HOW DOES THE RNA POLYMERASE KNOW WHERE TO INITIATE TRANSCRIPTION?
IN EUKARYOTES, TERMINATION IS COUPLED TO RNA PROCESSING
WHERE IS TRANSCRIPTION INITIATED?
IT IS INITIATED AT PROMOTERS
RNA POLYMERASES RECOGNIZE AND BIND TO SPECIFIC DNA SEQUENCES CALLED PROMOTERS
THE 1ST NUCLEOTIDE TRANSCRIBED IS DESIGNATED +1
THE PROMOTER SEQUENCE IS ASYMMETRIC, ALLOWING RNAP TO BIND ONLY IN THE CORRECT ORIENTATION
NOTE THAT THE PROMOTER SEQUENCE ITSELF IS NOT TRANSCRIBED INTO RNA
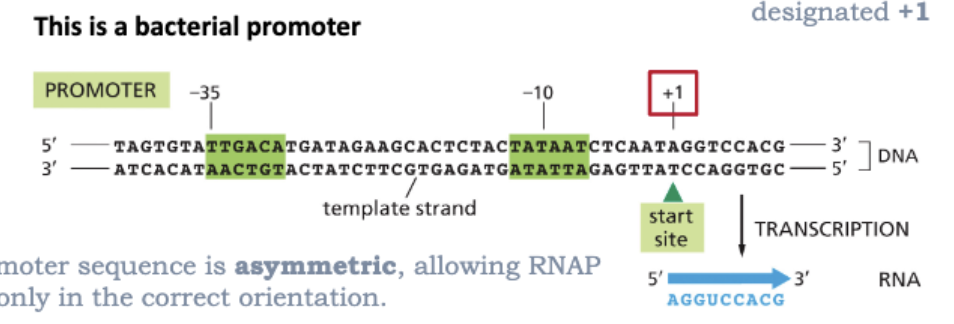
WHAT IS TRANSCRIPTION IN BACTERIA?
RNA POLYMERASE (RNAP) SCANS DNA AND BINDS TIGHTLY TO PROMOTORS
PROMOTOR RECOGNITION AND BINDING REQUIRES A SIGMA FACTOR
TRANSCRIPTION INITIATES AT THE START SITE (+1)
RNAP TRANSITIONS TO ELONGATION WITH RELEASE OF SIGMA FACTOR
ELONGATION CONTINUES UNTIL RNAP ENCOUNTERS A TERMINATION SIGNAL
RNAP RELEASES BOTH THE DNA TEMPLATE AND THE RNA TRANSCRIPT
FREE RNAP ASSOCIATES WITH A FREE SIGMA FACTOR TO BEGIN AGAIN
ALTHOUGH THE PROMOTER SEQUENCE IS NOT TRANSCRIBED INTO RNA, THE TERMINATION SEQUENCE IS PART OF THE RNA TRANSCRIPT
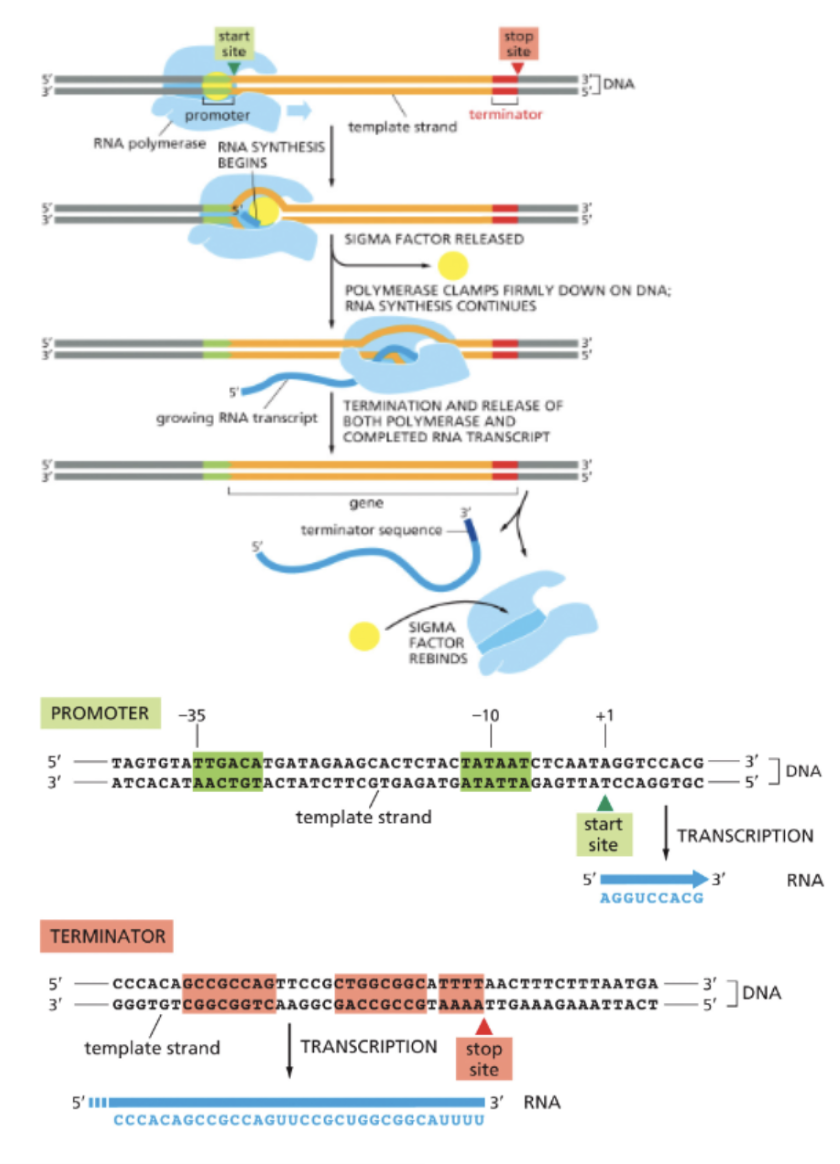
GENES CAN BE TRANSCRIBED…
FROM EITHER STRAND OF DNA
THE PROMOTER ORIENTATION DETERMINES WHICH STRAND IS TRANSCRIBED
RNA POLYMERASE ALWAYS MOVES IN THE 3’—→5’ DIRECTION WITH RESPECT TO THE TEMPLATE DNA STRAND
WHAT IS TRANSCRIPTION IN EUKARYOTES LIKE?
MUCH MORE COMPLEX
EUKARYOTES HAVE 3 DIFFERENT RNA POLYMERASES
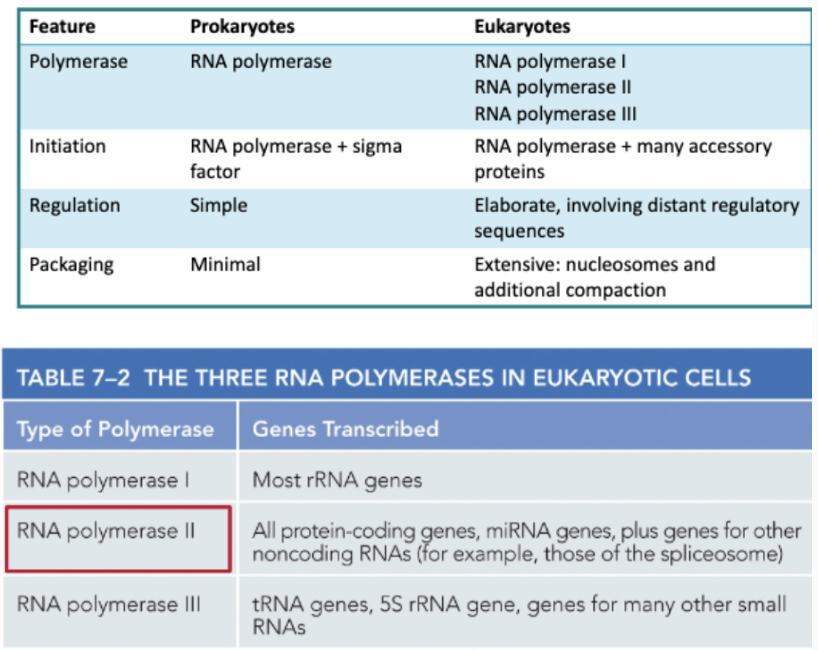
WHAT IS REQUIRED TO INITIATE TRANSCRIPTION IN EUKARYOTES?
A SET OF GENERAL TRANSCRIPTION FACTORS
GENERAL TRANSCRIPTION FACTORS ARE ACCESSORY PROTEINS THAT POSITION THE POLYMERASE AND PULL APART THE DNA
THESE FACTORS + RNA POLYMERASE II MAKE UP THE TRANSCRIPTION+INITIATION COMPLEX
MANY EUKARYOTIC PROMOTERS CONTAIN A RECOGNITION SEQUENCE CALLED A TATA BOX ABOUT 30 NUCLEOTIDES UPSTREAM OF THE START SITE
TBP (TATA-BINDING PROTEIN), A SUBUNIT. OF TFIID, RECOGNIZES THE TATA BOX
TFIIH UNWINDS THE HELIX AND PHOSPHORYLATES THE TAIL OF RNA POLYMERASE II (RNAPII)
PHOSPHORYLATED RNAP II IS RELEASED TO BEGIN TRANSCRIPTION ELONGATION
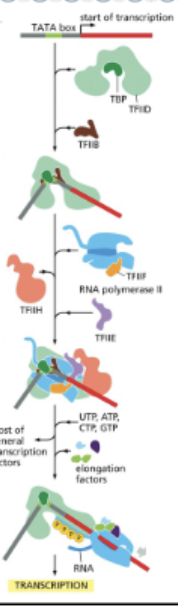
EUKARYOTIC PROMOTERS CONTAIN…
SEQUENCES THAT PROMOTE BINDING OF GENERAL TRANSCRIPTION FACTORS
THE PROMOTER SEQUENCE IS ASYMMETRIC, ALLOWING RNAP TO BIND ONLY IN THE CORRECT ORIENTATION
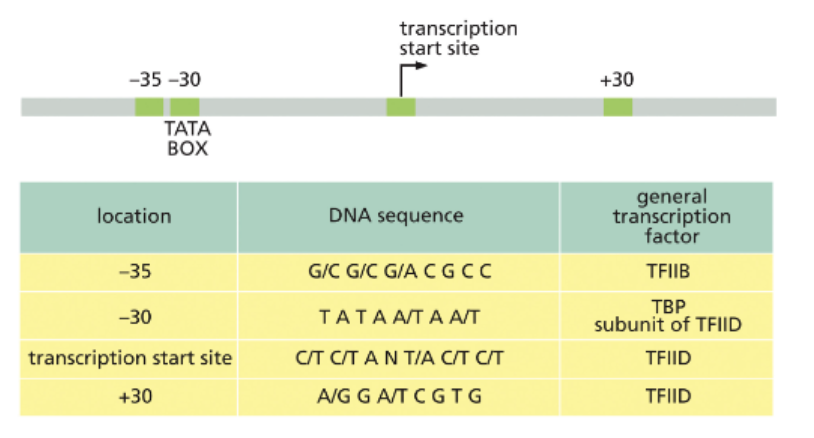
WHAT IS TRANSCRIPTION: ELONGATION?
PHOSPHORYLATION ON RNAPII BY TFIIH PROMOTES RELEASE OF THE GENERAL TRANSCRIPTION FACTORS
ELONGATION FACTORS ALLOW RNAPII TO MOVE THROUGH DNA THAT IS PACKAGED INTO NUCLEOSOMES
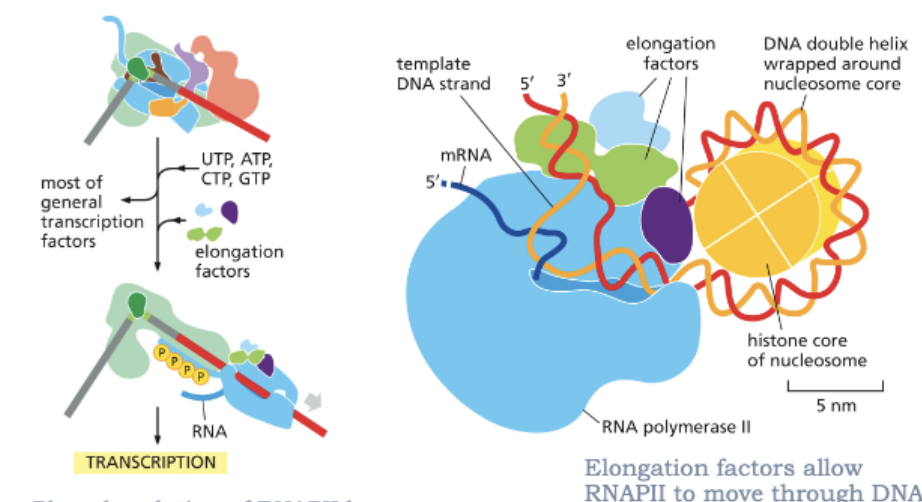
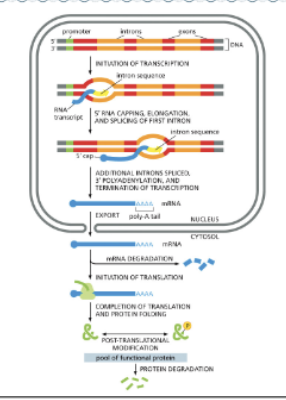
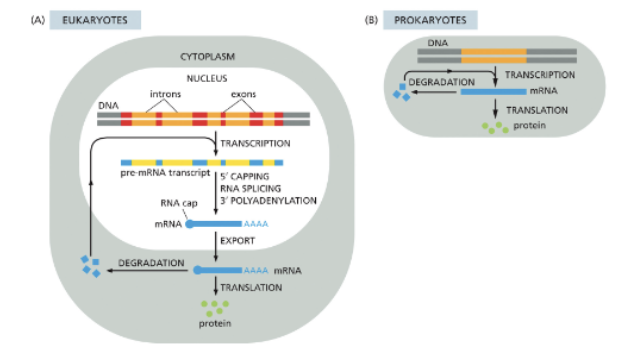
WHAT IS PROCESSED IN THE NUCLEUS?
EUKARYOTIC RNAS ARE PROCESSED IN THE NUCLEUS
IN PROKARYOTES, RIBOSOMES IMMEDIATELY ATTACH TO THE 5’ ENDS OF TRANSCRIPTS TO BEGIN TRANSLATION
EUKARYOTIC RNAS MUST BE TRANSPORTED OUT OF THE NUCLEUS THROUGH NUCLEAR PORES (ARROWS)
FIRST, THE RNAS MUST BE PROCESSED…
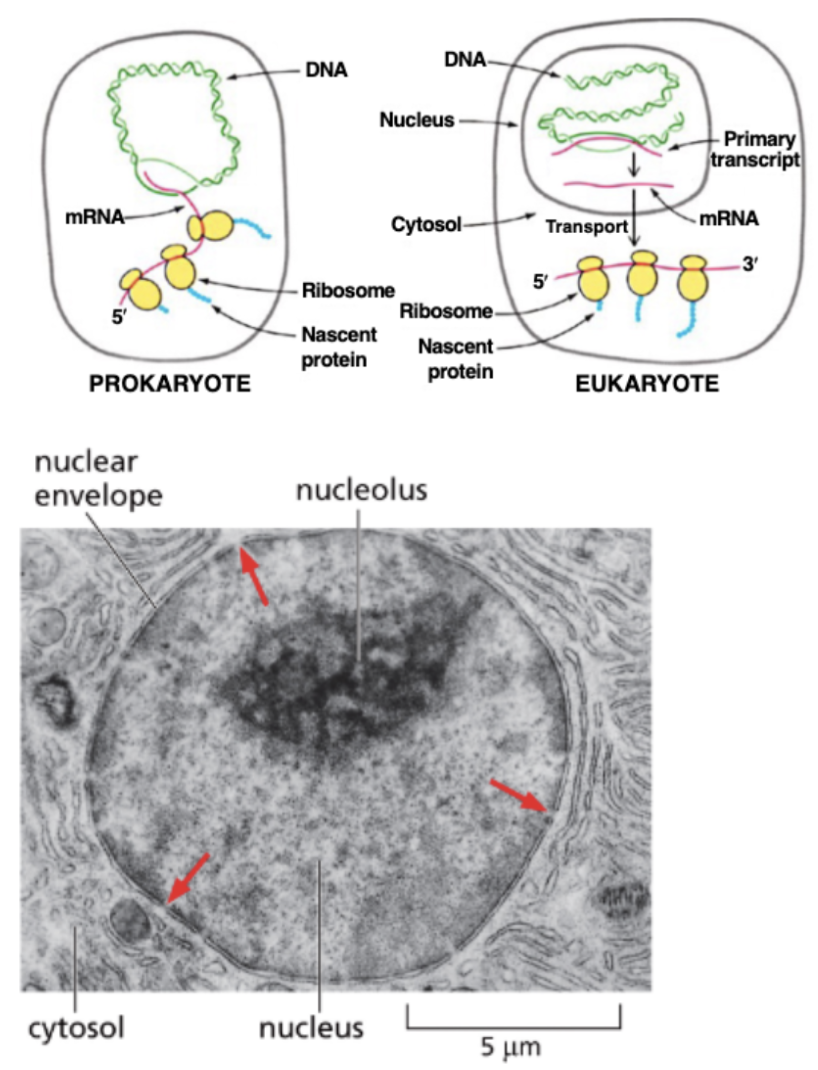
WHAT IS EUKARYOTIC RNA PROCESSING?
3 RNA PROCESSING STEPS:
CAPPING
SPLICING
POLYADENYLATION
RNA PROCESSING FACTORS ASSEMBLE ON THE PHOSPHORYLATED TAIL OF RNAP11
RNA PROCESSING IS CO-TRANSCRIPTIONAL
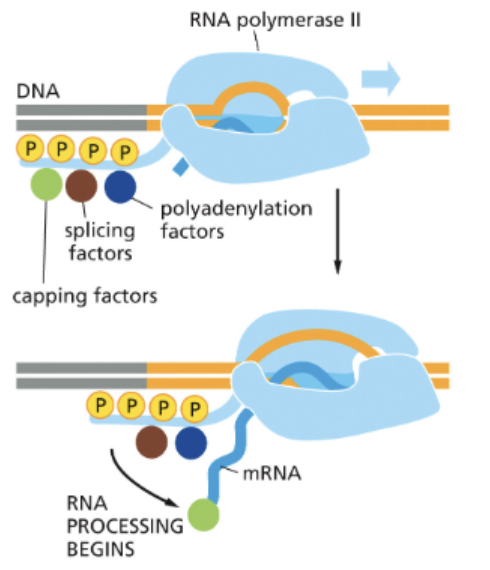
WHAT IS 5’ CAPPING AND 3’ POLYADENYLATION?
CAPPING MODIFIES THE 5’ END OF THE MRNA
THE CAP IS MODIFIED GUANINE NUCLEOTIDE
CAPPING OCCURS AFTER ~25 NTS HAVE BEEN TRANSCRIBED
POLYADENYLATION MODIFIES THE 3’ END
THE 3’ END OF THE MRNA IS FIRST TRIMMED
A SECOND ENZYME ADDS ADENINE(A) REPEATS TO THE 3’ END
THE RESULT IS POLY-A TAIL
IMPORTANT FOR:
EXPORT
STABILITY
TRANSLATION
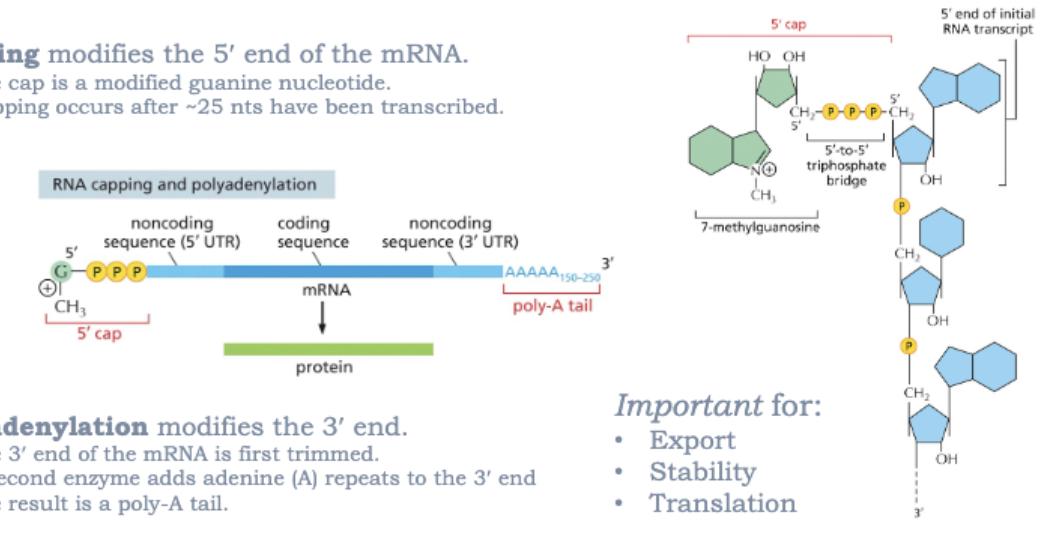
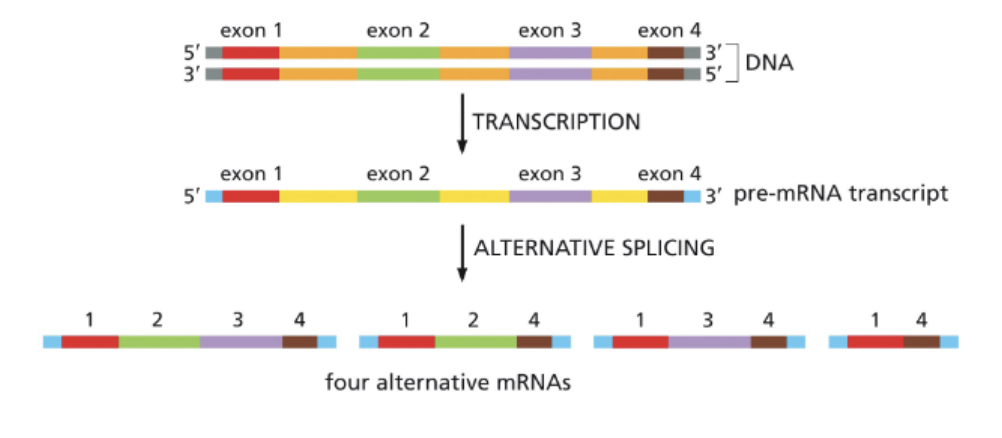
WHAT DO MOST EUKARYOTIC GENES CONTAIN?
INTRONS!
EXONS: CODING (EXPRESSED) SEQUENCES ARE SPLICED TOGETHER
INTRONS: NONCODING (INTERVENING) SEQUENCES ARE REMOVED DURING SPLICING
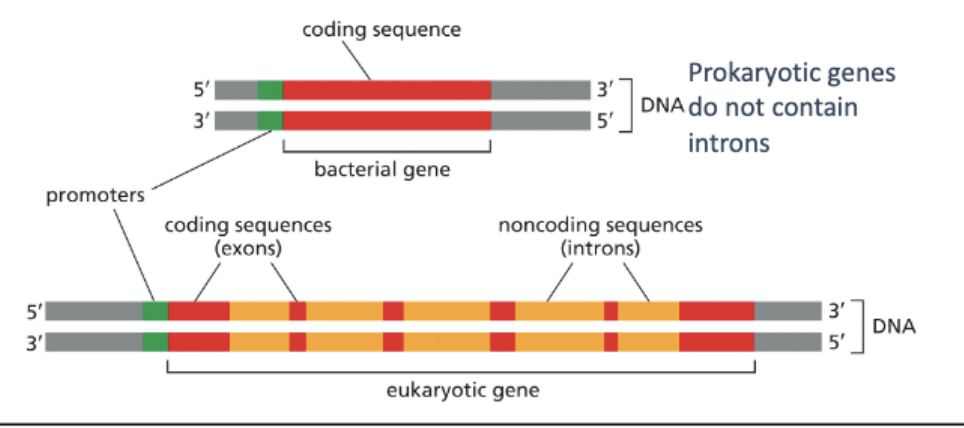
WHAT ARE REMOVED BY SPLICING?
INTRONS!
BEFORE SPLICING, THE RNA IS CONSIDERED A PRE-MRNA
SPLICING IS PERFORMED BY SPLICING MACHINERY THAT RECOGNIZES SPECIFIC SEQUENCES IN THE PRE-MRNA
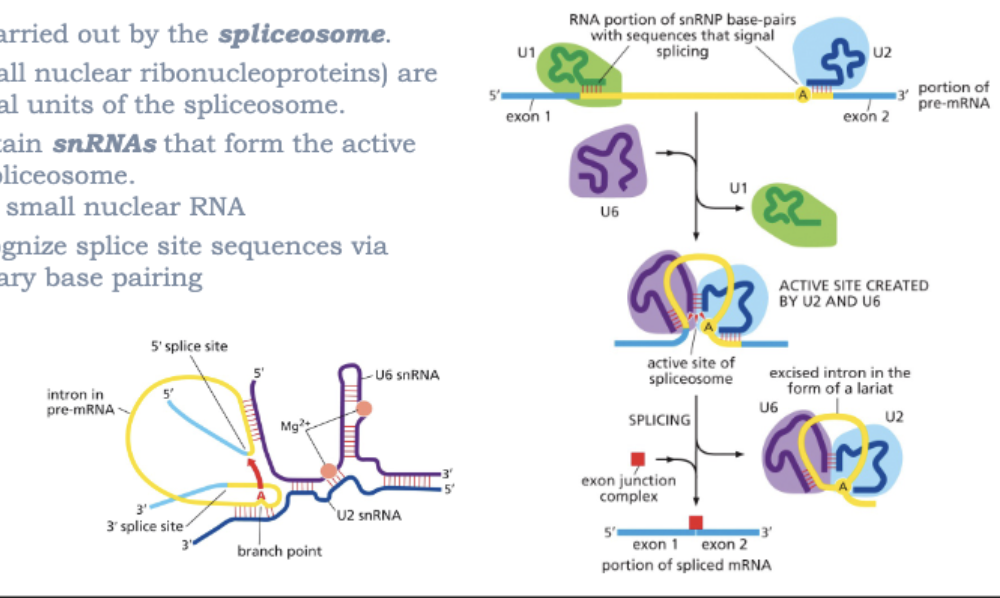
SPLICING INVOLVES 2 TRANSESTERIFICATION REACTIONS. THE 2’ OH OF THE BRANCH-POINT ADENOSINE (A) ATTACKS THE PHOPHODIESTER BOND AT THE 5’ SPLICE STRUCTURE
THE 3’ OH AT THE 3’ END OF THE EXON ATTACKS THE PHOSPHODIESTER BOND AT THE 3’ END OF THE INTRON TO JOIN THE EXONS
THE LARIAT STRUCTURE IS RELEASED
WHAT IS THE SPLICING MACHINERY?
SPLICING IS CARRIED OUT BY THE SPLICEOSOME
snRNPs (SMALL NUCLEAR RIBONUCLEOPROTEINS) ARE FUNCTIONAL UNITS OF THE SPLICEOSOME
snRNPs CONTAIN snRNAs THAT FORM THE ACTIVE SITE OF THE SPLICEOSOME
snRNA=SMALL NUCLEAR RNA
snRNAs RECOGNIZE SPLICE SITE SEQUENCES VIA COMPLEMENTARY BASE PAIRING
WHAT ALLOWS DIFFERENT PROTEINS TO BE PRODUCED FROM A SINGLE GENE?
ALTERNATIVE SPLICING!
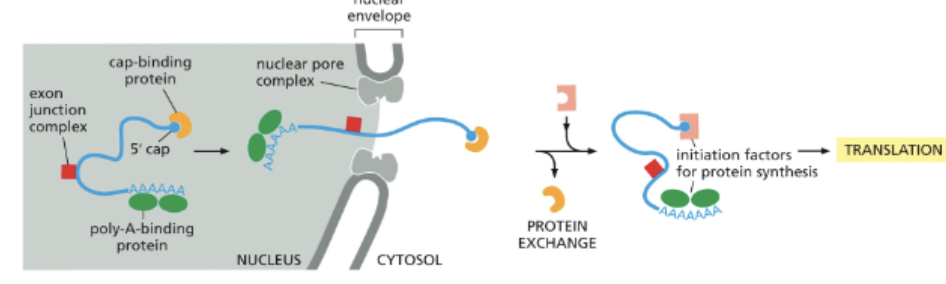
WHAT ARE EXPORTED TO THE CYTOPLASM THROUGH NUCLEAR PORES?
MATURE MRNAS
ONLY CORRECTLY PROCESSED MRNAS ARE EXPORTED. THESE RNAS ARE BOUND TO:
CAP-BINDING PROTEIN
POLY-A-BINDING PROTEIN
EXON JUNCTION COMPLEX
NUCLEAR PORE COMPLEXES CONNECT THE NUCLEOPLASM WITH THE CYTOSOL
MRNAS ARE TRANSLATED IN THE CYTOSOL AND EVENTUALLY DEGRADED
THE LIFETIME OF A EUKARYOTIC MRNA IS VARIABLE (30 MIN-10 HOURS)
THE 3’UTR CONTRIBUTES TO THE STABILITY OF THE MRNA
WHAT IS MORE COMPLEX IN EUKARYOTES THAN IN PROKARYOTES?
GENE EXPRESSION!
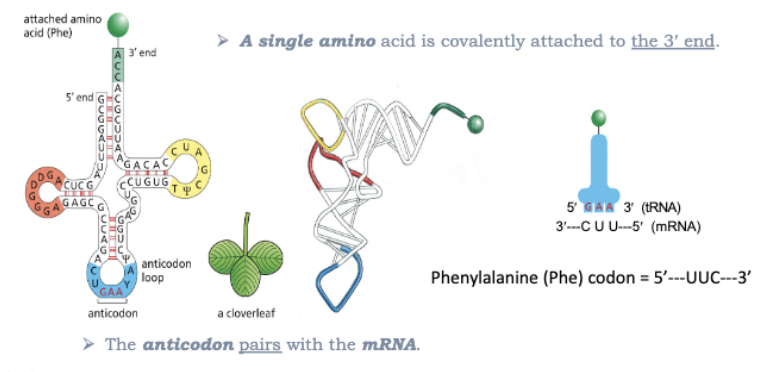
WHAT IS THE ADAPTOR MOLECULE?
TRNA !
A SINGLE AMINO ACID IS COVALENTLY ATTACHED TO THE 3’ END
THE ANTICODON PAIRS WITH THE MRNA
WHAT IS THE GENETIC CODE?
IS A NONOVERLAPPING TRIPLET CODE
FOR 20 AMINO ACIDS, AT LEAST 3 NUCLEOTIDES ARE NECESSARY PER CODON
BECAUSE THE CODE IS NONOVERLAPPING, EACH FRAME GIVES A DIFFERENT SET OF TRIPLETS
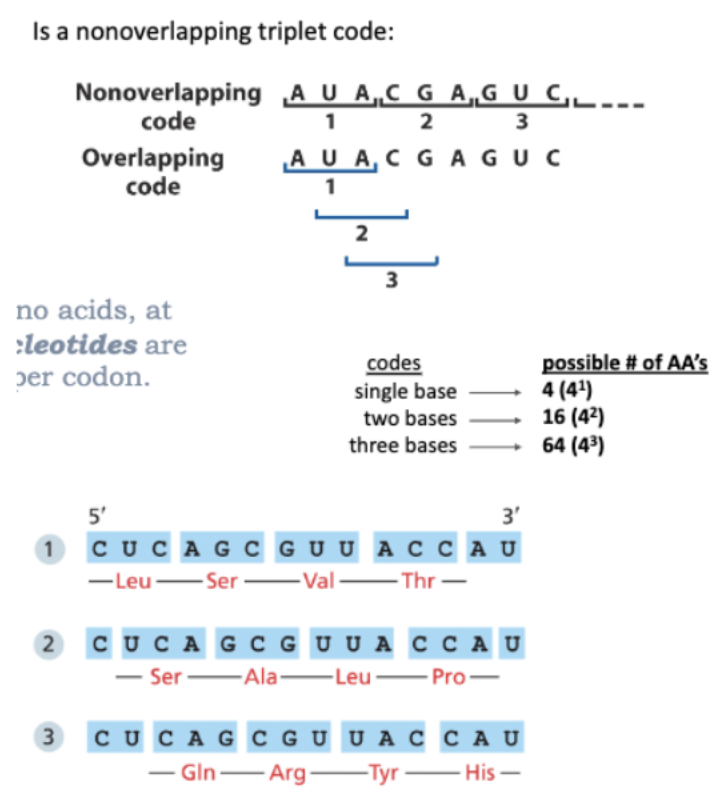
WHAT IS THE READING FRAME?
IN HOW MANY WAYS CAN WE READ THIS?

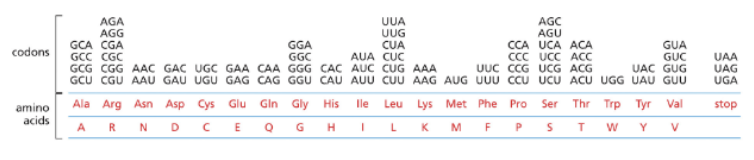
WHAT DOES THE GENETIC CODE DEFINE?
THE GENETIC CODE DEFINES THE RULES BY WHICH THE NUCLEOTIDE SEQUENCE OF A GENE IS TRANSLATED INTO THE AMINO ACID SEQUENCE OF THE PROTEIN
THE GENETIC CODE IS A NONOVERLAPPING TRIPLET CODE-EACH TRIPLET IS CALLED A CODON
CODONS CODE FOR AMINO ACIDS, EXCEPT FOR THREE STOP CODONS
THERE ARE 64 CODONS BUT ONLY 20 AMINO ACIDS-THE CODE IS REDUNDANT
THERE IS ONLY 1 CODON THAT START ALL PROTEINS: AUG-THE START CODON
HOW DOES THE TRNA RECOGNIZE THE SPECIFIC AMINO ACID?
REMEMBER THIS?
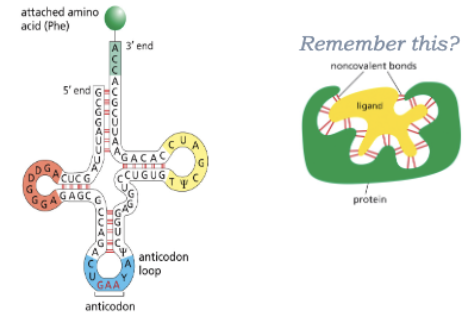
WHAT IS THE GENETIC CODE TRANSLATED BY?
BY AMINOACYL-TRNA SYNTHESASES AND TRNAS!
EACH AMINO ACID HAS A DIFFERENT SYNTHETASE ENZYME-A PROTEIN! (20 IN TOTAL)
SYNTHETASES RECOGNIZE SEQUENCES IN THE ANTICODON AND THE AMINO ACID-ACCEPTING ARM
TRNAS COVALENTLY LINKED TO AMINO ACIDS AT THEIR 3’ ENDS ARE CHARGED
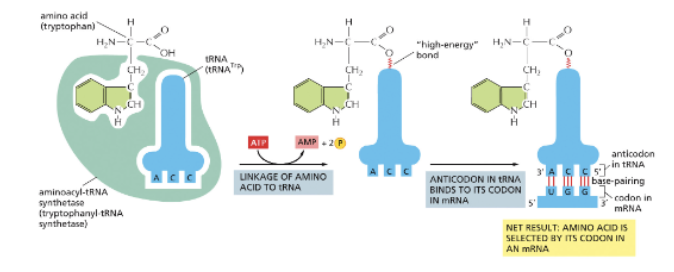
WHAT MESSAGE IS DECODED ON RIBOSOMES?
THE MRNA MESSAGE
MOST EUKARYOTIC CELLS HAVE MILLIONS OF RIBOSOMES IN THE CYTOPLASM
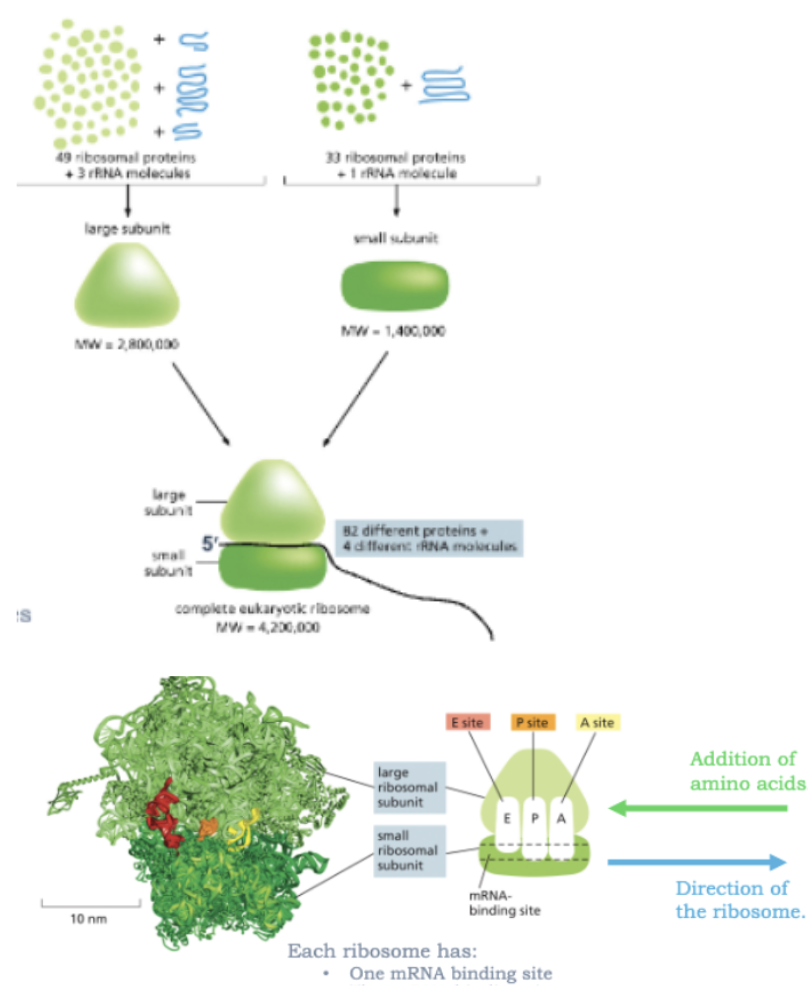
THE RIBOSOME IS A LARGE COMPLEX OF rRNAS AND RIBOSOMAL PROTEINS
THE RIBOSOME HAS TWO SUBUNITS: A LARGE SUBUNIT AND A SMALL SUBUNIT
4 STAGES OF TRANSLATION:
CHARGING
INITIATION
ELONGATION
TERMINATION
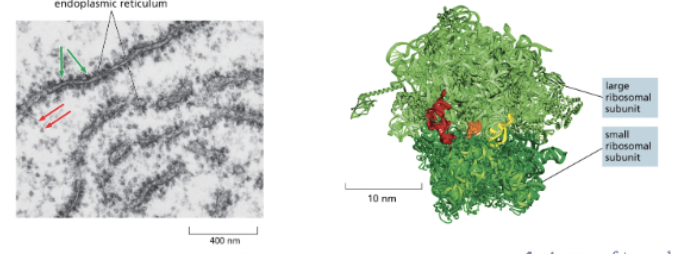
WHAT IS THE EUKARYOTIC RIBOSOME?
SMALL SUBUNIT
BINDS THE FIRST MRNA
MATCHES TRNAS TO CODONS
LARGE SUBUNIT
CATALYZES PEPTIDE BOND FORMATION
INITIATION: THE SUBUNITS COME TOGETHER NEAR THE 5’ END OF THE MRNA
ELONGATION: THE MRNA IS PULLED THROUGH THE RIBOSOME AND TRANSLATED ONE CODON AT A TIME USING TRNAS AS ADAPTORS
TERMINATION: THE SUBUNITS SEPARATE WHEN TRANSLATION IS COMPLETE
PROKARYOTIC RIBOSOMES ALSO HAVE A SMALL AND A LARGE SUBUNIT
RATES:
2 AMINO ACIDS/SEC IN EUKARYOTES
20 AMINO ACIDS/SEC IN PROKARYOTES
EACH RIBOSOME HAS:
1 MRNA BINDING SITE
3 TRNA BINIDNG SITES
A=AMINOACYL-TRNA SITE
P=PEPTIDYL-TRNA SITE
E=EXIT SITE
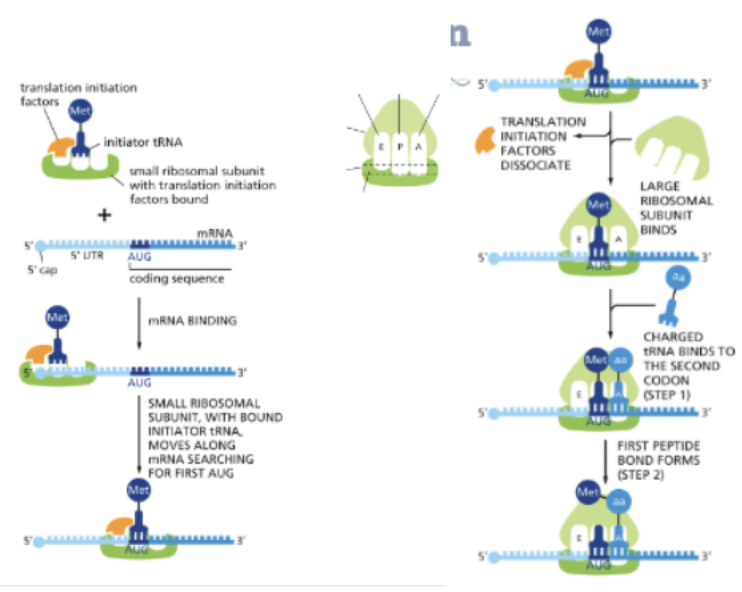
WHAT IS THE INITIATION OF TRANSLATION?
TRANSLATION BEGINS WITH AN AUG CODON AND ESTABLISHES A READING FRAME
INITIATOR TRNA IS A SPECIALLY CHARGED TRNA THAT CARRIES METHIONINE
INITIATOR TRNA AND INITIATION FACTORS ARE LOADED INTO THE P SITE OF THE SMALL SUBUNIT
THE COMPLEX BINDS TO THE 5’ CAP OF THE MRNA
THE COMPLEX MOVES ALONG THE MRNA “SCANNING” FOR THE FIRST AUG CODON
THE LARGE SUBUIT BINDS, AND THE INITIATION FACTORS DISSOCIATE
A CHARGED TRNA BINDS IN THE A SITE, AND IF THE CODON-ANTICODON BASE PAIRING IS CORRECT, PEPTIDE BOND FORMATION OCCURS
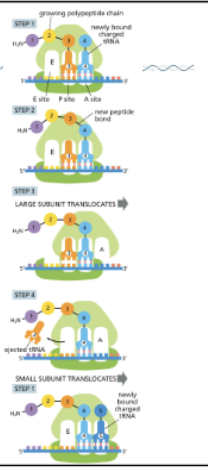
WHAT IS ELONGATION?
ELONGATION IS A 4-STEP CYCLE
CHARGED TRNAS ENTER THE A SITE AND BASE PAIR WITH THE CODON ON THE MRNA
PEPTIDE BOND FORMATION OCCURS BETWEEN THE NEW AMINO ACID AND THE CHAIN HELD BY THE TRNA IN THE P SITE
THE LARGE SUBUNIT SHIFTS BY ONE CODON, MOVING THE UNCHARGED TRNA TO THE E SITE
THE SMALL SUBUNIT TRANSLOCATES AND THE UNCHARGED TRNAS IS RELEASED
THIS CYCLE IS REPEATED UNTIL A STOP CODON IS REACHED
MRNA IS READ 5’ —> 3’
PROTEIN IS MADE N—>C
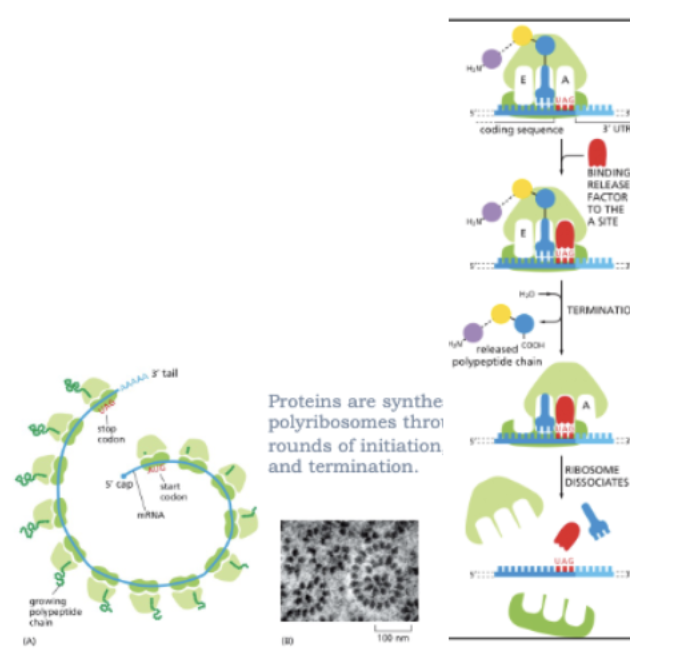
WHAT IS THE TERMINATION OF TRANSLATION?
THE TRANSLATION IS TERMINATED AT A STOP CODON
UAA, UAG, UGA
RELEASE FACTOR BINDS THE A SITE AT STOP CODONS INSTEAD OF A CHARGED TRNA
WATER IS ADDED TO THE POLYPEPTIDE INSTEAD OF AN AMINO ACID
THE SYNTHESIZED POLYPEPTIDE IS RELEASED AND THE RIBOSOME DISSOCIATES
PROTEINS ARE SYNTHESIZED ON POLYRIBOSOMES THROUGH MULTIPLE ROUNDS OF INITIATION, ELONGATION, AND TERMINATION
WHAT IS POST-TRANSLATIONAL MODIFICATIONS OF PROTEINS?
POST-TRANSLATIONAL MODIFICATIONS INCLUDE
METHYLATION
ACETYLATION
PHOSPHORYLATION
UBIQUITINATION
GLYCOSYLATION
MODIFICATIONS CAN AFFECT BINDING TO CO-FACTORS, STABILITY, AND SUBCELLULAR LOCALIZATION

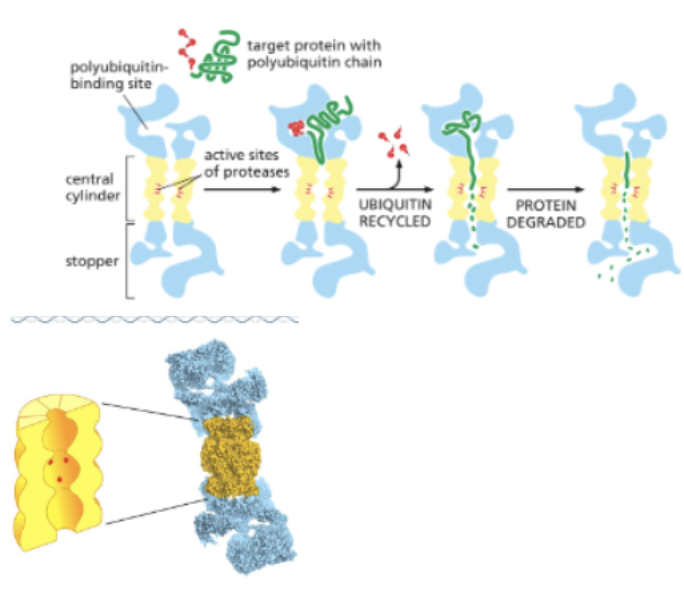
WHAT IS REGULATION OF PROTEIN DEGRADATION?
PROTEOLYSIS: THE BREAKDOWN OF PROTEINS INTO THEIR CONSTITUENT AMINO ACIDS
PROTEASES: CUT PEPTIDE BONDS
PROTEASOME: A MULTI-SUBUNIT MACHINE THAT DEGRADES PROTEINS
PROTEINS TO BE DEGRADED ARE MARKED BY THE COVALENT ADDITION OF UBIQUITIN
WHAT IS THE OVERVIEW OF EUKARYOTIC GENE EXPRESSION?
IMAGE!