Unit 11 - Gene Expression
1/35
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
36 Terms
Gene Expression
The determination of the amount and type of exact proteins in a cell at a given time.
Gene Expression includes
if a gene gets read or not
how transcript is spliced
how long transcript will last in the cytoplasm
how many times transcript will be translated
is the transcript be translated right away or delayed
how the protein is modified after its made
when the protein is activated (if made initially in the wrong shape)
how long the protein lasts before being destroyed
Why do cells need to control gene expression?
Different stages of development
Differentiation
Change during the cell cycle
Meet demands of the organ
Respond to changing environment
Respond to outside signals
Make sure you are not wasting energy making things you don’t need
Epigenetics
Epi means on top of
epigenetics means on top of the genome
It involves several types of signals that either loosen or tighten the DNA
If the DNA is super tight (not as tight as in chromosomes) it cannot be read
Remember that DNA can only be read in G1 and G2 anyway, so DNA is in chromatin form at this time
Euchromatin
true chromatin that isn’t methylated and is loose enough to be read
Heterochromatin
super tight highly methylated chromatin
How do epigenetics work?
Methyl groups added to the DNA make it tighten and make it unreadable – the more methyl groups on an area of DNA, the less readable – RNA polymerase II can’t get in to make an mRNA copy.
How does the cell know where to put the methyl groups in any given cell at any give time? Enzymes add other tags to the DNA based on signals received that tell the methylating enzymes where to add the methyl groups.
Acetyl groups can be added to the histone proteins, causing the DNA to unattach from the histones and unwind and loosen allowing a gene to be read
When do epigenetic patterns get added to DNA? When can gene expression change?
Most epigenetic patterns get wiped off in the sperm and egg
This makes sense because the zygote cell has to be able to divide and then each cell needs an epigenetic pattern to make it a certain type of cell.
In about 80 genes, the genes are methylated while in the sperm and egg – this is called imprinting. That means a gene inherited from the egg will have a different methylation pattern than that same gene inherited from the sperm
Example – same mutated gene inherited from the father – Prader Willi, from mother – Angelman’s
When are genes methylated to control gene expression?
Methylation tags added during embryonic development to help cells become specific types (e.g., neuron vs. muscle), even though all cells have the same DNA.
Mom’s diet affects methylation — like in mice where certain foods turned off the agouti gene.
Early life experiences can change methylation, like rat pups getting licked more → demethylated GR gene → more relaxed for life.
Methylation patterns shift over time, especially during big changes like puberty or menopause → explains aging effects (e.g., hair color changes).
Lifestyle choices (food, stress, etc.) affect methylation, which influences gene expression and long-term health.
Some methylation patterns survive in sperm/egg, passing traits to children (transgenerational epigenetics).
Examples:
Mice scared of cherry blossom smell → their babies feared it too.
Insects given antibiotics grew eye bristles → passed to offspring.
Men starved/gluttonous in puberty → sons/grandsons died decades earlier.
Foxy Gene
🧠 Foxo is a special gene that helps control which other genes are turned on or off inside a cell.
🧬 It acts like a master switch for important genes that control cell survival, repair, and stress response.
⚙ When Foxo is active, it tells the cell to protect itself, slow down growth, or fix damage.
🚨 If there's stress (like too little food or too much damage), Foxo turns on helpful genes to keep the cell safe.
🔒 If Foxo gets turned off, the cell might grow too fast or stop fixing itself — this can lead to aging or even cancer.
🌱 That’s why Foxo has such a big effect on the cell — it controls many genes that keep the cell healthy and balanced.
Activators or Repressors – Bind to Distal Control Elements
🚀 Activators turn genes on by boosting transcription.
🛑 Repressors turn genes off by blocking transcription.
📍 They attach to distal control elements, like:
Enhancers (for activators)
Silencers (for repressors)
📏 These are often far from the gene but still affect it by looping the DNA.
silencer
🚫 A silencer is a DNA element where a repressor binds.
🔕 It shuts down gene expression, like pressing a mute button.
Transcription Factors – Bind to Proximal Control Elements AND to the Promoter
🧩 These are proteins that help RNA polymerase know where to start.
🎯 They bind to:
Proximal control elements (close to the gene)
The promoter (right before the gene)
🔐 They’re key helpers in turning a gene on.
DNA Parts
Proximal Control Elements:
🎚 Short DNA sequences near the promoter.
Help fine-tune when a gene is turned on.
Promoter:
🏁 Starting line where transcription begins.
RNA polymerase binds here.
Distal Control Elements:
🌉 Far-away DNA sections like enhancers and silencers.
Still control gene activity through DNA looping.
Steroid Hormones
Steroids bind to a cytoplasmic receptor and translocate into nucleus.
Steroid/receptor complex binds to DNA in upstream regulatory elements to switch on genes.
Bacterial Gene Expression
Allows bacteria to live in a changing environment
Operons – a whole gene unit – all genes necessary for an enzymatic pathway are lined up behind a promoter and operator
Promoter Operator Genes Term. Seq.
Bacterial Operons
Operator – controls access to promoter for RNA polymerase
Operator is always “on” unless a protein is bound to it
Repressor – binds to operator and blocks RNA polymerase from binding – specific to the operon.
Repressors for Operators
For Anabolic Operons
Repressible Operon
Product shuts off operon by activating the repressor
For Catabolic Operons
Inducible Operon
Substrate turns on operon by deactivating the repressor
In order to read a gene, what must be true?
It can’t be part of a nucleosome
If it is part of a nucleosome, the histones must be acetylated
It can’t be highly methylated
It must be attached to the nuclear matrix
It needs transcription factors – both specific and general – bound to the promoter and proximal control elements
It needs activators and not repressors bond to the distal control elements
Post Transcriptional regulation of Gene expression
RNA splicing - alternate splicing creates diff proteins from same gene
RNA degredation - things bind to 3’ UTR controlling when mRNA gets chewed up
make it last longer by preventning it from chewing up, also can make it get chewed up by removing itself
Control of translation proteins bind to the 5’ UTR and allow or block translation
Post translational modifications - can modify protein differently in the RER to make protein diff and can change the shape later by activating a protein
Protein degredation proteins tagged for destruction
Control of RNA Processing
Alternative Splicing – spliceosomes bind to ends of introns – identified by the specific and general mRNA sequences that snRNP’s bind to - proteins can bind to repress the binding of snRNP’s
every intron has 2 snerps that identify the ends
if you have different snerps then you splice out different introns
Transposons “Jumping Genes”
May copy and move or just move. May jump into a gene and disrupt it; may jump into a regulatory area and increase or decrease production of that protein
Contains a gene for an enzyme that when expressed, cuts the gene out and inserts it somewhere else
May contain just the gene for that enzyme or may contain additional genes
Retrotransposons – code for RNA which is then copied into DNA and inserted somewhere else in genome
Gene Rearrangements
Example: antibodies
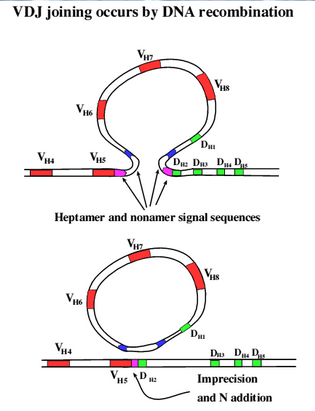
Like mRNA splicing except done in embryonic cells that will become immune cells
Done in DNA and the DNA removed is destroyed so this is gene splicing not mRNA splicing, and it is permanent
Gene Expression and Embryonic Development
How does one cell become many types of cells?
If it takes different signals to cause differential gene expression, how do cells make or receive different signals if they start out all the same?
What kinds of proteins will make the biggest difference in gene expression cell to cell?
have to have different signals for different gene expressions
transcription factors are the proteins that affect gene expression the most
Embryonic Development 1
After the zygote divides – cells need to express different proteins based on location – How?
Step #1: Cytoplasmic Determinants – RNA and proteins distributed and anchored in the egg that signal development – when the cell divides – different cells end up with different mRNA and proteins that it got from the egg. This leads to step 2
pre made proteins
anchored in the cells in a particular place so that when it divides its in one cell and not the other
whole program for your whole body is in the egg
Embryonic Development 2
After the zygote divides – cells need to express different proteins based on location – How?
Step #2: Induction – cells receive different signals from surrounding cells that control development (must also have diff receptors to receive the signals
some cells make certain messengers and some cells make certain recpetors
Embryonic Development 3
After the zygote divides – cells need to express different proteins based on location – How?
Step #3: Turn on master regulatory genes which turn on tissue specific genes to make tissue specific proteins
How do cytoplasmic determinants lead to steps 2 & 3
These determinants (RNA’s and proteins) code for signals, receptors, and transcription factors (most master control genes)
Cells will make different signals and receive different signals and turn on different genes based on what determinants they have
The determinants a cell has is based on its location, so eventually a cell will “know” what it should be by its location.
signals and receptors are just as important as transcription factors
can make and receive different signals
ex: fruit fly early embryonic dev.
Example Early Embryonic Development in the Fruit Fly
mRNA of bicoid gene is anchored to cytoskeleton in anterior portion of the fruit fly egg
after division – only the cells that will become the front of the fly make the bicoid protein (only cells that got the mRNA when the cell divided)
front cells with bicoid protein (transcription factor) develop the front structures because the bicoid protein turns on genes to give those cells the right receptors and cause them to make the right messengers and make the right transcription factors to develop the head
If inject the bicoid protein other places develop anterior structures in other places
Body Plan Patterning
Both cytoplasmic determinants and inductive signals help set up positional information
Homeotic/Homeobox/Hox Genes
Are highly conserved and have same sequences within the genes called homeoboxes.
Code for transcription factors and are master control genes
Hox genes found in clusters on the chromosomes
Genes are lined up in order of what part of the body they control the formation of.
basically determine the body plan
“program for it” — master control genes
Oncogenes
cancer causing genes induced by viruses
Protooncogenes
promote cell division, moving through cell cycle, normal genes that become cancer causing when they are:
mutated
amplified
or move into an area with an active promoter (jumping gene)
examples- growth factors, cell cycle proteins
Proto-oncogene Example
Ras gene – codes for a G-Protein (activated by a receptor when messenger binds and sets off a series of chemical reactions leading to increase in cell division thru the activation of transcription factors)
Mutated Ras gene – G protein is always on even when nothing is bound to receptor
Found in 20-25% of all human tumors
Tumor Supressor Genes - restrict cell div, stimulate apoptosis
Repair damaged DNA
Control cell adhesion
Inhibit the cell cycle
Activate cell suicide if damage is unfixable
Example – p53 gene (p53 mutation found in over 50% of human tumors)
much more like to get cancer from having p53 but p53 doesn’t cause it
In response to damage it :
Halts the cell cycle
Turns on DNA repair enzymes
Activated apoptosis if damage cannot be repaired
Cancer Cells vs. Normal
Cancer cells can be identified because they look different —
cytoskeleton misshapen
nucleus misshapen
The less round the nucleus, the more aggressive the cancer – therefore cancer due to messed up gene expression