Microbiology - Bacterial and functional genomics
1/36
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
37 Terms
The e.coli chromosome
Circular
oriC
Genes are densely packed
Contains operons
Only ~118 b between genes
Sizes range from 112kbp to 14 Mbp
Haploid
No introns
Common mobile genetic elements
Plasmids
Transposons (jumping) and insertion sequences (both often have resistance genes)
Integrons (accumulate 'useful;'genes e.g resistance))
Bacteriophage (lytic/lysogenic)
Plasmids are only 2.5% of the size of the bacterial chromosome
Extrachromosomal - conjugation
Plasmids confer diverse phenotypes:
Extrachromosomal but can be integrated
Varied size range
Often mobile (conjugation)
Can confer accessory functions
Bacterial genomes vary between strains of the same species:
Core genome - all strains of a species will contain these genes
Accessory genome - includes mobile elements (vary)
All the genes found across all different isolates of a species is called the pangenome
Pathogenic bacteria frequently possess pathogenicity islands - clusters of genes of foreign consent present in certain strains (not core) and correlated with virulence
types of genome sequencing
Sanger - slow and expensive but very accurate
Illumina - highly scalable sequencing of short fragments of DNA
Sequence assembly: bioinformatics
Overlapping sequences are aligned
However repetitive sequences interfere with assembly
Complete genome assembly:
Compare with known sequences
Use long-read sequencing methods (oxford nanopore)
Infectious disease and epidemiology and sequencing:
Pre-screen patients for problem organisms
Treat or quarantine patients before hospitalisation is necessary
Can identify all the virulence factors and antibiotic resistances in a single patient sample and you can prescribe appropriate treatment using combination (precision medicine)
Identification of new threats and rapid response
Sequence isolates from multiple patents
Determine epidemics
Track spread of disease and identify the origin of new variants
Metagenomics
allowing for new ways of studying the diversity of bacteria because many bacteria are unculturable difficult to isolate
The study of genetic material recovered directly from environmental samples
Gut microbiome
skin microbiome
Ways we use bacteria everyday (3)
Food industry: bacteria generate useful properties such as flavour and texture in every food products
Water treatment facilities:
Activated sludge in water treatment plants contain microbes that metabolise organic matter to CO2
Therefore cleaning out waste organic material from the water
Can be reused to "seed" the aeration tank
Composting is a natural process of decomposing heterogenous wastes in a controlled, microbially active environment - can be highly optimised or rudimentary - basic ingredients are carbon, nitrogen, oxygen and water
Bioremediation
Bacteria used in environmental applications
A process that uses mainly microorganisms, planta or microbial/plant waste to detoxify contaminants in the soil or other environments
Rapidly form spherical biofilms (to increase SA) around oil droplets which expedite degradation
Hydrocarbon degrading bacteria occur naturally
Xenobiotic pollutants often are resistant to natural degradation
Microbes that can deal with them can often be found and exploited
For example, pesticide de-chlorination using oxygenases
New bacteria is being discovered and characterised - particularly with a focus on methods to degrade plastics
Biotechnology
Bacterial cell factories - living organisms which produce medically or commercially useful biomolecules a(e.g antibiotics, enzymes, drugs, hormones)
The main advantages is that cell does the hard work
Bacterial cell cultures grow quickly and easily, input material often cheap and environmentally friendly, whereas chemical synthesis can be difficult and expensive
Antibiotics
Naturally occurring secondary metabolites produced naturally by bacteria
Ongoing quest for new biosynthetic gene clusters which may produce new drugs
Discovery of previously "unculturable" microbes may lead to new drug discovery
E.g tetracycline/gentamicin
vitamins (e.g b12)
Essential for DNA synthesis and metabolism
Very complex
Only produced by certain bacteria and archaea and in certain foods e.g
Hence a popular supplement
hard to produce synthetically
Genetic modification of bacteria:
Expressing a mammalian or a mutant protein
Using recombinant DNA technologies, these proteins can be made in large quantities bny factorial fermentation too
First, we clone the gene of interest into an expression vector, then transform into E.coli
Generates a microbial "factory" for the gene of interest
E.coli factories are grown by large scale fermentation
Expression of the gene of interest is induced
Cells are lysed and the expressed protein is purified for research, commercial or medical
recombinant therapies
Huge range of actual and potential application
Well understood theoretical and practical path to commercialisation
Small scale, high value products and high value, low cost commodities
Synthetic biology and bio-engineering:
Synthetic biology is the design and construction of new biological parts, devices and systems, and the re-design of existing, natural biological systems for useful processes
Wide ranging definition - Multidisciplinary field
often uses engineering concepts
Biological components of bio-engineering:
Components need to be modular and well-characterised
Can be put together in any order
Allow for complex designs and predictable outcomes
Components can be replicated by symbols (like in electrical engineering)
Gene regulation recap: (brief)
Units of DNA encoding genes are controlled by the activity of genetic elements including the promoter, the RBS, the terminator and the DNA binding transcription factors
The classic example is the lac operon- the lac repressor and it switching "on" in the presence of lactose (allolactose preventing the repressing characteristic)
Optogenetics:
Optogenetic circuits from MIT
Different wavelengths of light trigger the production of different coloured compounds by E.coli
Control of gene expression though light
Induction of bacterial gene expression in tissue via small molecule signalling
Potential therapeutic delivery tool
Temperature-controlled gene expression
Gene expression can respond to temperature
E.g Piraner et al - genes expression turned on between 40-45 degrees
The circuits dictate the colour response on the e.coli
Potential applications in temperature triggered therapeutics (e.g inflammation and infection)
Protein engineering: (brief)
Adding new functions to proteins or improving their current functions
These proteins will change the behaviour/functionality of bacteria which express them
E.g PETase (from plastic degrading bacteria) by active site mutation leads to increased enzyme activity
Metabolic engineering:
Gene circuits and engineered proteins can be combined to alter or create new metabolic processes in bacteria
Complex, multi-system
Improves efficiency of current metabolic processes, or allow bacteria to synthesise new compounds
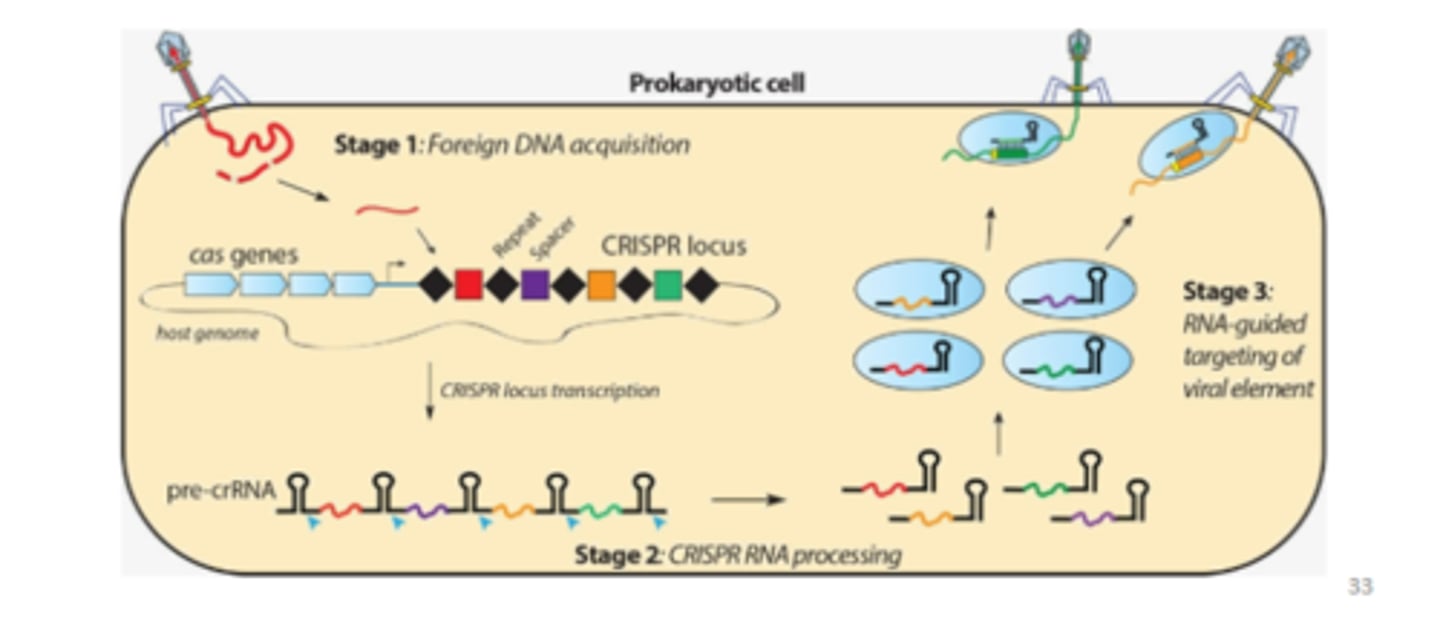
CRISPR DNA editing technology
Evolved as a bacterial anti-viral immune systems
Can be modified to allow precise editing of any genome
A highly powerful tool with wide ranging applications from synthetic biology to basic research to gene therapy (ethics)
If the cell gets infected by the same virus again, guide-RNAs will detect if the same virus is infecting them again due to those regions of homology. The loop attracts the cas enzymes to that interaction and will physically chop up the virus before it can infect the cell once more (a memory system - similar to the immune system). You can engineer these enzyme systems to knock-in/out genes by utilising these guide-RNAs
Detecting our microbes: (2 methods)
Culturing
Grow 'live' microbes than can be tested in lab studies (mechanistic)
Required for drug/therapy development
DNA/RNA sequencing:
Determine microbial community 'fingerprint'
Rapid and high throughput
NGS microbiome profiling approach to profiling our microbes :
Genus level profiling (16s rRNA)
Shotgun approaches:
Metagenomics (DNA)
Metatranscriptomics (RNA)
Metabolomics
Proteomics
Species vary person to person but function coded for stays the same (conserved)
Culturomics
Previously estimated could only culture <10% of the human microbiome
BUT culturomics developed to culture and identify 'unknown' microbes
rebirth of culture techniques in microbiology
very important for:
- improving reference databases for further NGS approaches
- phenotypic/mechanistic studies
- culture collections
- therapy development
The gut microbiota:
Most densely colonised microbial habitat found in nature
2500+ species
Broad range of physiological conditions creates niches along the gut
4 main bacterial phyla
Proteobacteria
Actinobacteria
Firmicutes
Bacteroidetes
Major functions of the microbiota: (2)
Breaking down complex sugars
97 genes for carbohydrate degradation
Energy
Microbiota breaks things down by fermentation, producing resistant dairy components (e.g fibre) which then can be used in energy metabolism and metabolite generation
Energy ends up in adipose tissue as fat (uptake and storage), being used in the brain for appetite regulation and for sugar metabolism in the liver
Ingestible dietary components are broken down by 'primary' degraders (an ecological network of cross-feeders)
Types of dietary fibre (soluble and insoluble) and how they are metabolised:
(only some starch is metabolised by humans)
Mammalian glycans serve as a primary nutrient source for gut bacteria
When gut bacteria degrade complex fibres that humans cannot otherwise digest, it produces short-chain fatty acids through fermentation
Plant foods > dietary fibres > gut microbial metabolism (complex carbs > monosaccharides > phosphoenolpyruvate) > metabolites
What if there's no fibre in the diet?
Healthy gut:
Bacteria cannot contact host
Expansion of the mucolytic bacteria
A fibre-deficient diet
Dietary emulsifiers
Increased osmolarity
Leads to a eroded mucus:
Mucus layer depletion
Bacterial contact, sentinel goblet cell death
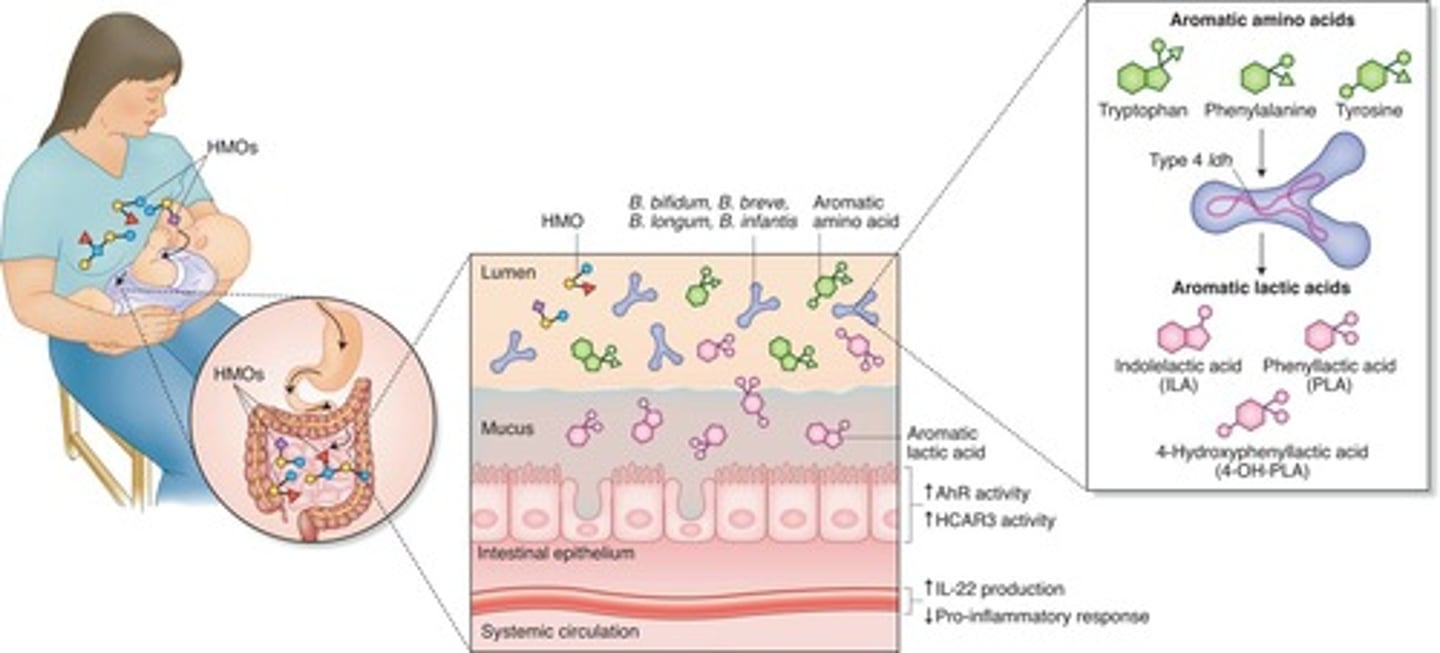
Human milk oligosaccharides (HMOs)
Carbohydrates with multiple simple sugars that are found in human milk
The composition of our microbiota evolves throughout our entire lives...
Unborn:
sterile?
Baby:
Pioneer microbes
High instability
Low diversity
Toddler:
New species
Competition
Increase in diversity (diet)
High instability
Adult:
Distinct and diverse
May still change but at a much slower rate
Elderly:
Different microbiota to adult
Lower diversity
May be limited by diet
Early life factors influencing the microbiome:
During pregnancy:
Intra-uterine environment
Bacteria in amniotic fluid
Maternal exposures
Stress, weight gain, antibiotics, smoking
Length of gestation
Term vs preterm
Weight at birth
During birth:
Mode of delivery
Contact with mother/healthcare
Environment straight after birth
After birth:
Feeding modality
Breast-milk vs formula
Antibiotics
Weaning or food supplementation
Home or family setting
Rural vs urban
Home structure
Siblings or pets
^early microbiota disturbances linked to numerous diseases (e.g autoimmune and IBS)
Viral colonisation from birth
Month 0 = pioneering bacteria and phage induction
Month 1 = viruses of human cells
Alongside = breast milk = immune cells, maternal antibodies, HMOs, lactoferrin, mucin
Microbiota and colonisation resistance
Resistance to colonisation by ingested bacteria, or inhibition of overgrowth of resident bacteria normally present at low levels within the interstitial tract is called colonisation resistance
Commensal or pathogenic bacteria
Successful colonisation of the intestinal tract is very hard both for invading or beneficial bacteria
(competition for niches)
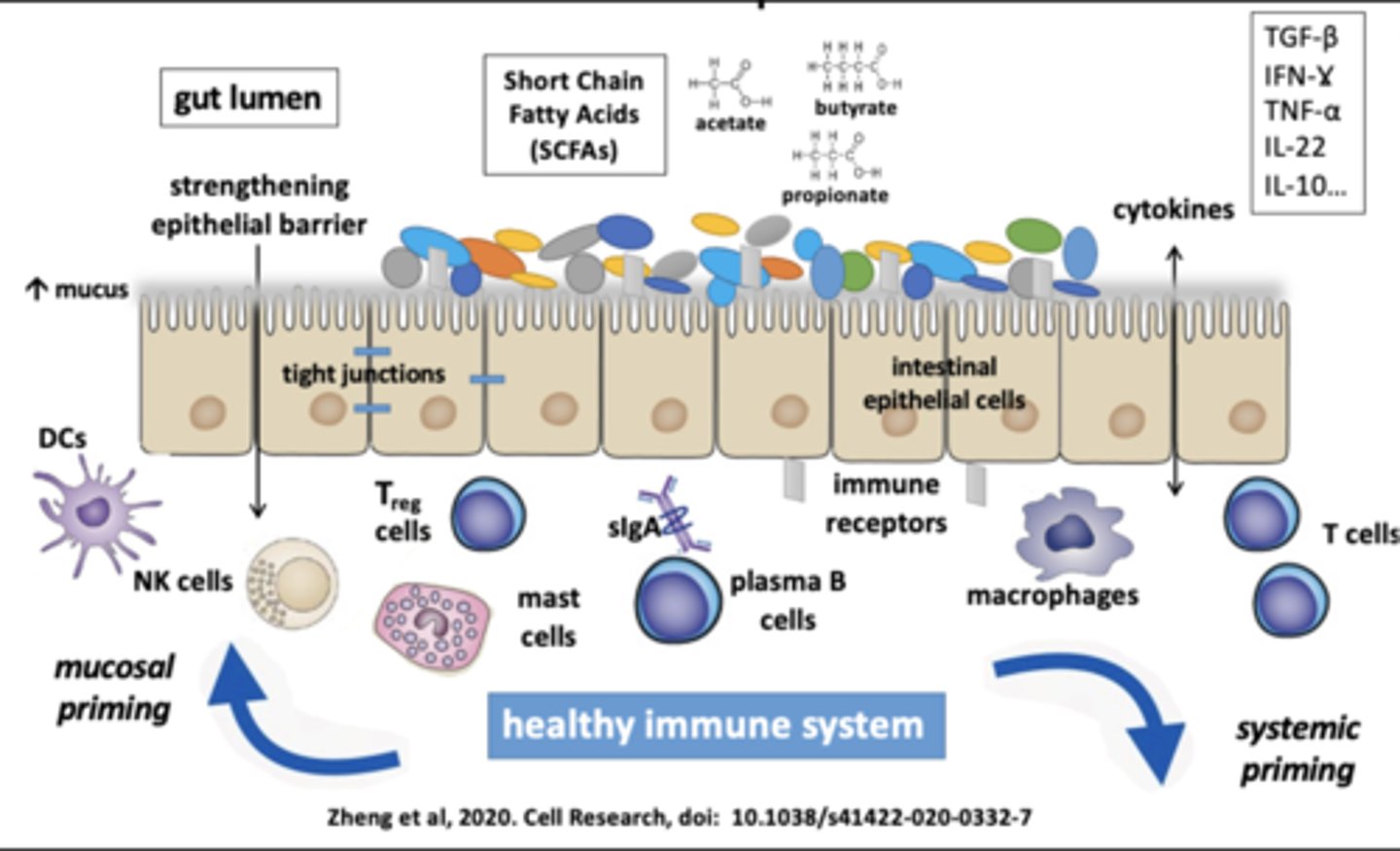
The microbiota plays a critical immune programming: (2)
1) Mucosal and systemic immune compartments
2) Stimulates both tolerance and boosts specific responses
Microbiota disturbances and disease links: (loads like way too many)
Autoimmune diseases:
Arthritis
SLE
Type I diabetes
MS
Brain-linked conditions:
Depression
Anxiety
IBS
Autism
Intestinal diseases:
Crohn's
Ulcer colitis
Colon cancer
Metabolic diseases:
Diabetes
Obesity
Malnutrition
Immune disease:
Asthma
Eczema
Rhinoconjunctivitis
Food allergies