Biochem SI Session 3- Protein Structure III
1/46
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
47 Terms
Describe the structure of myoglobin. (number of subunits/AAs/a-helices)
Monomeric polypeptide of 153 amino acids
Secondary structure contains 8 a-helices. These fold into a compact globular shape to form the tertiary structure.
Myoglobin contains a non-AA heme “prosthetic” group. Describe the heme group.
The heme group contains iron that binds oxygen and is found buried in the folded polypeptide core
How does the hydrophobic effect influence tertiary protein structure?
Hydrophilic amino acids are typically found on the surface exposed to aqueous solvent, while hydrophobic amino acids cluster into the core away from water
The secondary structural elements of a protein are typically amphipathic. What does this mean? Provide an example.
This means they have both hydrophilic as well as hydrophobic character
Example: α-helix will cluster hydrophilic residue R groups on one side and hydrophobic R groups on the other side
Christian Anfinsen performed an experiment to examine how proteins fold and assume a particular tertiary structure. What polypeptide did he use? Describe it.
Ribonuclease
-a monomeric polypeptide (124 aa) that degrades RNA in cells
-Ribonuclease has eight cysteine groups that form four disulfide linkages. These are not random and have specific pairings.
Anfinsen used two reagents that disrupt tertiary structure. Which one disrupted non-covalent interactions and what did it do?
Urea-chaotropic agent
This causes unfolding into a “random coil”
This is called “protein denaturation”
Anfinsen used two reagents that disrupt tertiary structure. One broke covalent bonds in the molecule. Which one was it and how would this affect the polypeptide?
β-ME
This was responsible for breaking disulfide bonds in ribonuclease, which greatly reduced stability and aided in the denaturation process.
When removing β-ME and urea from the denatured and reduced ribonuclease (0% active) and allowing it to reoxidize, what happened?
It returned to its native structure and became 100% active
When removing only β-ME (leaving urea) from the denatured and reduced ribonuclease (0% active) and allowing it to reoxidize in urea, what happened?
It became a “scrambled” form of ribonuclease that was 1% active
When urea was removed and trace amounts of β-ME were added to the scrambled ribonuclease (1% active) what happened?
Ribonuclease returned to its native structure and was 100% active
What do Anfinsen’s findings demonstrate regarding protein structure?
He proved that the primary amino acid sequence of a protein contains the information necessary to fold into its 3D structure and that this folding is a spontaneous process. This also helped establish the “sequence→structure→function” principle.
What did Cyrus Levinthal’s Paradox seek to determine?
Theoretically, how long would it take a protein to fold from a random structure to a folded active structure?
How many conformations did Levinthal assume were possible for each peptide bond?
3
Given Levinthal’s assumption, how many protein conformations would ribonuclease have?
124 AA, so 3^124 conformations
Given Levinthal’s assumption that each peptide bond takes 0.1 picoseconds to form, how long would it take if the folding process was random?
1.45 × 10^46 sec
Clearly false, since the universe is estimated to only by 1.38 × 10^10 years old
Protein folding actually takes seconds or less
Describe the nucleation condensation model.
Protein folding involves the formation of several stable intermediates:
Begins with denatured then goes through multiple intermediates before becoming a “molten globule.” This is the proteins final intermediate before returning to its native state
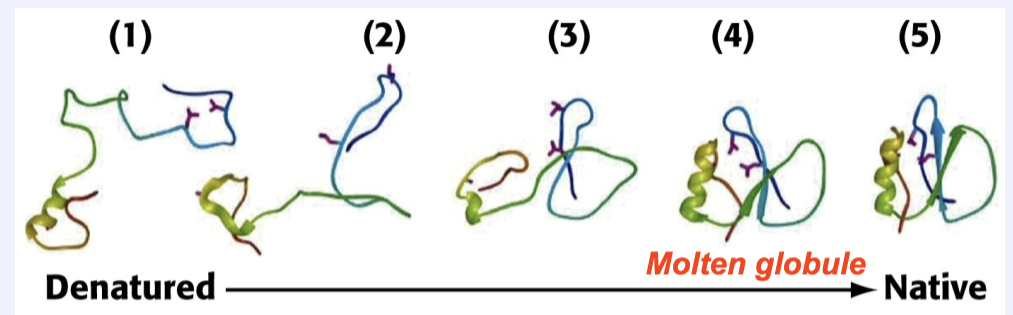
What is the molten globule? How does it form and why is it important?
Its formation is a key event in protein folding
It is a globular mobile shape→This is driven by the hydrophobic effect and clustering of nonpolar R-groups into the core of the protein
It is similar to the final native structure→allows the rapid formation of additional non-covalent interactions that lead to the active final conformation
Protein folding is a co-operative process-represented by an “energy folding funnel.” Describe what this looks like.
As folded intermediates form, entropy and energy of the protein decrease. This is because lower energy conformations are more energetically favorable, and the entropy of the protein decreases as it becomes more organized. The energetically favored native state is ultimately attained.
How is protein folding a spontaneous process if the native state has the lowest entropy?
Because the organization of proteins into a more compact structure allows for the release of solvent back into the overall system. This releasing of solvent increases the overall entropy, making protein folding a spontaneous and energetically favorable process.
What happens when proteins misfold? Provide an example.
Protein misfolding causes proteins to be unable to function and interact properly.
Example: Aβ peptide and Alzheimer’s disease
-Accumulation of Aβ in brain tissue leads to formation of β-sheet structures that interact
-Eventually these form large aggregates known as amyloid fibrils
(Monomer→(eq)←Oligomer→(eq)←Fibril)
-This leads to Alzheimer’s plaques
What misfolded protein causes Alzheimer’s?
β-amyloid
What misfolded protein causes CTE?
tau
What misfolded protein causes Huntington’s?
huntingtin
What misfolded protein causes Parkinson’s?
α-synuclein
What misfolded protein causes Creutzfeld-Jakob Disease, BSE (Mad Cow Disease), Kuru?
PrP (prions)
What misfolded protein causes cystic fibrosis?
CTFR (Cl- channel)
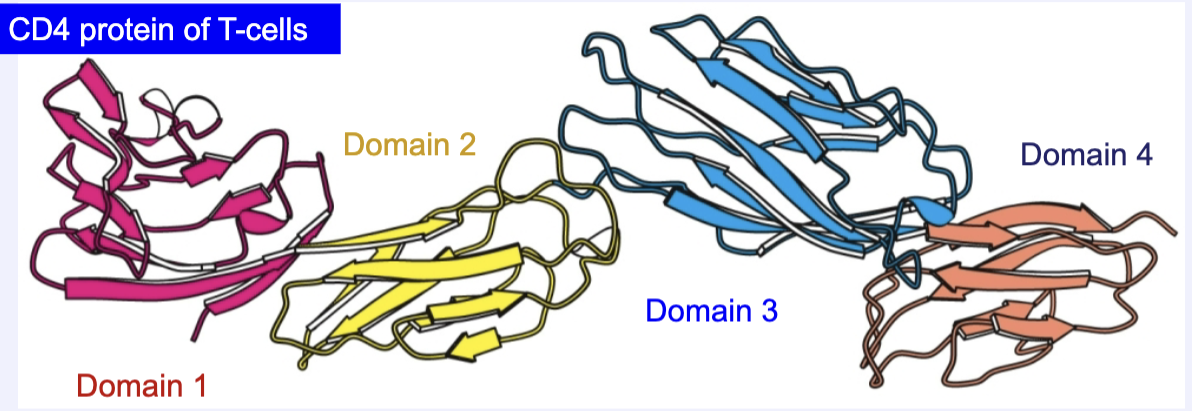
Some proteins are made up of multiple domains. What are domains?
-Domains are functional three dimensional units that are folded separately
-Connected by flexible segments like “beads on a string”
-Component of tertiary structure
Briefly describe the structure and function of hemoglobin.
-Oxygen carrier of blood
-Similar to myoglobin, but has 4 subunits/polypeptide chains (tetrameric)
-Each subunit (a/b) has a heme prosthetic group that binds to O
b subunit is 5 AA longer than a in hemoglobin
-The tetrameric structure of hemoglobin allows additional regulation of oxygen binding that is not possible with myoglobin
If the heme group of myoglobin were placed closer to the protein surface instead of buried in the hydrophobic core, how would this impact oxygen binding and protein stability?
If the heme group were placed closer to the surface, oxygen binding would become less efficient as a result of the oxidation of the iron atom, and stability would decrease because moving the heme group would disrupt the hydrophobic core.
If ribonuclease refolded correctly in the presence of urea but without β-mercaptoethanol, what would you predict about the status of its disulfide bonds and its activity?
The disulfide bonds would remain scrambled, with cross-links potentially holding cysteines in non-native disulfide bonds. Without β-ME, the disulfide bonds can’t reshuffle into their native pattern, which will likely result in an inactive protein.
Levinthal’s paradox suggests folding can’t occur by random search. Explain how the “molten globule” model resolves this paradox. How does the energy funnel concept illustrate this process?
The molten globule is a result of hydrophobic collapse, and is very close to the native state. The energy funnel demonstrates how proteins go through multiple intermediates that become lower in energy and are more organized. This demonstrates that, rather than going through a random process of trial and error, the energy funnel shows a protein being guided through intermediates of decreasing energy before reaching its native state. The funnel shape also indicates that many pathways are possible, but all lead to the native state
Consider Aβ peptide forming β-sheet amyloid fibrils. Why do these misfolded states often form large aggregates that are more thermodynamically stable than the native folded protein?
Because the extended hydrogen bonding and stacking interactions allow them to be more stable and energetically favorable than the native protein.
Compared to a native folded protein, which balances structure with flexibility, these fibrils maximize hydrogen bonding and stacking interactions. That extended network makes them more energetically favorable, even though they’re pathological. Amyloid fibrils outcompete native folding because their stacked β-sheets lock into an ultra-stable, low-energy structure.
The CD4 protein has multiple domains connected by flexible linkers. What functional advantage does a multi-domain structure provide compared to a single compact globular protein?
This allows the protein to be more flexible, perform a wider range of functions, be involved in more interactions, and they have greater evolutionary adaptability.
Hemoglobin can regulate oxygen binding while myoglobin cannot. Using structural reasoning, explain how quaternary structure contributes to this regulatory function.
Multiple subunits allow for cooperative binding (subunits interact through interfaces) and allosteric regulation. Myoglobin does not have multiple subunits (is a monomer), so it does not have cooperative binding.
The spike protein of SARS-CoV-2 can exist as a monomer or trimer. Predict how trimerization could affect viral binding to host cells compared to the monomer form.
Trimerization increases binding avidity by presenting multiple receptor-binding sites, and strengthening attachment to host cells compared to the monomer. The subunits can also influence each other (cooperativity) and trimers tend to be more stable than monomers.
Folding is a cooperative process. How does this property make proteins more sensitive to denaturing agents (like urea) than if folding occurred in independent, additive steps?
Because this means that a disruption or unfolding of one region will cause the whole protein to collapse, whereas, if folding occurred independently, denaturing agents would only affect small segments at a time.
a. ____ orientation of amino acids is preferred due to minimizing _______.
Trans, steric clash
b. ____ orientation of amino acids is NOT preferred due to high amounts of ______.
Cis, steric clash
All amino acids appear in the trans orientation. _______ is an exception to this rule
Proline (not necessarily always cis, just most likely AA to be in cis orientation)
The Ramachandran plot shows what about Psi and Phi bonds?
It shows the sterically allowed combinations of Φ and Ψ bond angles in the polypeptide backbone, which define possible secondary structures.
In an α helix, where are the side chains located? Where are the hydrogen bonding groups located?
The side chains point outwards from the helix, while hydrogen bonding groups are located in the center.
Where would you expect to find an α-helix on a Ramachandran plot? What about a β-sheet?
Right-handed a-helices are located in the lower-left quadrant because they are more bent
β-sheets are located in the upper-left quadrant because they are more flat
I have an R-group that forms covalent bonds that are reduced by β-mercaptoethanol_____
Cysteine
My side chain can form hydrogen bonds and be negatively charged ______
Glutamate/Aspartate
I am a hydrophobic amino acid that can have a charge in the side chain _____
Tyrosine
I am a large amino acid that strongly disrupts the formation of α-helices _____
Proline
I am an amino acid that has an aromatic ring on the side chain _____
Tyrosine/Phenylalanine/Tryptophan