Chapter 4: Basic Plasmids and Advanced Vectors
1/71
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
72 Terms
Plasmid
A plasmid is a small, circular, double-stranded DNA molecule found in bacterial cells. It is independent of the chromosomal DNA.
Size range of plasmids
Between 2 kb and 100 kb
Examples of properties that plasmids can confer to their host cell.
Antibiotic resistance
Antibiotic production
Degradation of aromatic compounds
Sugar fermentation
Heavy-metal resistance
Induction of plant tumours
Bacteriocin and enterotoxin production
These properties are often linked to either fertility, resistance, metabolism, or virulence.
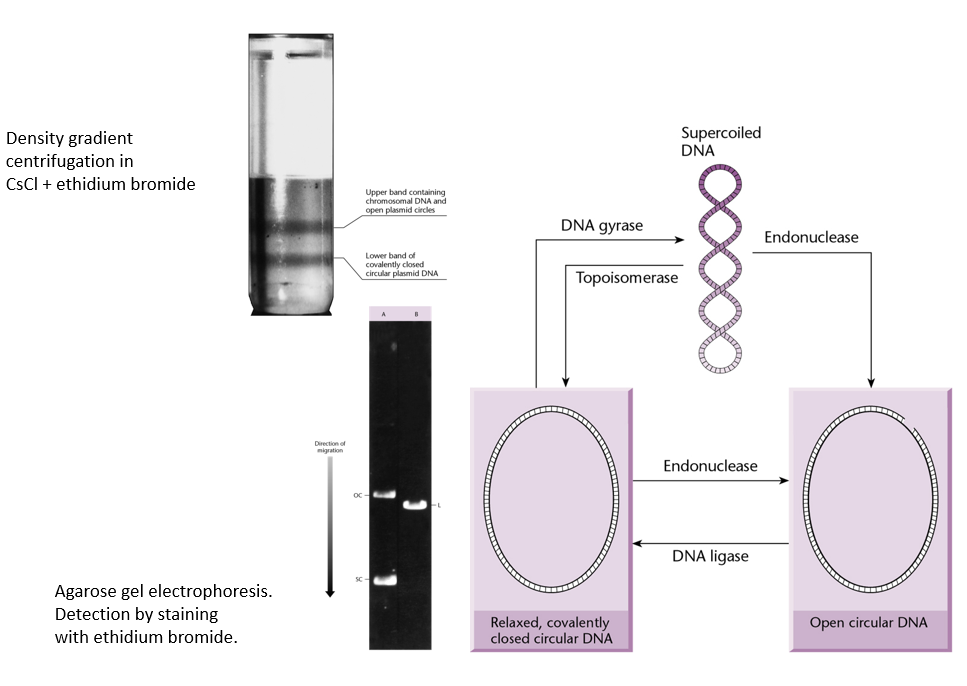
Explain the difference between supercoiled plasmids, relaxed closed circular DNA, and open circular DNA. Which migrate faster/slower in gel electrophoresis?
Supercoiled DNA migrate much faster than the other two.
Open circular DNA migrates the slowest.
Conjugation
The unidirectional transfer of genetic information between cells by cell-to-cell contact. It is a form of horizontal transmission.
Vertical VS Horizontal Transmission
Vertical → Transfer of genetic material from parent to offspring during reproduction.
Horizontal → Transfer of genetic material between organisms that are not in a parent-offspring relationship (e.g. bacteria to bacteria).
Why is conjugation said to be unidirectional?
Unidirectional because there is a donor cell and a recipient cell.
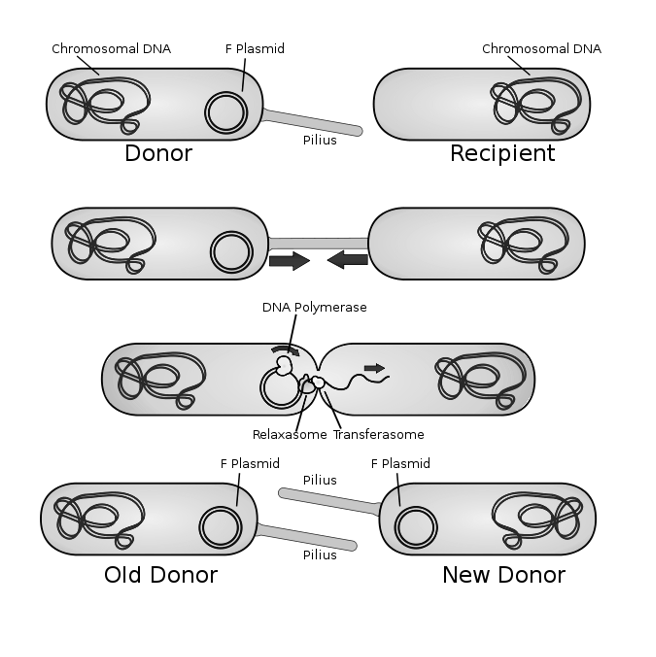
Explain how conjugation works.
Donor Cell Preparation: A donor bacterium with an F (fertility) plasmid forms a pilus.
Attachment: The pilus attaches to the recipient cell. The region of membrane contact is the conjugation bridge.
DNA Transfer: The plasmid is nicked (by relaxosome), and a single strand is transferred to the recipient through the conjugation bridge (by transferosome).
Replication: Both cells replicate the DNA strand, forming double-stranded plasmids.
Conjugative plasmids
Definition: These plasmids contain all the necessary genes for conjugation, including:
tra genes (for pilus formation and DNA transfer machinery).
oriT (origin of transfer).
Capabilities:
Can independently initiate and execute the process of conjugation.
Transfer themselves to recipient cells without requiring any help.
Examples: F plasmid (fertility plasmid) in E. coli.
Mobilizable plasmids
Definition: These plasmids lack some or all tra genes but still have an oriT.
Capabilities:
Cannot perform conjugation on their own.
Rely on the conjugative plasmids in the same cell to provide the transfer machinery (e.g., pilus and transfer proteins).
A plasmid lacking both the tra functions and oriT functions is …
non-conjugative and non-mobilizable.
Classification of plasmids based on their capacity to transfer to other bacteria.
Conjugative plasmids: plasmids that can transfer themselves to other bacterial cell through the pilus.
Non-conjugative plasmids: plasmids that cannot transfer themselves to other bacterial cells.
Mobilizable plasmids: plasmids that can be transferred to another bacterial cell with the help of another conjugative plasmid. Mobilizable plasmids are non-conjugative.
Classification of plasmids based on their functions.
Fertility (F) plasmids: plasmids that are capable of conjugation.
Resistance (R) plasmids: plasmids that encode genes that confer resistance to one or more antibiotics/toxins.
Col-plasmids: plasmids that encode colicines, which are proteins that kill other bacteria.
Degradation plasmids: plasmids that can degrade compounds like toluene or salicylic acid.
Virulence plasmids: plasmids that make their host bacterium pathogenic.
Addiction system: plasmids that make both a stable toxin and an unstable antitoxin. Called “addicting” because it forces the bacteria to keep the plasmid intact to survive: if the plasmid is lost or segregates poorly, the antitoxin is degraded more quickly than the toxin, leading to cell death.
Transformation
Transformation is the process by which a bacterium takes up foreign DNA (often from the environment) and incorporates it into its own genome.
Competent cells
Competent cells are bacterial cells that have been treated to become permeable to foreign DNA, allowing them to take up DNA molecules (such as plasmids) during processes like transformation.
2 techniques for bacterial transformation
Transformation by electroporation
Chemical transformation
What is the most efficient technique for bacterial transformation? Explain how this technique works.
Electroporation
Technique used to introduce foreign DNA into cells (i.e. transformation) by applying a brief electrical pulse that temporarily disrupts the cell membrane, creating pores that allow DNA to enter the cell. The electrical pulses are very short so that the membrane opens and close quickly and stays intact.
The DNA can then be integrated into the genome or remain as plasmid DNA.
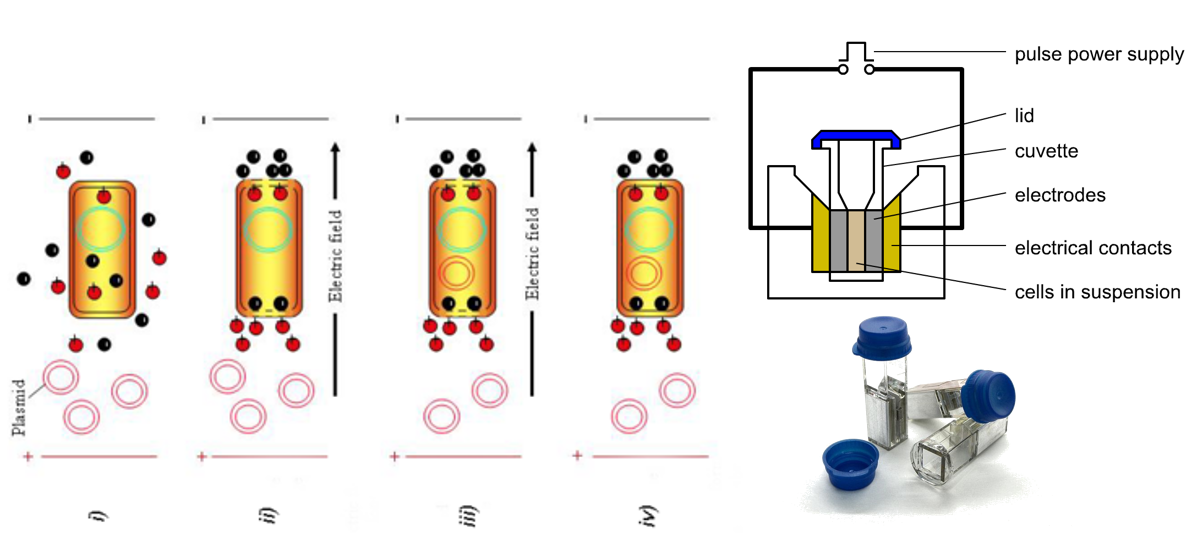
Give the main steps of a E. coli transformation protocol.
Centrifuge E. coli culture and resuspend bacterial pellet in sterile water.
Chill on ice.
Centrifuge and resuspend pellet in sterile water. This results in competent cells.
Mix competent cells and plasmids.
Electroporation.
Add transformed cell to saline buffer.
Plate the transformed cell on selective media to only get the cells that have taken up the plasmids (for example, if the inserted plasmid gives antibiotic resistance, hen the transformed cells are plated with the corresponding antibiotic
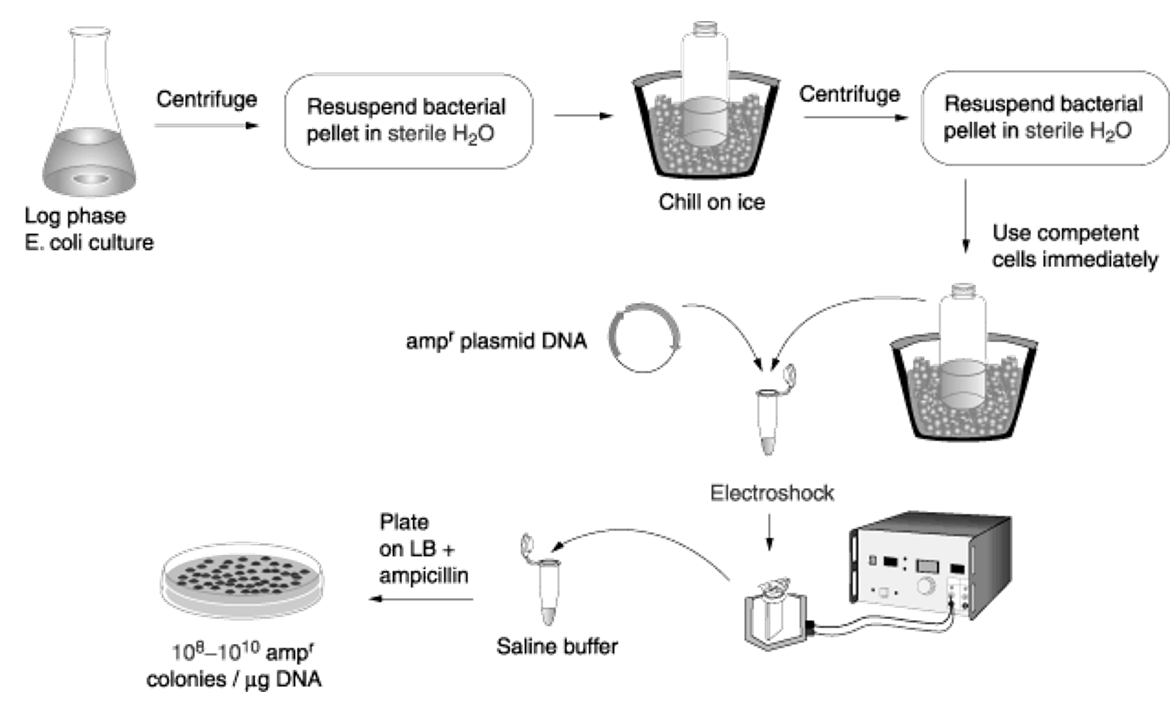
Explain chemical transformation in E. coli
Calcium + heat shock opens temporary pores in cell membrane and allows plasmids to enter the cell.
Less efficient than electroporation.
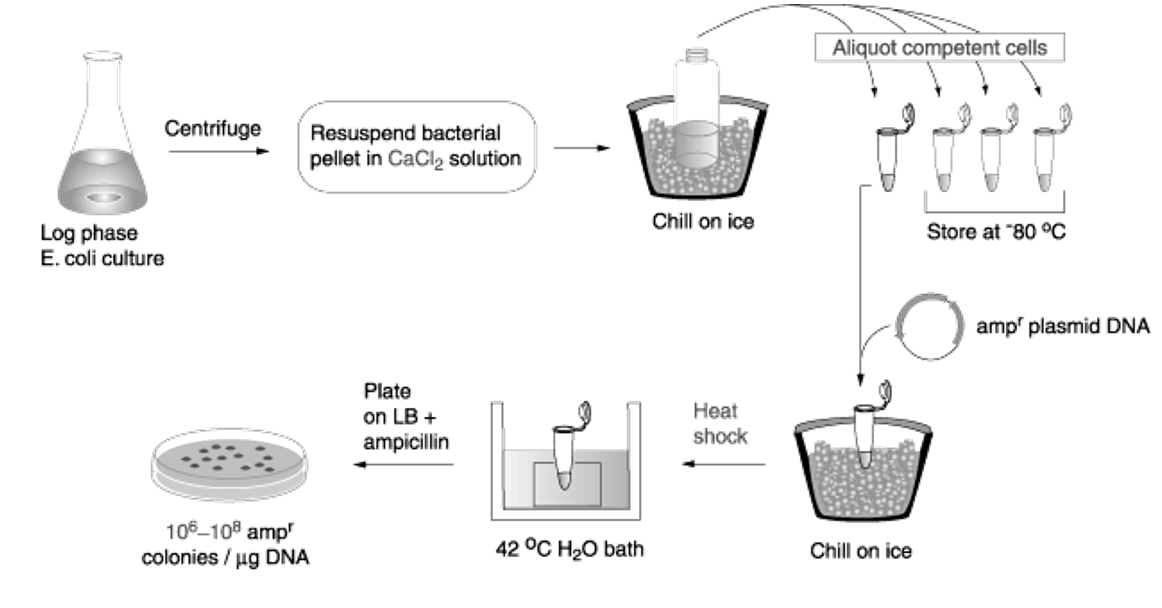
Important components of chemical transformation in S. cerevisiae.
Lithium acetate ensures the permeabilization of the cell membrane.
Heat shock: combination of lithium acetate with heat allows the plasmids to entre the cell.
Polyethylene glycol is a viscous substance that ensures that the aqueous phase containing the transformation components (plasmids) are kept close to the cell membrane → crowding agent.
Single-stranded DNA needs to be added in addition to the plasmid DNA. This is because the cell surface is positively charged, which means that the negatively charged plasmids will stuck to the outside of the cell rather than going inside. The ssDNA binds more strongly than dsDNA to the well surface, ensuring that the double-stranded plasmids do not get stuck on the cell wall and can entre the cell.
Selectable marker
A selectable marker is a gene included in a plasmid or vector that enables the identification of cells that have successfully taken up the plasmid by providing resistance to a specific selection pressure (usually an antibiotic).
Plasmid vector
A plasmid vector is a small, circular DNA molecule that is used to carry foreign genetic material into a host cell for the purposes of cloning, gene expression, or genetic modification.
Main properties of plasmid vectors
Modular
Limited in size (“the smaller the better”, because the smaller the plasmid vector, the lesser the burden will be for the host cell).
Contains unique cleavage sites to allow to cut and paste fragments inside the vector.
Contain a selection or identification (screening) marker.
4 main elements of a plasmid vector
Origin of replication (Ori)
Selection marker
Regulatory elements
Multiple cloning site
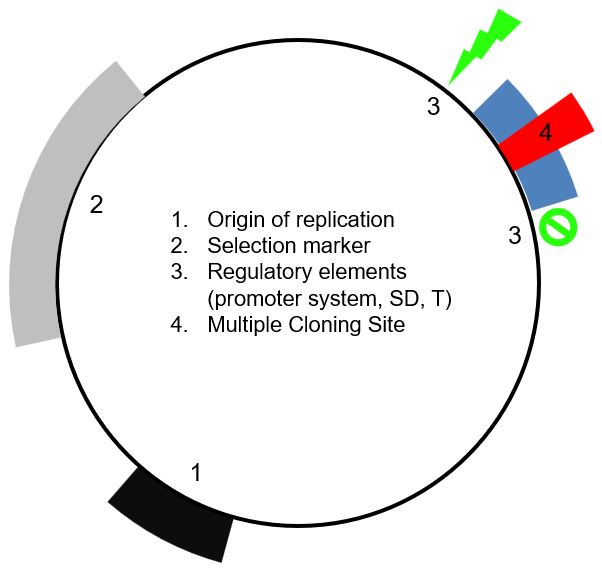
Origin of replication
Particular sequence at which replication is initiated. It usually has a high AT content (so that separation of the two strands for replication is easier)
Three important characteristics of a plasmid that are determined by the origin of replication of the plasmid.
Range
Narrow range: the plasmid does not work for many species bacterial
Broad range: the plasmid works for many bacterial species (e.g. all of the gram-negative bacteria).
Copy number
High copy number (i.e. the plasmid is present in high quantity in the host cell)
Low copy number (i.e. the plasmid is present in low quantity in the host cell)
Incompatibility
Determines if two plasmids can exist together or if one cannot exist if another plasmid is present (i.e. the plasmids interfere with each other).
Explain how the origin of replication of the ColE1 plasmid works.
Replication of the ColE1 plasmid depends on its copy number in the cell, as well as on two RNA molecules (RNA I and RNA II) produced from the plasmid's origin of replication, and on the Rop dimer, synthetised by the plasmid:
At low copy number, RNA II is synthetised to initiate replication of the plasmid:
RNA II is transcribed from the plasmid near the origin of replication.
It hybridizes to a complementary sequence in the origin and forms an RNA-DNA hybrid.
This hybrid structure is recognized and processed by RNase H, which cleaves RNA II to produce a free 3′-OH end.
This 3′-OH end acts as a primer for DNA polymerase I, which initiates DNA synthesis and starts plasmid replication.
At high copy number, replication by RNA II is repressed by RNA I:
More RNA I is synthetised.
RNA I binds to RNA II, which prevents RNA II from binding to DNA and start replication.
Production of the Rop protein increases the binding affinity between RNA I and RNA II, thus reducing the chances that RNA II can start replication.
Replication of ColE1 does not require translation, as the replication is done by RNA molecules directly.
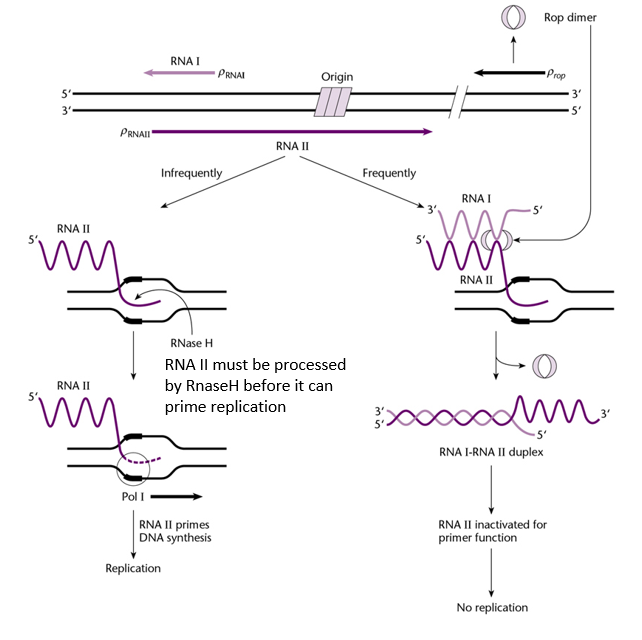
Elements required for regulation of the replication of the ColE1 plasmid.
RNA II: to initiate replication
RNase H: to initiate replication
DNA polymerase I: to perform replication
RNA I: to inhibit replication
Rop: to inhibit replication
Plasmid curing
Plasmid curing is the process of eliminating plasmids from bacterial cells. It is used to study plasmid biology, remove unwanted plasmids, or ensure cells are plasmid-free for specific applications.
Molecule used for plasmid curing
Acridine orange specifically inhibits plasmid replication but not chromosomal DNA replication.
Give a mechanism that prevents plasmid curing, and explain how it works.
Partition
Partition systems ensure the proper segregation of plasmids during bacterial cell division, so each daughter cell inherits at least one copy of the plasmid. This prevents plasmid loss and ensures stable inheritance, especially for low-copy-number plasmids.
Plasmids have a specific site called par which is involved in partitioning.
Why are different plasmids with the same replication mechanism incompatible?
Different plasmids with the same replication mechanism cannot function properly together, as their replication mechanism will interfere with each other, and their copy number cannot be regulated properly.
Example with ColE1:
Consider two different plasmids which both have the ColE1 origin of replication.
If one plasmid has a low copy number, RNA II will be synthetised to initiate replication of that plasmid. However, if the other plasmid has a high copy number, the presence of RNA II will interfere with the replication inhibition of that second high copy plasmid. On the other hand, the RNA I may also interfere with the replication of the low-copy plasmid, thus preventing its replication.
For two different plasmids to coexist in a cell, they should have … replication mechanisms (and thus origin of replication), and … par sites.
For two different plasmids to coexist in a cell, they should have distinct replication mechanisms (and thus origin of replication), and distinct par sites.
Shuttle plasmids
A shuttle plasmid is a plasmid vector designed to replicate and function in multiple host organisms (e.g. bacteria and yeast). They contain at least two origin of replication, which is what gives them the ability to work in different organisms.
Selection marker
Genes introduced into an organism which is required for the survival of said organism. Thus, if a selection marker is placed in a plasmid, only the cells that contain the plasmids with the selection marker can survive.
4 types of selection markers and if they perform positive, negative, or no selection.
Drug resistance (e.g. antibiotic resistance) → positive selection
Auxotrophy markers → positive selection
Visual markers → no selection (thus not really a selection marker)
Inactivation markers → negative selection
Auxotrophic marker
An auxotrophic marker is a specific type of selection marker used in molecular biology, particularly in yeast, to select for cells that have been successfully transformed with a particular gene or plasmid. These markers exploit auxotrophy, which is the inability of an organism to synthesize a specific essential compound required for growth (e.g. essential amino acids).
How does it work?
The host organism (e.g. yeast) is engineered to have a mutation in a gene that is essential for synthetising an essential element, such as an essential amino acid. As a result, the organism becomes auxotrophic for that compound.
A plasmid vector with a functional copy of the mutated gene is transformed into cells.
The transformed cells are then placed in media that lack the essential compound, and only the cells that have successfully incorporated the plasmid will survive.
Examples of auxotrophic markers
Leu2, which encodes a compound essential for the synthesis of the leucine amino acid.
His3, which encodes a compound essential for the synthesis of the histidine amino acid.
Visible markers
Visible markers are genes or markers that allow to visually identify transformed cells based on a detectable phenotype, such as colour, fluorescence, or pigment production.
Unlike selectable markers, visible markers do not confer a survival advantage but instead facilitate the easy observation of successful transformations or genetic modifications.
Different types of visual markers with examples
Fluorescent proteins
Green fluorescent protein (GFP), which emit green fluorescence after being excited by blue light.
Combination of three amino acids within the protein determines its excitation and emission spectra. Thus, we can have variants of GFP which are excited and emits at different wavelengths, and thus appear with different colours.
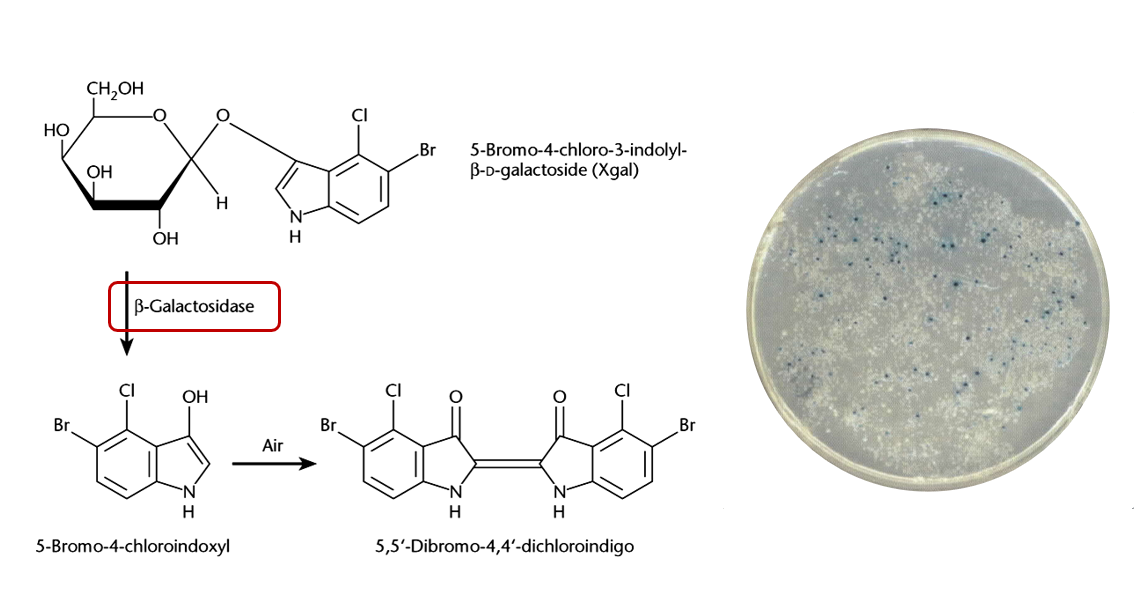
Enzymatic markers
Beta-galactosidase, encoded by lacZ gene. It hydrolyses X-gal to produce a blue-coloured product, making colonies appear blue.
Luciferase, which produces bioluminescence by catalysing reactions with luciferin
Negative selection markers
Negative selection markers are genes or sequences to eliminate cells or organisms that contain the marker, allowing only those without the marker to survive.
These markers are essential for counter-selection, where researchers want to remove unwanted cells that still carry a specific gene or plasmid after a desired genetic event.
Examples of negative selection markers
sacB: encodes levansucrase, whose activity is lethal on bacterial cells growing on medium containing 7% glucose.
ccdB: this gene is lethal when expressed in host cells , unless these carry a specific mutation in their DNA gyrase.
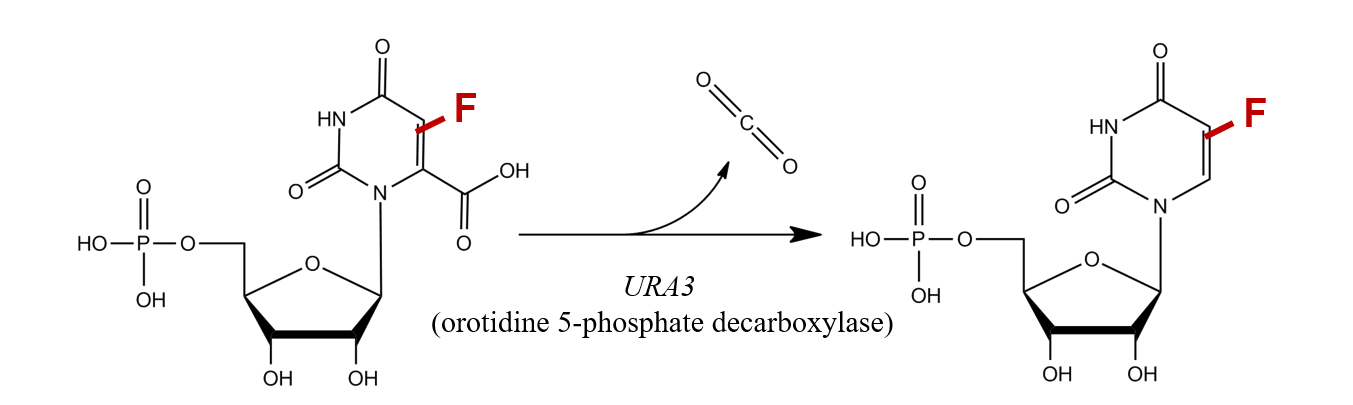
Give an example of a marker that can be used as either a positive or a negative selection marker.
URA3 can be used as either a positive or a negative selection marker depending on the growth and environmental conditions.
It is derived from the URA3 gene, which encodes orotidine-5'-phosphate decarboxylase. This enzyme is essential for the biosynthesis of uracil.
Positive selection
URA3 can work as an auxotrophic marker. Plasmids carrying this marker can be transformed into cells which have a mutant URA3 gene (meaning they can’t synthetise uracil). The cells are then placed on a media lacking uracil, and only the cells which have successfully taken up the plasmid will survive.
Thus, we select for the cell that have the URA3 gene.
Negative selection
By adding 5-FOA (fluoroorotic acid) to the growth medium, we can select for the cells who DO NOT have functional UR3 gene. This is because in cells expressing a functional URA3 gene, 5-FOA is converted into a toxic compound that is lethal for the cell.
Thus, cells that lack a functional URA3 gene survive on media containing 5-FOA, while URA3-expressing cells die
Multiple cloning site (MCS)
A multiple cloning site (MCS), also known as a polylinker, is a short DNA sequence within a plasmid or other cloning vector that contains a series of closely spaced and unique restriction enzyme recognition sites. These sites allow to insert foreign DNA into the vector at a precise location using restriction enzymes and DNA ligase.
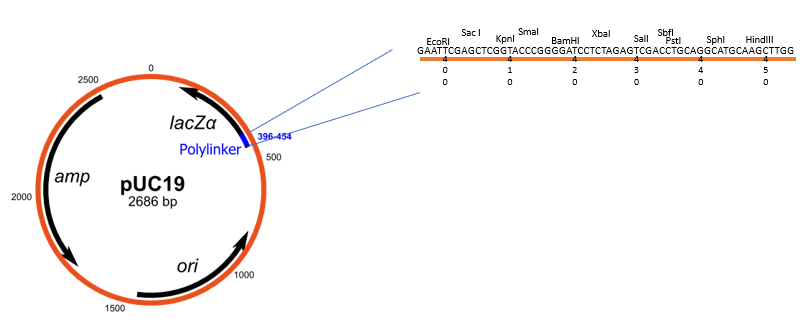
pUC vectors
Series of plasmid cloning vectors which are can be used for blue-white screening for the identification of recombinant clones.
Key components
pUC vectors have a segment called lacZα which encodes for the N-terminal fragment of beta-galactosidase (normally encoded by lacZ gene). This lacZα segment contains a multiple cloning site (MCS).
pUC vectors also have a ampicillin resistance gene.
Key variants of pUC vectors
pUC18: The MCS is in one orientation.
pUC19: The MCS is in the opposite orientation compared to pUC18.
The designation "pUC" is derived from the classical "p" prefix (denoting "plasmid") and the abbreviation for the University of California, where early work on the plasmid series had been conducted.
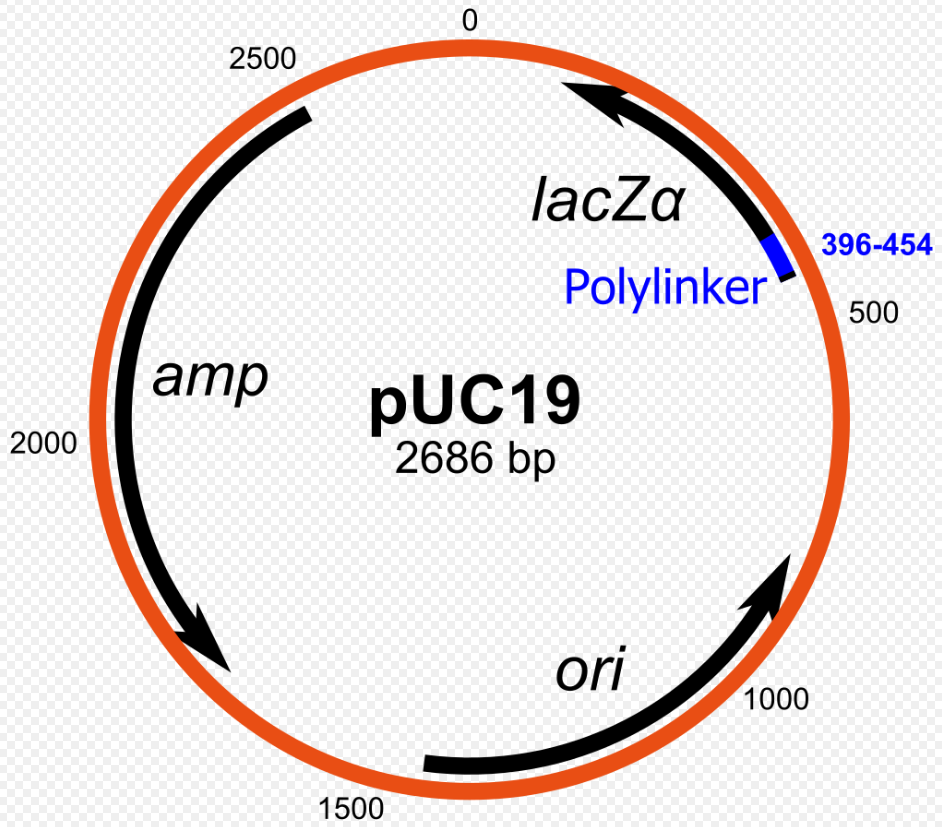
Explain how blue-white screening works with pUC vectors.
pUC vectors have a segment called lacZα which encodes for the N-terminal fragment of beta-galactosidase. Thus, if such plasmid is transformed into a host cell which encodes for the C-terminal fragment of beta-galactosidase, the host cell can then produce full beta-galactosidase. Beta-galactosidase can then convert X-gal into a pigment that make the colonies appear blue.
This lacZα region contains a MCS, which means that DNA fragments can be inserted into the lacZα sequence, disrupting it. Thus, colonies which have an insert in the lacZα region will not be able to produce the N-terminal of beta-galactosidase, and the blue pigment will not be produced, resulting in white colonies.
In summary
Blue Colonies: Bacteria that contain plasmids without inserts
White Colonies: Bacteria that contain plasmids with inserts
This is a popular technique to identify the colonies that have an inserts in the MCS.
cos sites in phage DNA
cos → Cohesive end sites
Phage DNA contains two cos sites, one at each end of the linear DNA molecule.
When the phage DNA is inserted in the host bacteria, the two cos sites can recognize each other and form a circular DNA molecule. In this conformation, the DNA can be replicated and participate in either the lysogenic cycle or the lytic cycle.
The cos sites also allow enzymes involved in the assembly of phage particles to package a single DNA molecule into each phage. This process happens in the host cell (e.g. bacteria).
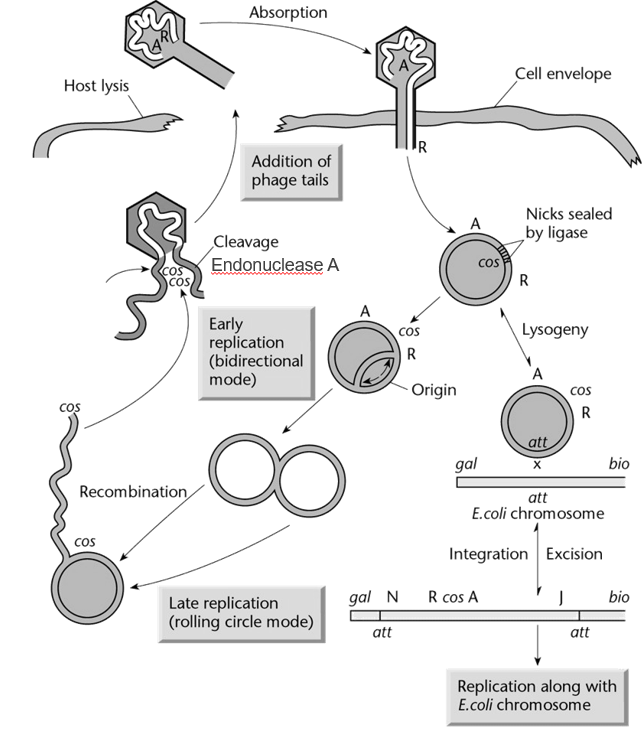
Key traits to keep in mind about phages.
Capacity of the head is between 37 and 52 kb
Proteins encoded by the phage DNA can self assemble to form other phage particles
The phage DNA contains two cos (cohesive end sites), one at end of the linear DNA. These allow the phage DNA to circularise, as well as to be packaged into new phage particles.
Transducing particles
Particles that can transfer bacterial DNA into cells, similarly to how bacteriophage insert their own viral DNA into cells.
For example, cosmids are packaged into transducing particles.
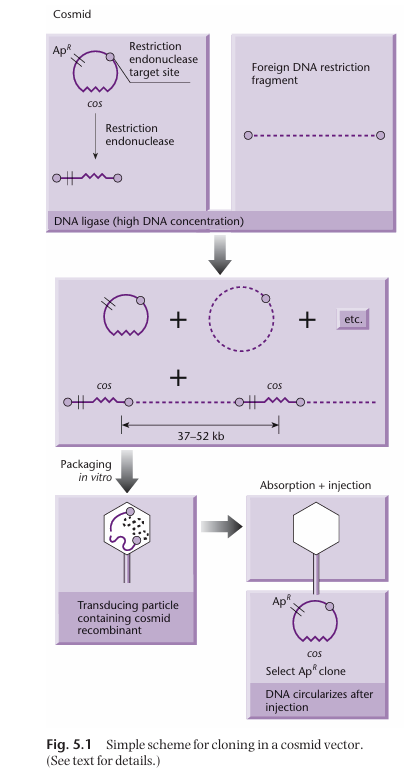
Cosmid vectors
Type of hybrid plasmid that contains a cos sequence (DNA sequence derived from bacteriophage lambda). These cos sequences allow the vector to be packaged into a transducing particle in the same way that phage DNA would be packaged in a phage.
Thus, cosmids allow foreign DNA to be transferred into cells by transduction.
Size of DNA insert a cosmid can accommodate.
37 to 52 kb
How are cosmid vectors built?
DNA we want to insert into bacteria (foreign DNA) is digested to create fragments.
Plasmid backbone with cos site, antibiotic resistance gene, and restriction endonuclease target site is digested by said restriction endonuclease, which result in a linear fragment.
Linearised plasmid backbone and broken down foreign DNA are mixed with DNA ligase. At high DNA concentration, the foreign DNA will ligate with the plasmid backbone to create concatemers (which contains the cos site and the antibiotic gene). Note that this will also results in unwanted circular products.
The concatemers are then packaged in vitro with all the necessary protein for phage particle formation.
How are the proteins necessary for the formation of the transducing particles obtained?
Two different bacterial strains are necessary to get all the proteins necessary for the assembly of the transducing particle.
One of the strain has a mutated gene E (which normally encodes for the head precursor). As a result, it can make all the necessary packaging proteins except the ones for making the head. Thus, this strain cannot package DNA on its own.
The other strain has a mutated D gene. As a result, it can make all the necessary packaging proteins except the ones encoded by gene D. Thus, this strain cannot package DNA on its own.
However, when the two strains are put together, they produce all the necessary gene product for the assembly of the transducing particles, in which the cosmids can be packaged.
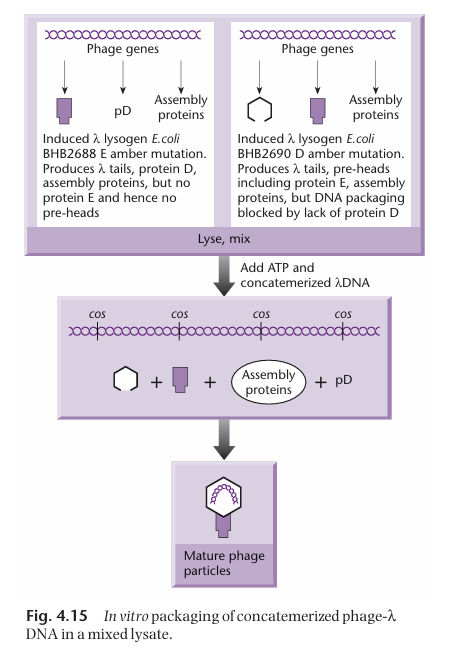
Why are two different bacterial strains necessary for the assembly of transducing particles?
This ensures that the strains cannot package DNA on their own.
What’s a major issue we want to avoid when making genome libraries using cosmids?
We want to avoid the formation of non-contiguous fragments.
When ligating the cos sites with the genomic DNA fragments, it is possible that two foreign DNA fragments from two completely different part of the genome ligate together and get flanked by cos sites. Thus, this result in a DNA fragment that is not representative of the genome anymore.
One major cosmid vector application
Thanks to their capacity for large DNA fragments, cosmids can be used for constructing libraries of eukaryotic genome fragments.
Briefly explain how E.coli phage P1 work.
It has a circular permuted genome:
Capsid 1 → ABCDEFAB
Capsid 2 → CDEFABCD
Capsid 3 → EFABCDEF
Unique DNA is around 90kb, but around 10kb is redundant on each end (total of 110kb).
Can undergo both the lysogenic or lytic cycle:
Lysogenic: unlike other phage, the DNA is not integrated in the bacterial chromosome. The DNA is kept in low copy number.
Lytic: raises copy number
It has a pac site followed by a loxP site:
The pac site encodes for an enzyme called pacase which cuts the DNA every 110kb so it can be packaged.
The loxP sites are around 90kb apart, which means that each DNA fragment cut by pacase has two loxP sites. These are then recognised by Cre recombinase which allows recircularization of the DNA once in the bacterial host.
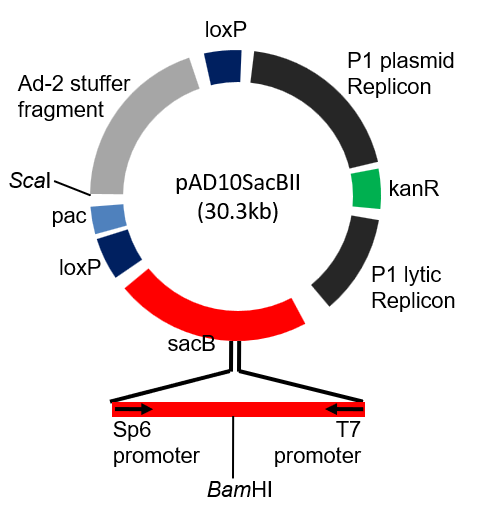
Bacteriophage P1 Vectors
Bacteriophage P1 vectors are cloning tools derived from the P1 bacteriophage, designed for cloning large DNA fragments (70-95 kb, almost twice as much as cosmids). They combine features of plasmids and P1 phage replication systems, making them highly efficient for maintaining large DNA inserts.
Different components of P1 vectors and their function
P1 plasmid replicon: contains the origin of replication for the lysogenic pathway. It keeps the copy number to around 1 per cell.
P1 lytic replicon: contains the origin of replication for the lytic pathway. It keeps the copy number higher (controlled by lac promoter)
Kanamycin resistance gene kanr
2 loxP sites necessary for circularisation of the vector once the foreign DNA has been inserted.
pac site which encodes the pacase.
Stuffer DNA fragment, which is meant to be cut and removed once the foreign DNA has been inserted (see card on how foreign DNA is inserted into P1 vector).
sacB, a positive selection marker to identify cells that have the foreign DNA. Expression of sacB in the presence of sucrose is detrimental to E.coli. It contains the BamH1 restriction site for insertion of the foreign DNA.
ScaI restriction site which is necessary for DNA insertion.
Explain how foreign DNA can be inserted in a Phage P1 vector system, and how this DNA is then incorporated into E. coli cells.
The P1 vector is digested with Sca1 and BamH1 restriction enzymes, which cut at their respective restriction site. This results in:
a short arm, which contains the pac site, one loxP site, and a part of the sacB selection marker (not relevant once cut). This arm is not viable as it does not have an origin of replication.
a long arm, which contains the stuffer DNA, the other loxP site, the two replicons, the kanamycin resistance gene kanr and the other part of the sacB selection marker (not relevant once cut). This arm cannot be packaged since it does not have a pac site.
The short and long arms, as well as the insert DNA, are mixed with ligase. This results in the insert DNA being flanked by the short arm on one side and the long arm on the other (the other combinations are filtered out cause not viable.
The pacase (encoded by pac site) cuts approximately 100 kb further from the pac site, in the stuffer DNA. This ensures that the DNA fragment has the correct size to be packaged. The stuffer DNA is here simply to be cut and prevent important sequences to be cut and made non-functional.
In the presence of the necessary packaging proteins, the cut DNA fragment can be packaged into the P1 phage head, and is ready for insertion into a bacterial cell.
The host cell must be able to encode a functional Cre recombinase protein, which is responsible for circularising the DNA with the help of the loxP sites. The circularisation results in the loss of the rest of the stuffer DNA.
This process results in E. coli cell with the P1 plasmid, which contains two replicons (one for lytic cycle and one for lysogenic cycle), the insert DNA, one loxP site, and the kanamycin resistance gene.
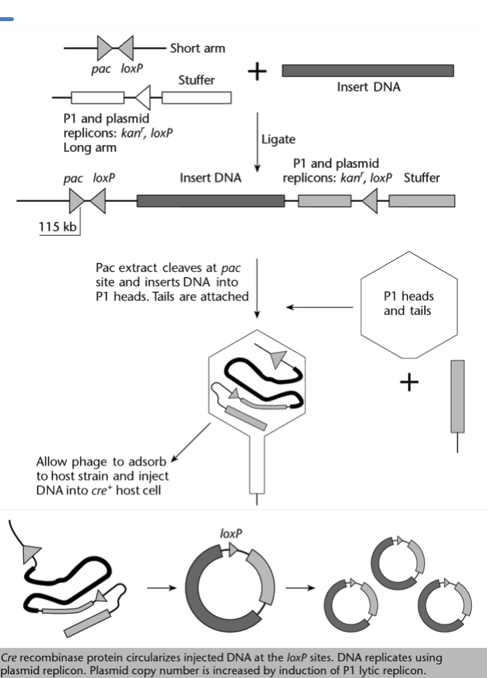
Why can’t the DNA insert in a Phage P1 vector be longer than 100 kb?
If the DNA insert is longer than 100kb, instead of cutting into the stuffer DNA, the pacase would cut before the loxP site, preventing the DNA molecule from circularising later on.
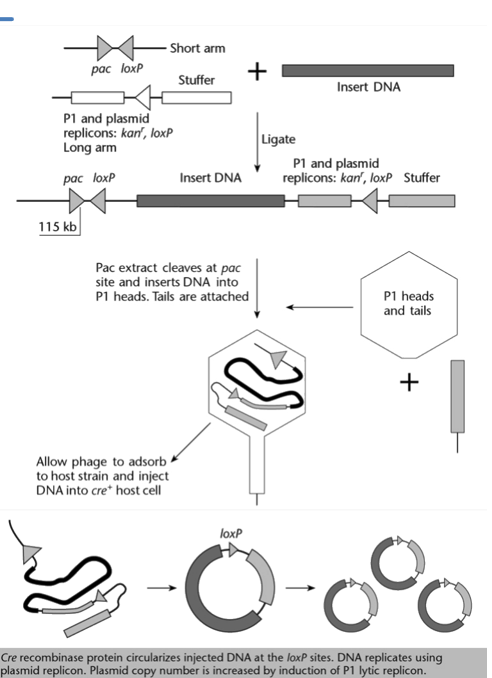
What is the point of having a short and a long arm in the phage P1 vector system?
Ensures that only the DNA consisting of the insert flanked by a shot arm and a long arm is viable.
Bacterial Artificial Chromosome (BAC)
A cloning vector propagated as a mini chromosome in a bacterial host. It allows DNA insertion of large fragments (200 to 500 kb, more than double of phage P1 vector system) thanks to the use of fertility (F) factors.
Elements of a BAC. Which elements are specific to BACs?
Origin of replication (OriS) derived from E. coli fertility plasmid (low copy vector, 1 copy/cell).
Antibiotic resistance gene chloramphenicol (Cmr)
Multiple cloning site for DNA insertion. It is within lacZ alpha or sacB (with lacZ, colonies with insert are white, with sacB, only colonies with insert will survive).
Elements specific to BACs:
Since there is only one copy per cell, partitioning of the BAC during cell replication is crucial. This is mediated by repE (encodes for helicase, enhances DNA replication) and parA, parB, and parC, which ensure correct distribution of the artificial chromosome during replication.
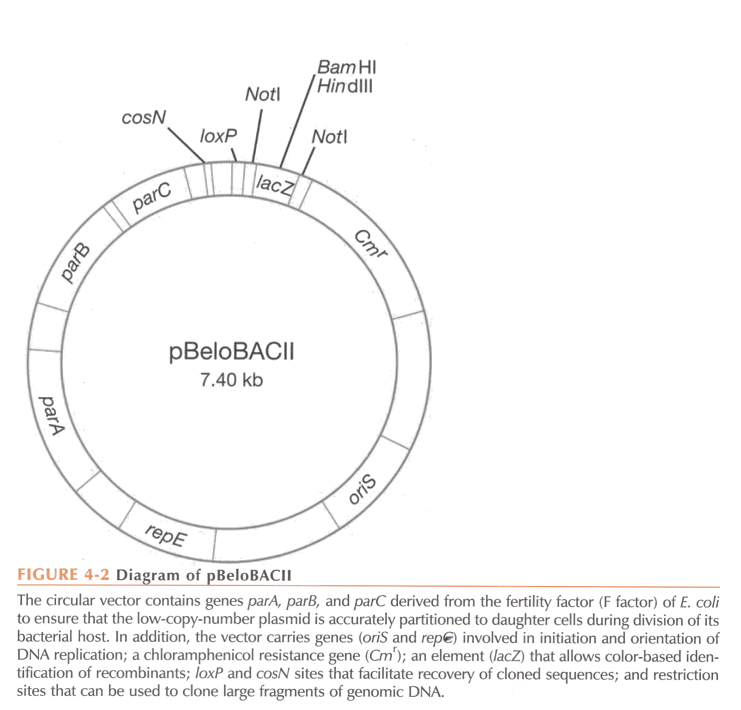
BACs are built through …
in vitro ligation.
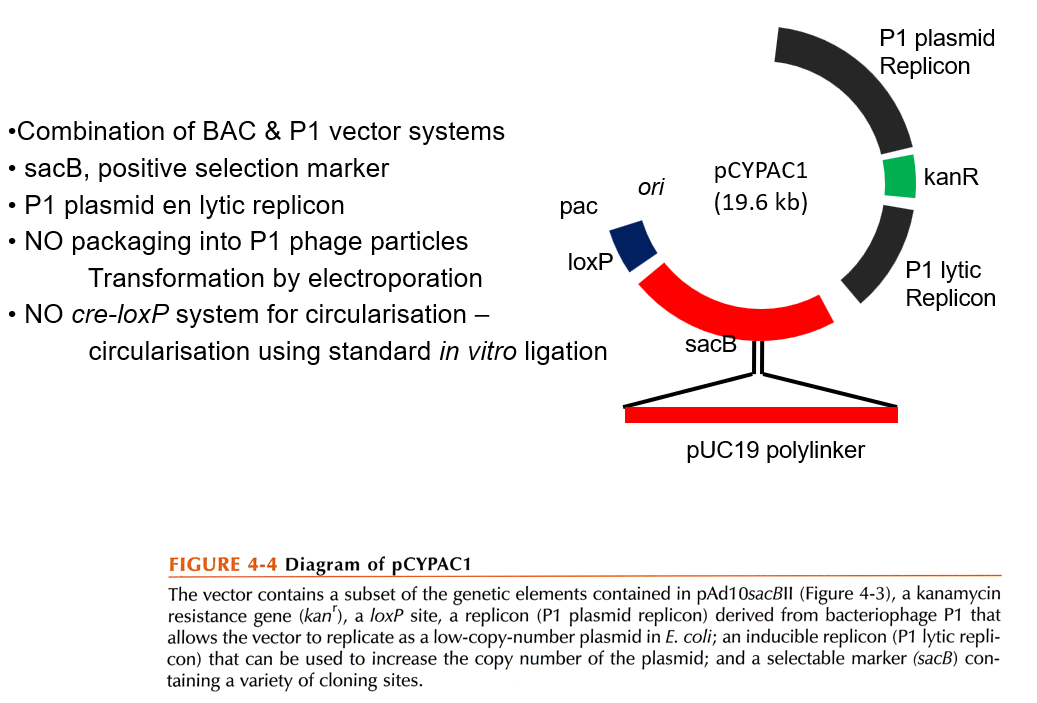
P1 Artificial Chromosome
Combination of BAC and P1 vector system.
Contains the P1 plasmid replicon (low copy number) and the P1 lytic replicon (high copy number).
No packaging into P1 phage particles
Yeast Artificial Chromosome (YAC)
A Yeast Artificial Chromosome (YAC) is a synthetic vector used for cloning large DNA fragments in yeast cells. It is particularly useful for studying large eukaryotic genomes due to its ability to carry DNA fragments up to 1 megabase (Mb) in size. It is a self replicating element.
Key elements of YACs
Autonomously Replicating Sequence (ARS):
A yeast origin of replication.
Ensures that the YAC replicates autonomously within the yeast cell.
Centromeric Sequence (CEN):
Enables proper segregation of the YAC during cell division by mimicking yeast chromosome behavior.
Telomeric Sequences (TEL):
Protect the ends of the YAC and prevent degradation, mimicking natural chromosome ends.
Selectable Markers:
Allow selection of yeast cells containing the YAC.
Common markers include genes conferring the ability to grow in specific nutrient-deficient media, i.e. auxotrophic markers (e.g., URA3, TRP1, LEU2).
Multiple Cloning Site (MCS):
Contains restriction enzyme recognition sites for the insertion of large DNA fragments.
Problems associated with YACs
Instability:
Large DNA inserts can sometimes rearrange or be lost during yeast cell replication.
Chimeric Clones:
Incorrect assembly of inserts may lead to hybrid clones containing unrelated DNA fragments.
Not easily separated from host DNA
Pulsed-field electrophoresis required to separate host DNA from YAC DNA.
Solution is to circularise the YAC so it is more easily identifiable.
BAC/YAC
Transformation associated recombination (TAR)
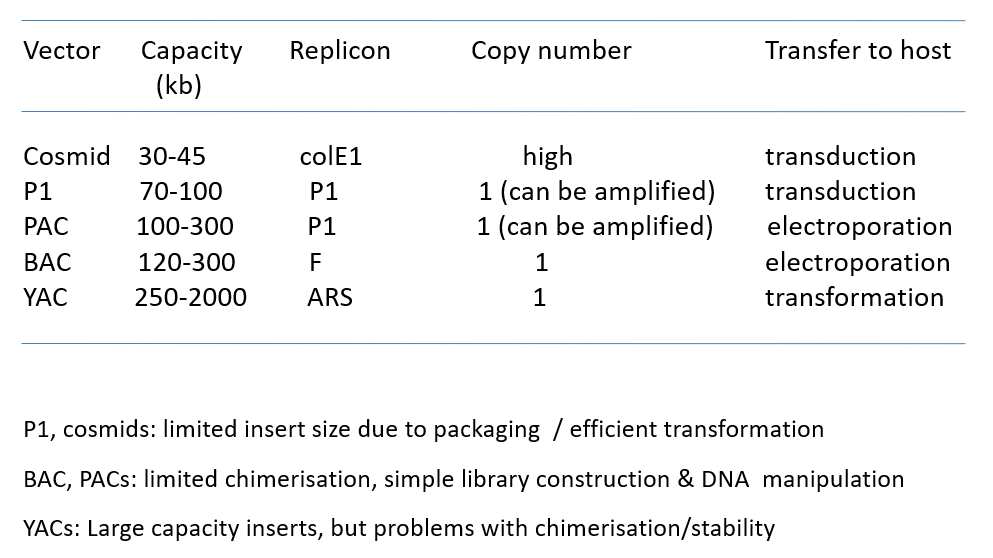
Differences between cosmids, P1 vector system, BAC, PAC, YAC.