L5: Exploring genetic diversity
1/36
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
37 Terms
- and - determine phenotypes
genotype, environment
how can we understand the function of a gene?
sequence/structural similarity
environmental perturbation
genetic perturbation
homeotic genes determine -
organ identity
perfect flowers have - whorls of organs
mutants revealed three sets of genes controlling -
four
floral organ identity
double flowered mutants are common in - flowers
ornamental
mutations can be artificially induced in a number of ways
chemical
EMS (alkylating agent)
physical
Xrays, gamma
biological
transposons, virus, bacteria
forward genetics
start from phenotype of mutants to find gene
backward genetics
mutation of gene to find phenotype
mutant libraries for reverse genetics
saturated collections of mutants can be screened for genes of interest, looking for mutant phenotype
tilling populations
large collections of mutated organisms, most commonly plants, created for research to study gene function
Salk T-DNA lines
a collection of genetically modified Arabidopsis thaliana plants used for genetic research, where a piece of DNA called T-DNA has been randomly inserted into the plant's genome
direct gene manipulation
it is possible through transgenesis, gene editing to directly inactivate or manipulate the activity of a gene but transformation/transfection of organism of interest is required
mutations in coding sequences or regulatory elements are more likely to - that mutations in intergenic regions
disrupt gene function
semi-dwarf mutants and the green revolution
mutations producing semi-dwarf plants increase yield and reduce lodging
impaired ability to produce or respond to plant hormone, gibberellin
mutation breeding for crop improvement
breeders commonly induce mutations to generate new varieties
gamma greenhouse
gene diversity allows organisms to adapt to -
different/changing environments
where does genetic diversity come from?
all new genetic variants arise from mutations
mutations are frequent, but only some are maintained in a population/species
whether a new variant is maintained or lost in a population depends on
selection: variants become more abundant or are lost due to their effect on the phenotype of individuals
drift: variants become more abundant or are lost due to chance events
genetic drift is when - determines the fate of an allele
chance
selection is when - determines the fate of an allele
phenotype
positive selection
the process where beneficial genetic variants spread through a population because they increase an organism's fitness
negative selection
the process of removing traits, alleles, or cells that are detrimental to an organism's survival or function
interaction between selection and drift
neutral variants?
non-neutral variants?
drift
selection
the effect of drift is much larger in - populations
deleterious mutations have a much higher chance to persist in - groups
smaller
small/isolated
two more sources of phenotypic and genetic diversity
sexual reproduction: generation of new genetic combinations through recombination
migration: introduction of new genetic variants through migration and/or hybridization
larger variants are important as well
SNV, SNP, inversions, translocations, indels, CNV
SNV
single nucleotide variant

SNP
single nucleotide polymorphism
CNV
copy number variation
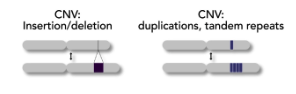
insertions/deletions
indels can introduce or remove parts of genes, complete genes, regulatory sequences
single nucleotide indels in gene can introduce frame-shifts in the coding sequence
larger indels can affect whole genes or gene clusters
several genetic disorders are due to very large chromosomal deletions
chromosomal inversions
effects of inversions
disrupt genes or gene regulation at their boundaries
suppress recombination between the two orientations
by locally suppressing recombination, inversions keep together combinations of adaptive alleles
inversions act as supergenes to maintain complex adaptations
Copy number variation
can affect short repeats or whole genes
can vary from zero, to few, to manychr
chromosomal translocations
generally bad for individual
can facilitate speciation
dispensable genes can be linked to
disease resistance
abiotic resistance
secondary metabolites
duplicated genes
etc
reference bias in genomics
single reference genomes represent only one version the genome of a species, resulting in “invisible” genes
pangenomes
provide more complete representation of a species diversity
super pangenomes compare variation across related species
graph pangenomes
a sequence graph is a compressed way to represent multiple genome assemblies
nodes: unique DNA sequences
edges: ways in which nodes are connected in different genomes