Lecture 1-8
1/90
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
91 Terms
List of Biomolecules
Proteins
Nucleic Acids/Nucleotides
Carbohydrates
Lipids
Other(any molecule found in organisms and needed for biological processes)
Proteins
Rhino Horn(source of keratin), Red Blood Cells(source of hemoglobin), Firefly luciferase
Nucleic acids/nucleotides
DNA double helix, ATP
Carbohydrates
Sugar(sucrose), Cotton(cellulose), Insect shells(chitin)
Lipids
Waxes, Fat droplets in adipose tissue
Other Biomolecules
Metabolites, Vitamins, Antibiotics, Penicillin
hundreds or thousands 630 listed in one database(KEGG)
thousands in other databases(human, yeast, E. coli, metabolome databases)
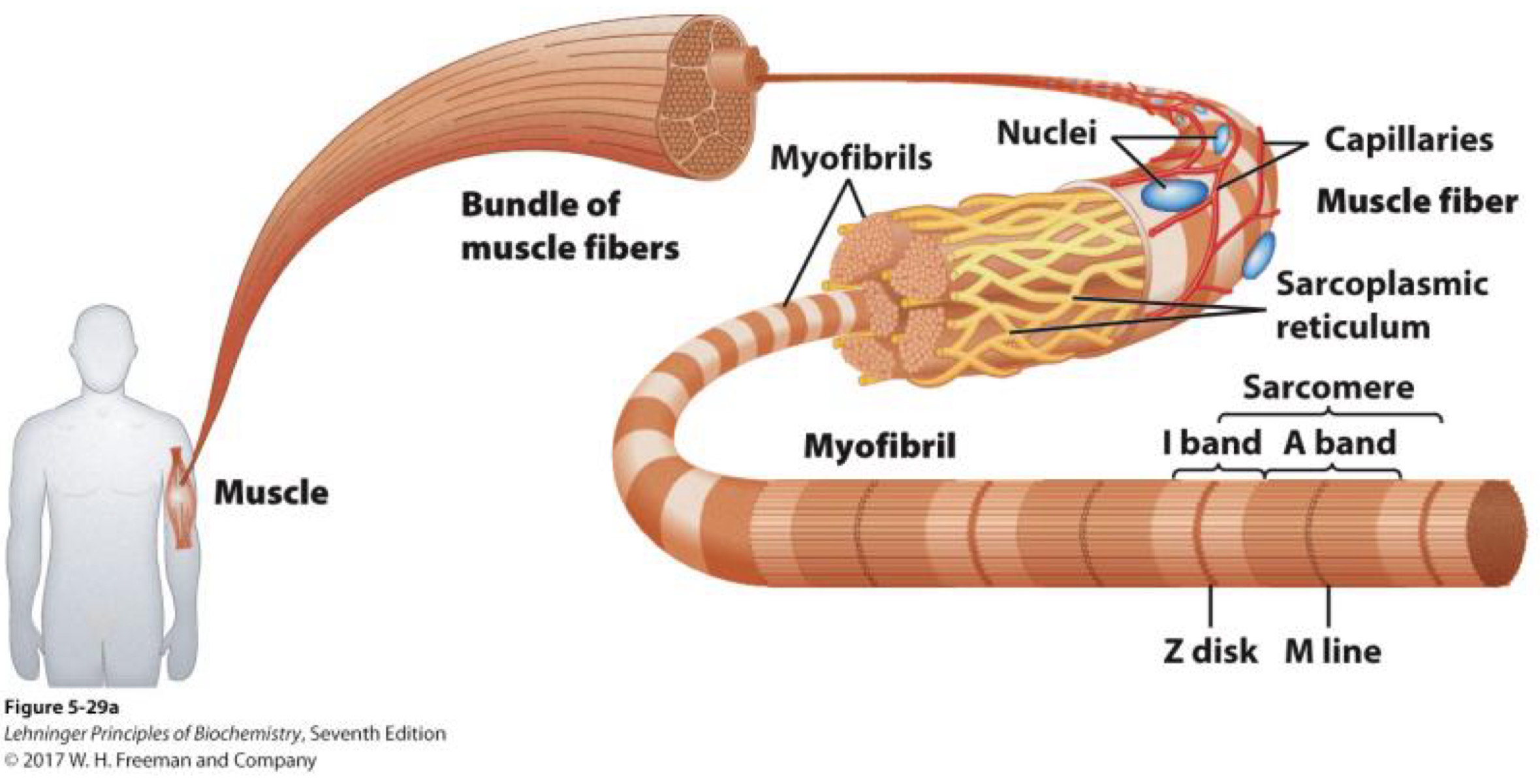
What do biomolecules do?
Building blocks of cells and life
Fuel for Life
Engine for Life
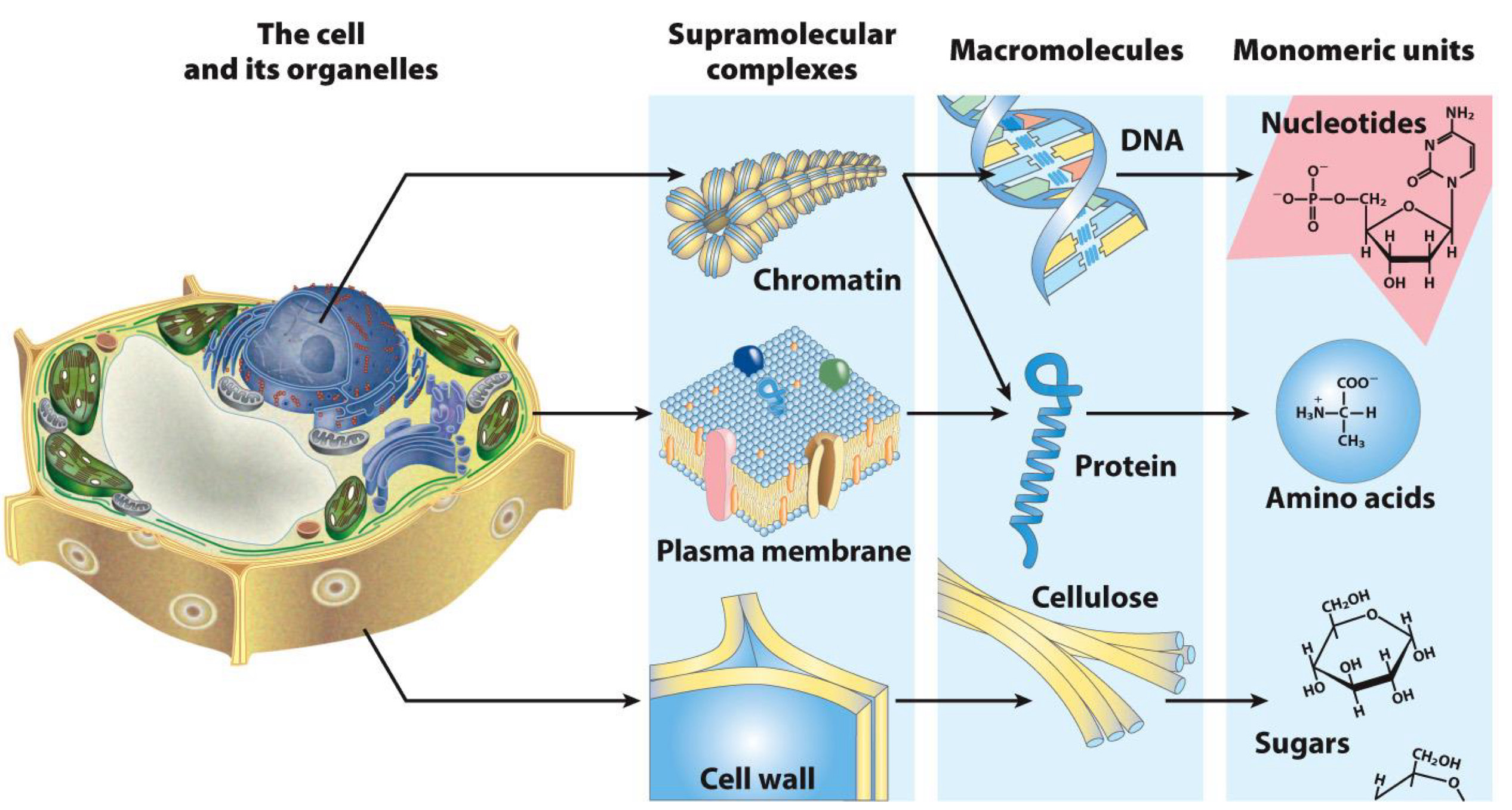
Importance of Amino Acids
Building Blocks of proteins
What are amino acids named by?
have similar structure
named by R substituent(group)
The Five Groups based on R substituent(group)
Nonpolar, aliphatic R groups
Aromatic R groups
Polar, uncharged R groups
Positively charged R groups
Negatively charged R groups
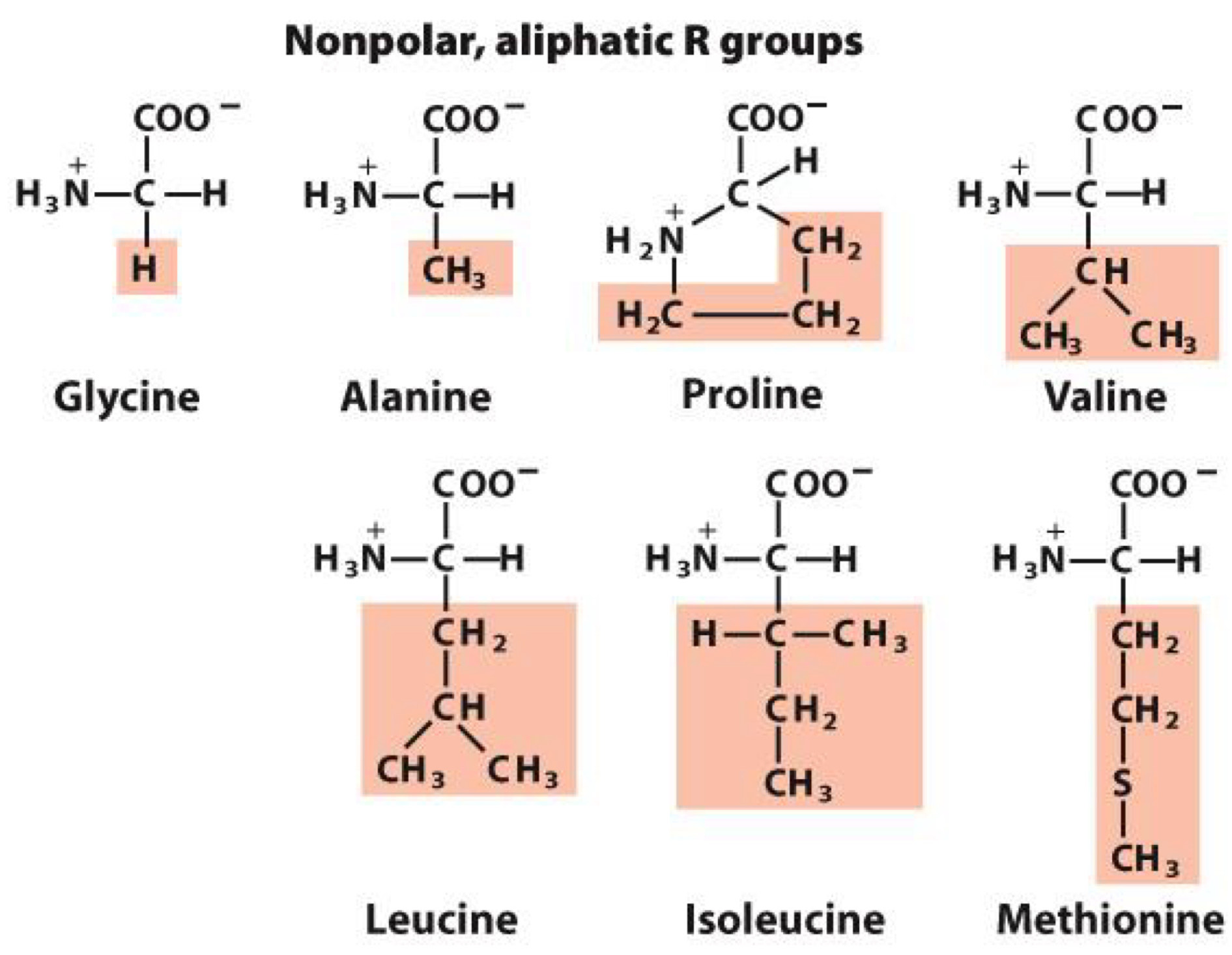
Nonpolar, aliphatic R groups
nonpolar group makes them hydrophobic
aliphatic means they are hydrocarbons with straight or branches chains(not aromatic)
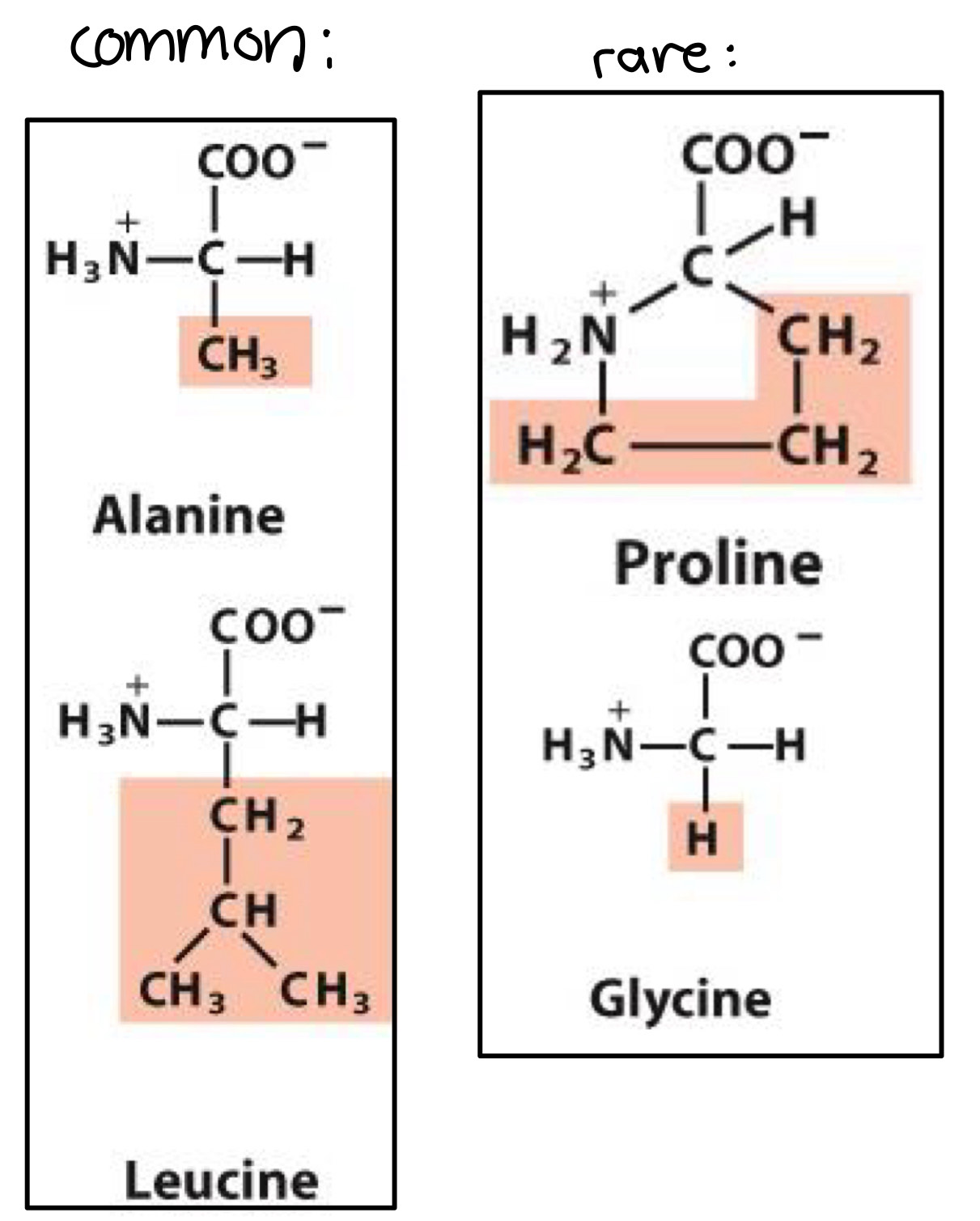
Glycine, Alamine, Proline, Valine, Leucine, Isoleucine, Methionine
members with simple structure R= -H, R= -CH3
Proline: cyclic structure that causes kinks in peptide chains
Methionine: contains sulfur
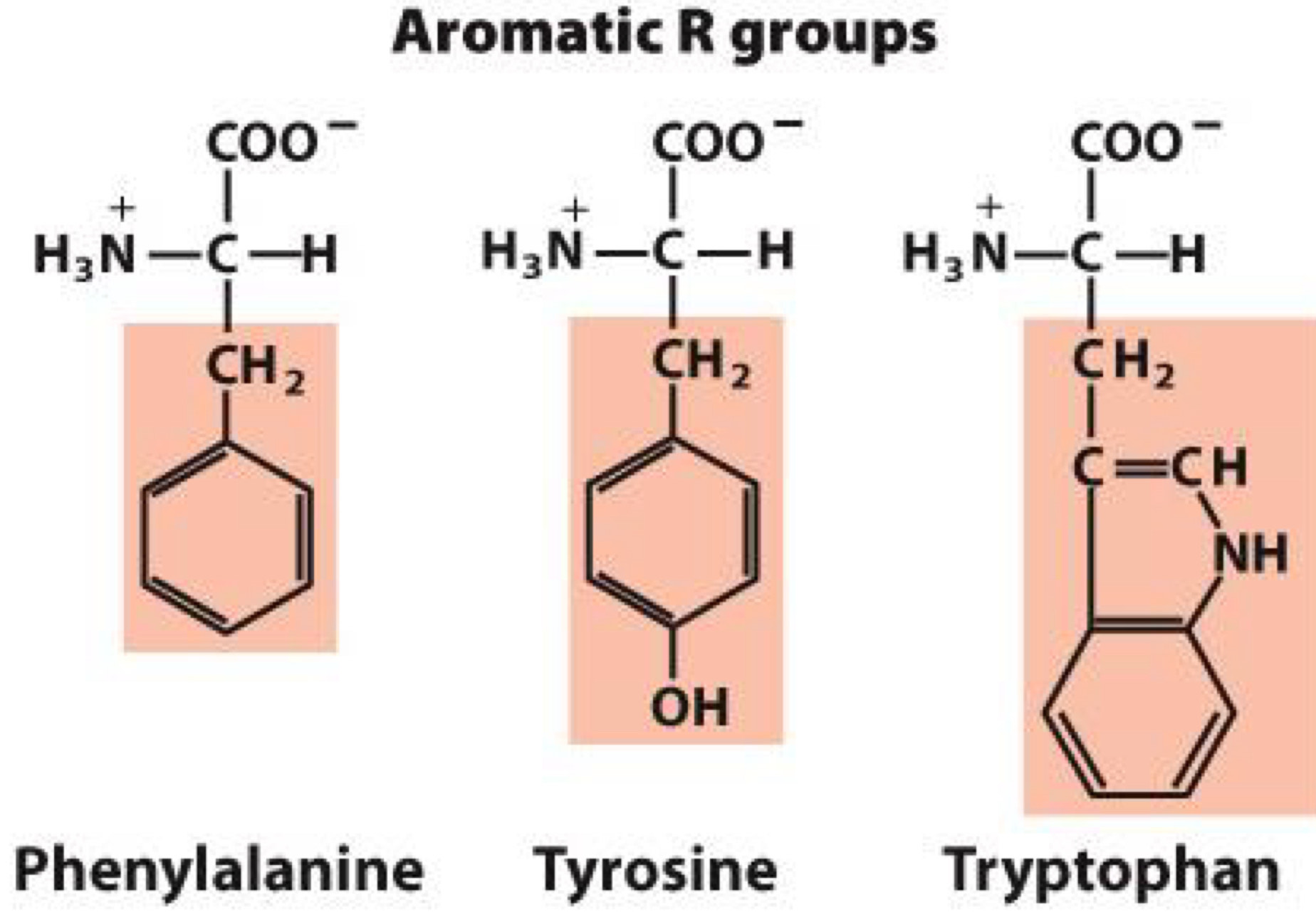
Aromatic R group
nonpolar, large aromatic groups
aromatic means they are cyclical(with bonds in resonance)
Phenylalanine, Tyrosine, Tryptophan
Tyrosine: Hydroxyl group forms hydrogen bonds and is important functional group in some enzymes
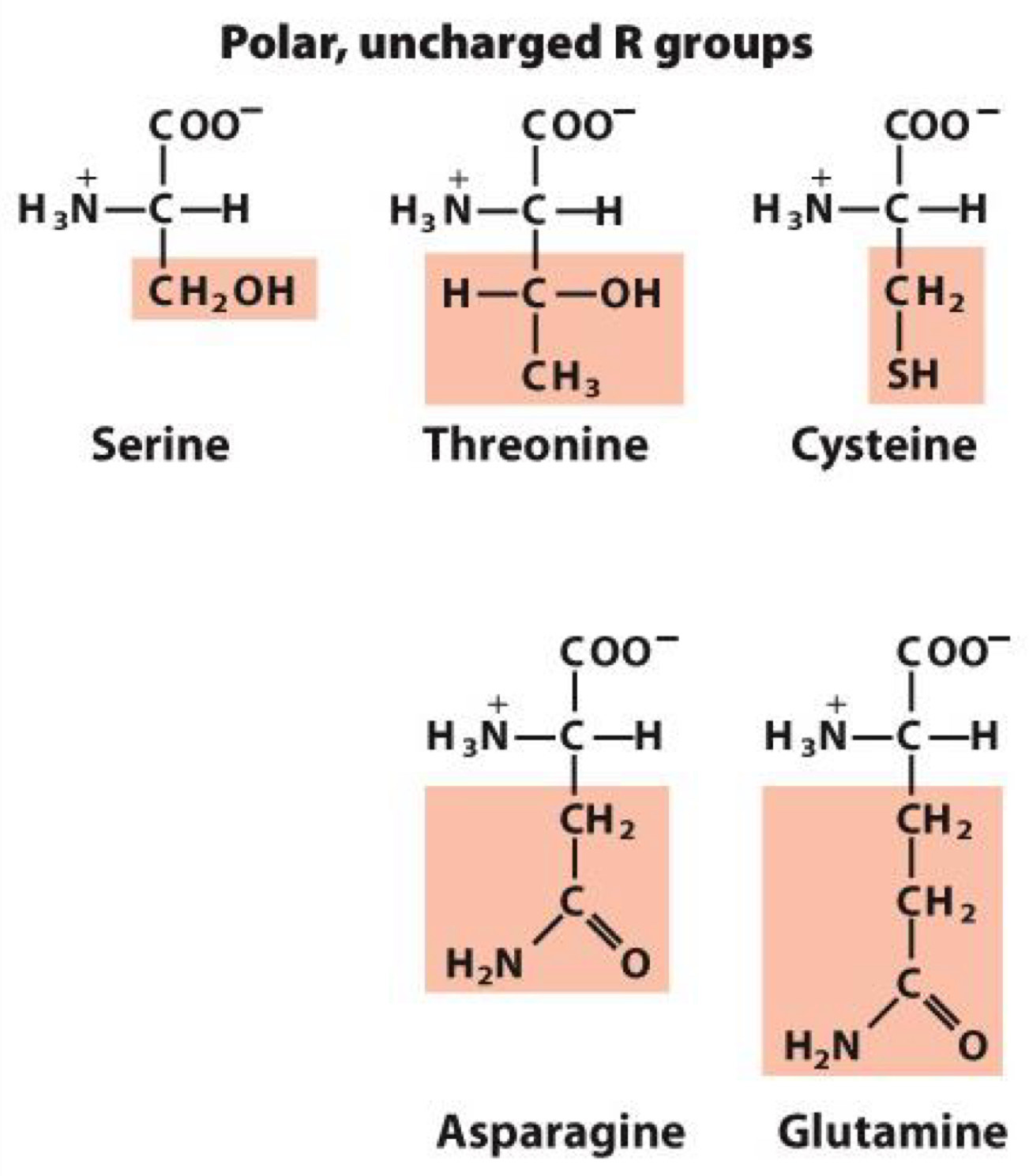
Polar, Uncharged R group
polar groups makes them hydrophilic
form hydrogen bonds
Serine, Threonine, Cysteine, Asparagine, Glutamine
Cysteine: contains sulfur and form disulfide bonds
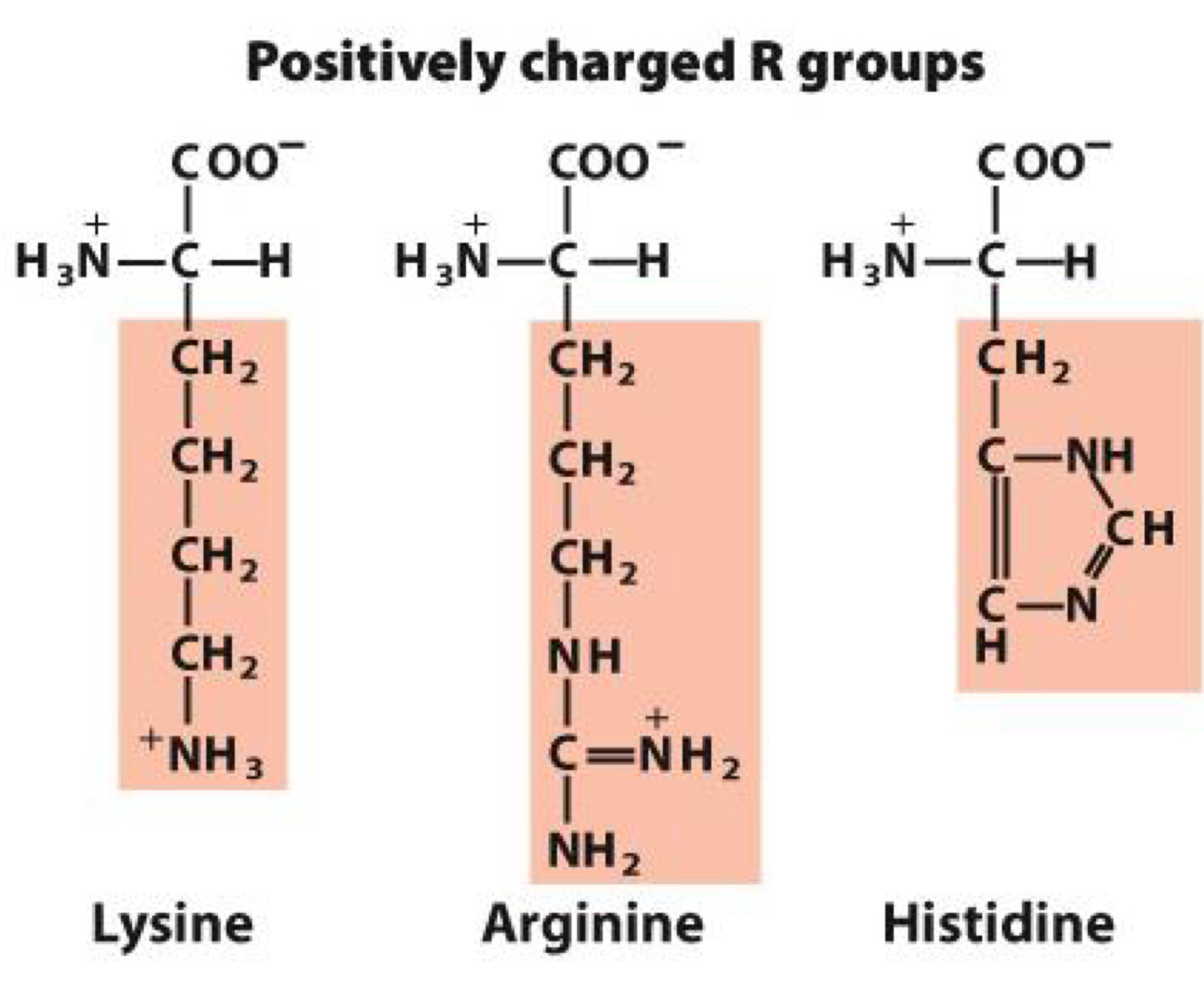
Positively charged R groups
Positive charge makes them hydrophilic
Lysine, Arginine, Histidine
Histidine: serves as proton donor or acceptor in many enzymes
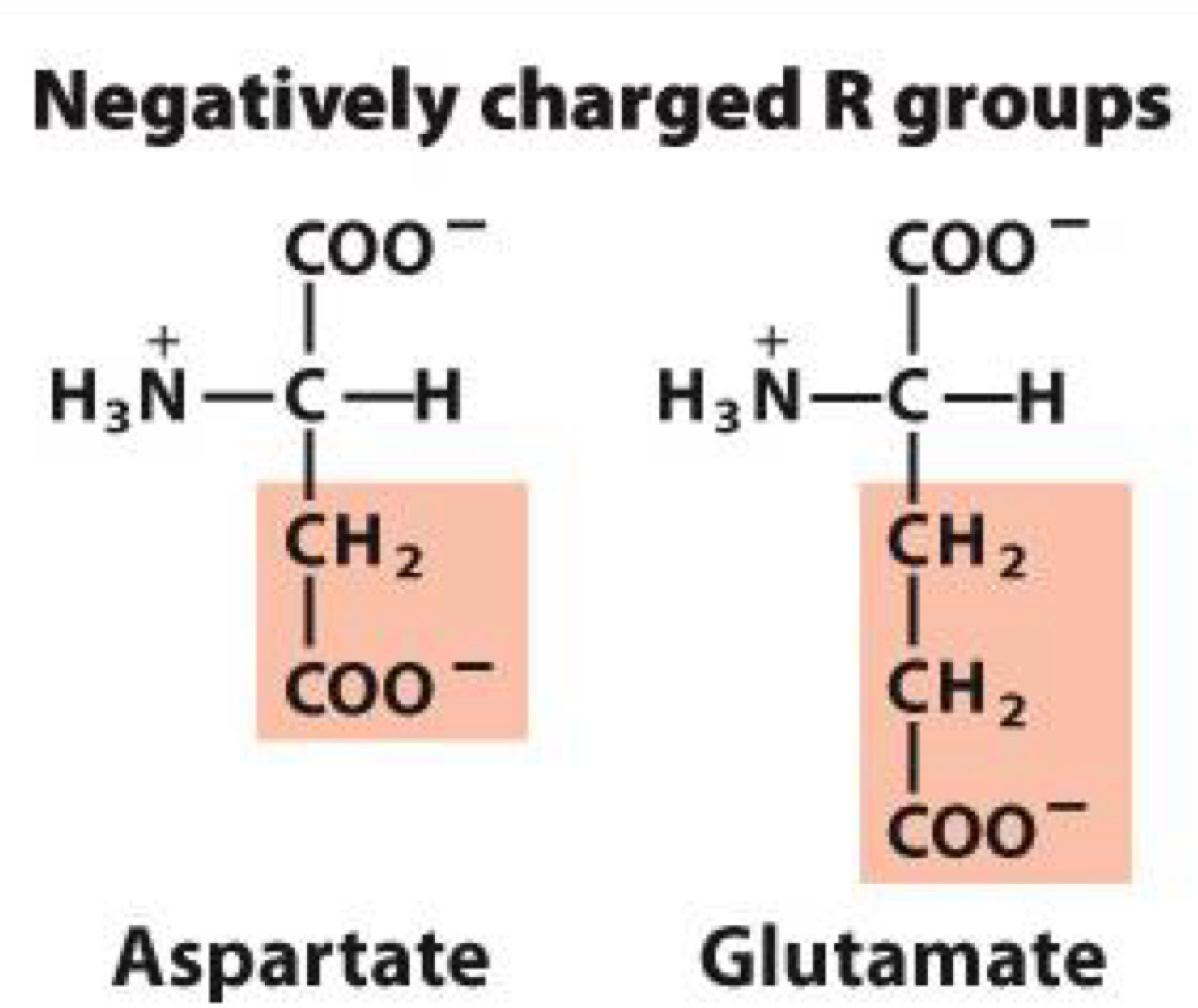
Negatively charged R groups
Negative charge makes them hydrophilic
Aspartate, Glutamate
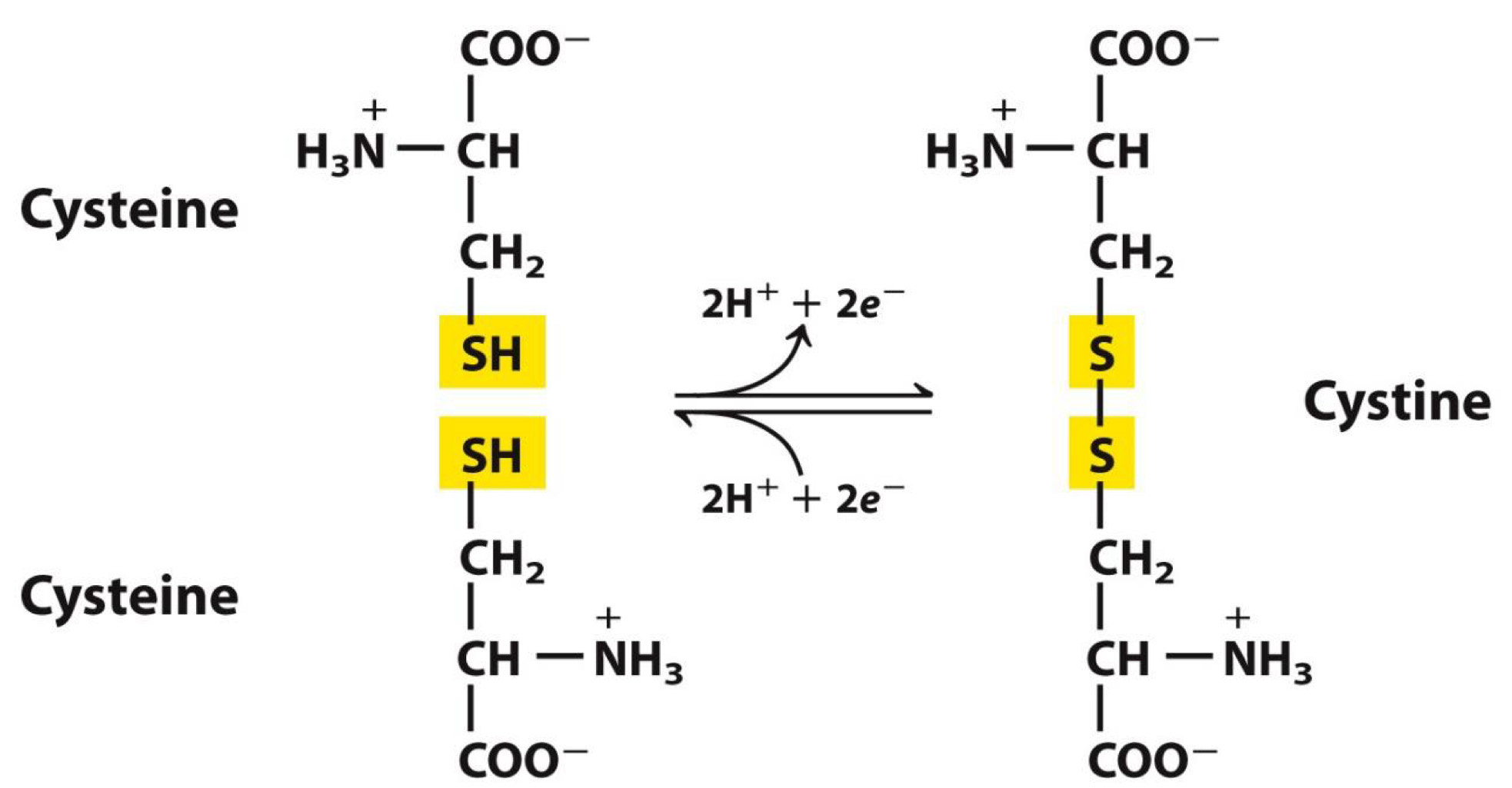
Polar, Uncharged R group-Cysteine →disulfide bonds
Sulfur atoms of two cysteines can form a bond
Bond is important to protein structure and folding
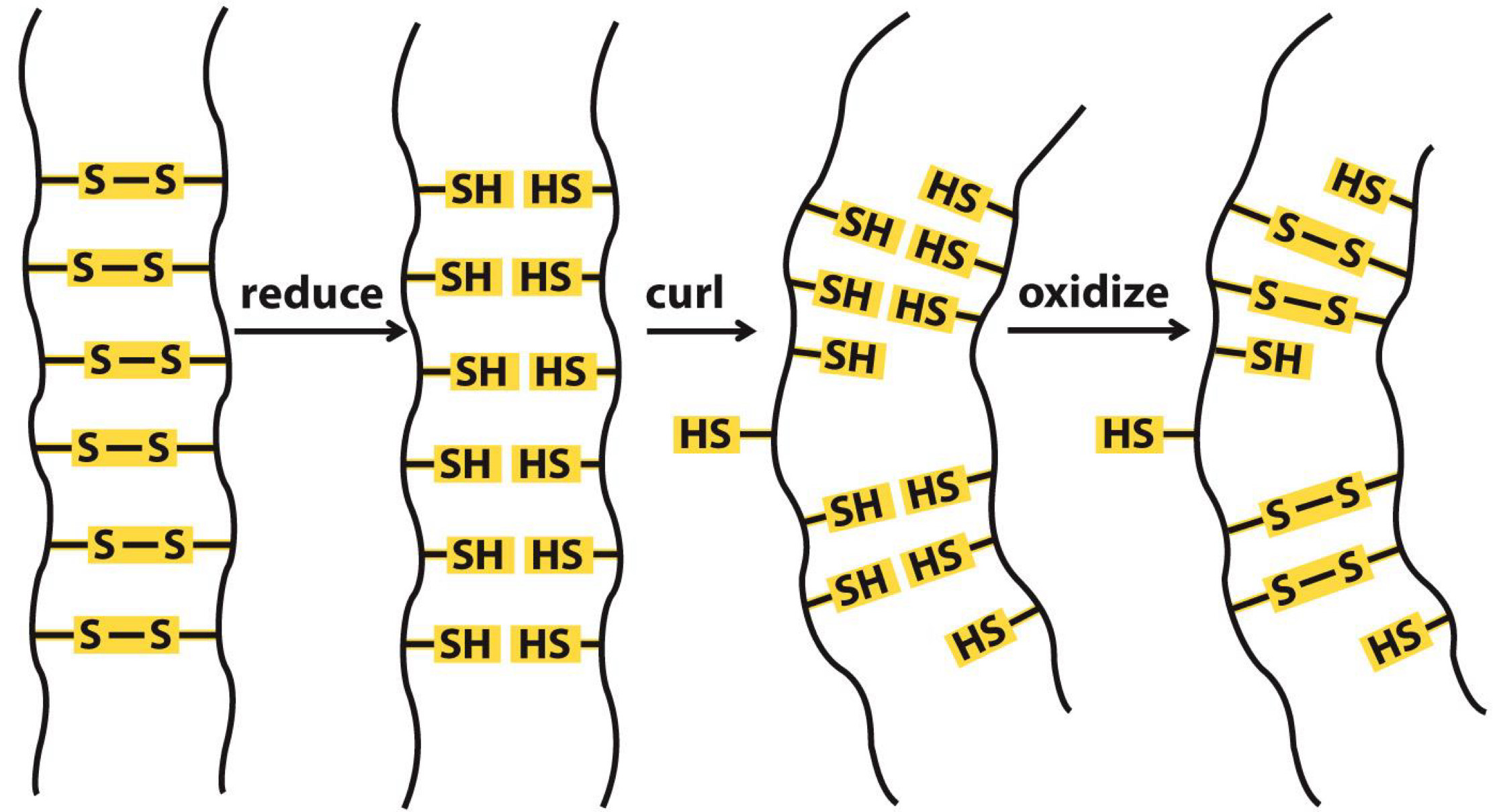
Curls in hair are due to disulfide bonds
Straight hair can be made curly by chemical treatment
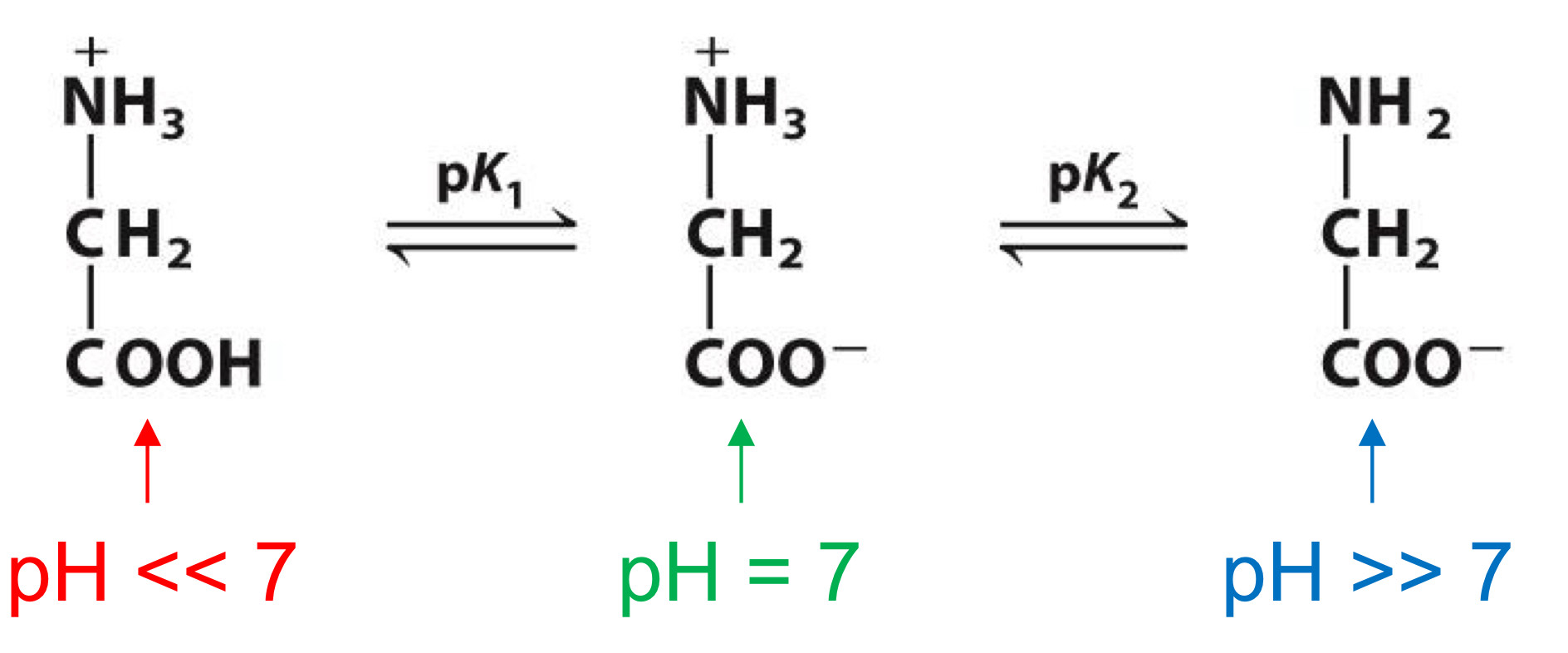
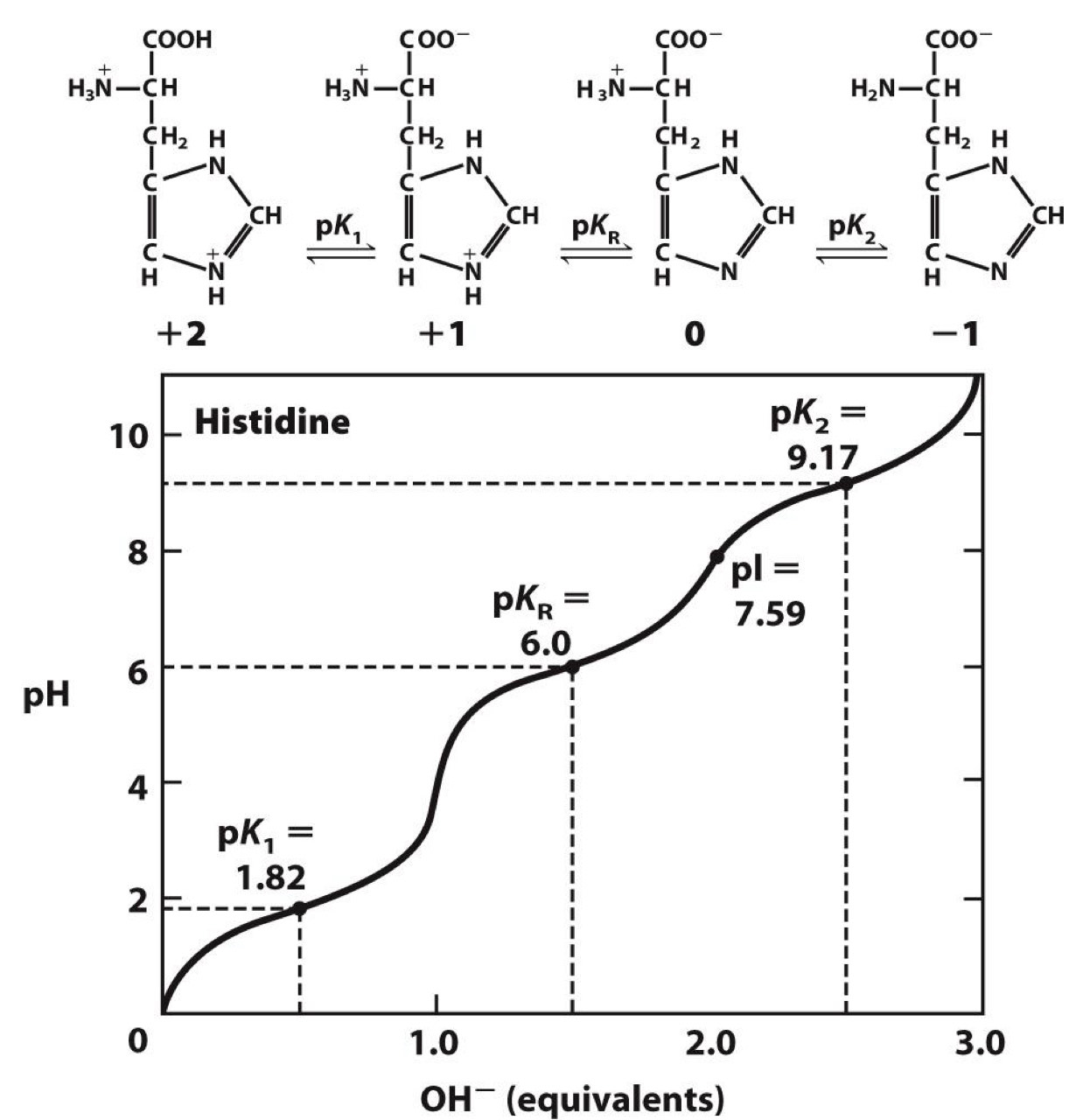
Titration curve
deprotonation of carboxylic group with increasing pH→shows pKa(pH where unprotonated and protonated forms are equal in concentration)
Same for amino group, but pH for deprotonation is higher
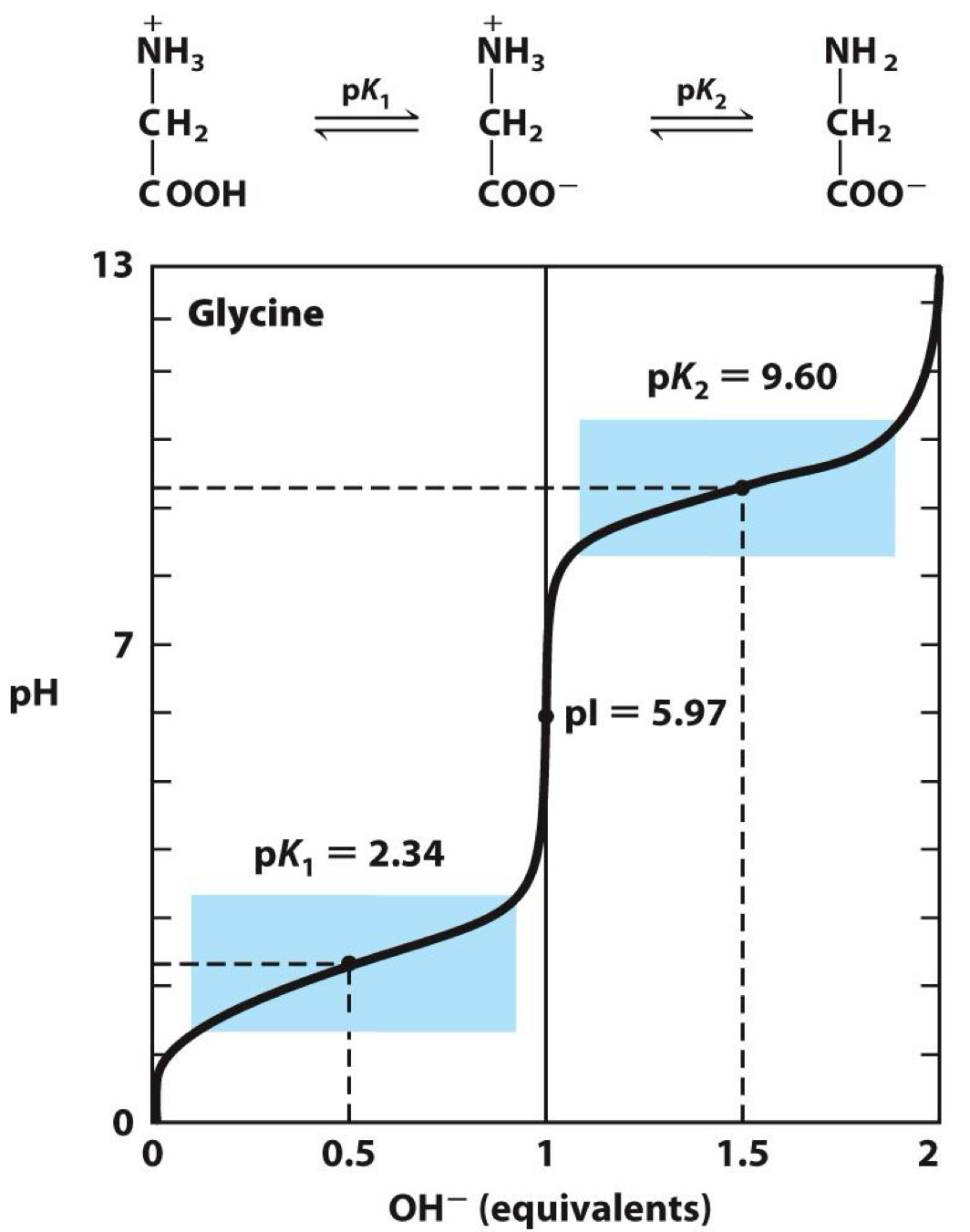
Carboxyl groups are ionizable
acidic pKa and is unprotonated at pH = 7
amino groups are ionizable
basic pKa and is protonated at pH = 7
R group can also be ionizable
Histidine has group with pKa = 6
-Near pH in cells (~ 7)
-Explains why it is a good proton donor or acceptor in enzymatic reactions
Positively charged R groups
◼ pKa >> 7 (except histidine)
Negatively charged R groups
◼ pKa << 7
Uncommon amino acids
many uncommon (nonstandard) amino acids
Usually not encoded directly by DNA
May or may not be part of protein
derived from common amino acids
Some are metabolites
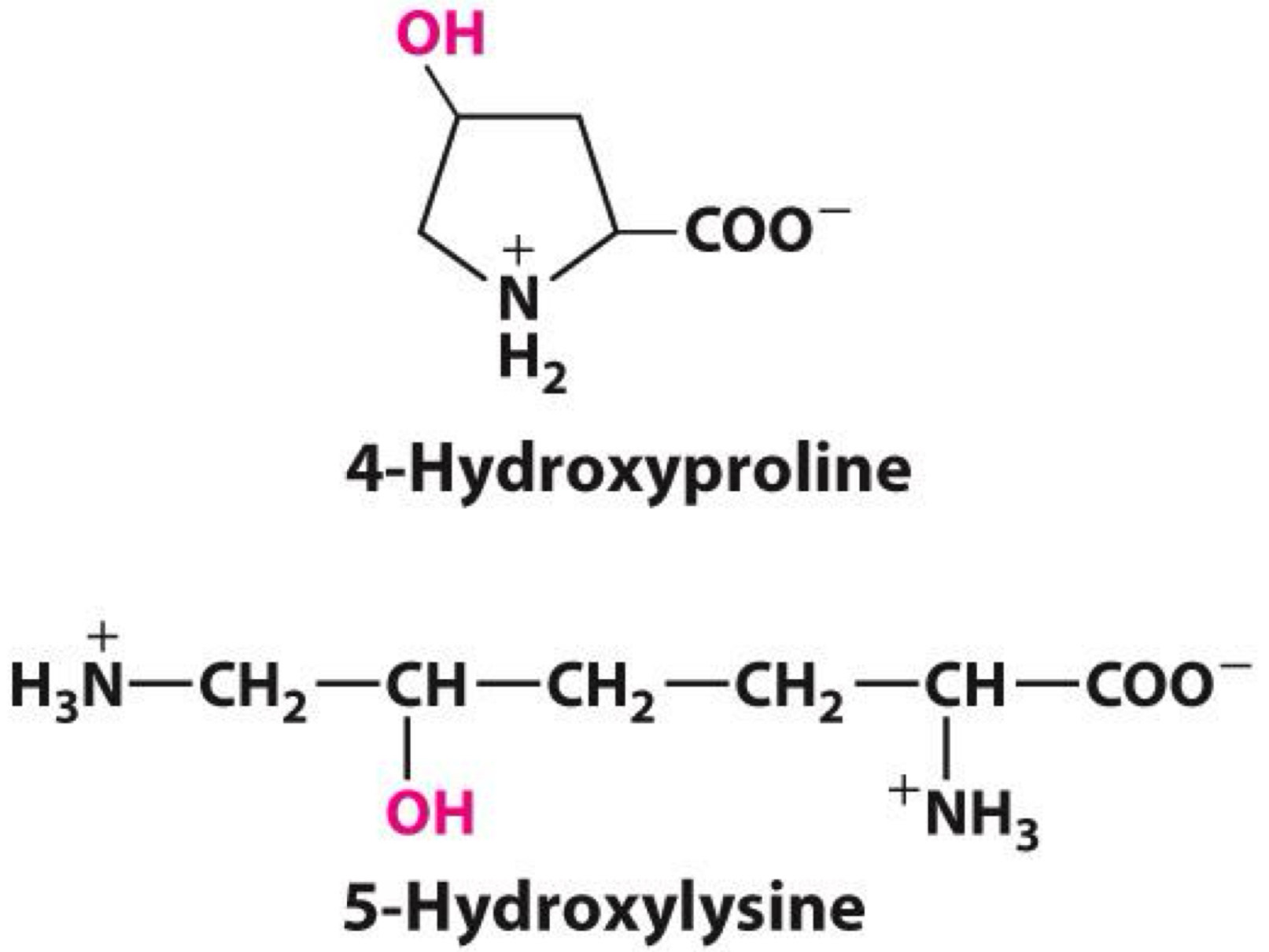
example of Uncommon amino acids
4-hydroxyproline and hydroxylysine
◼ Formed by hydroxylation of proline and lysine
◼ Occurs after translation
◼ Part of collagen
phosphoserine and other phosphorylated amino acids
◼ Formed by phosphorylation
◼ Modification is reversible
◼ Done to regulate activity of proteins
ornithine and citrulline
◼ Formed during urea cycle (conversion of ammonia to urea)
◼ Not part of proteins
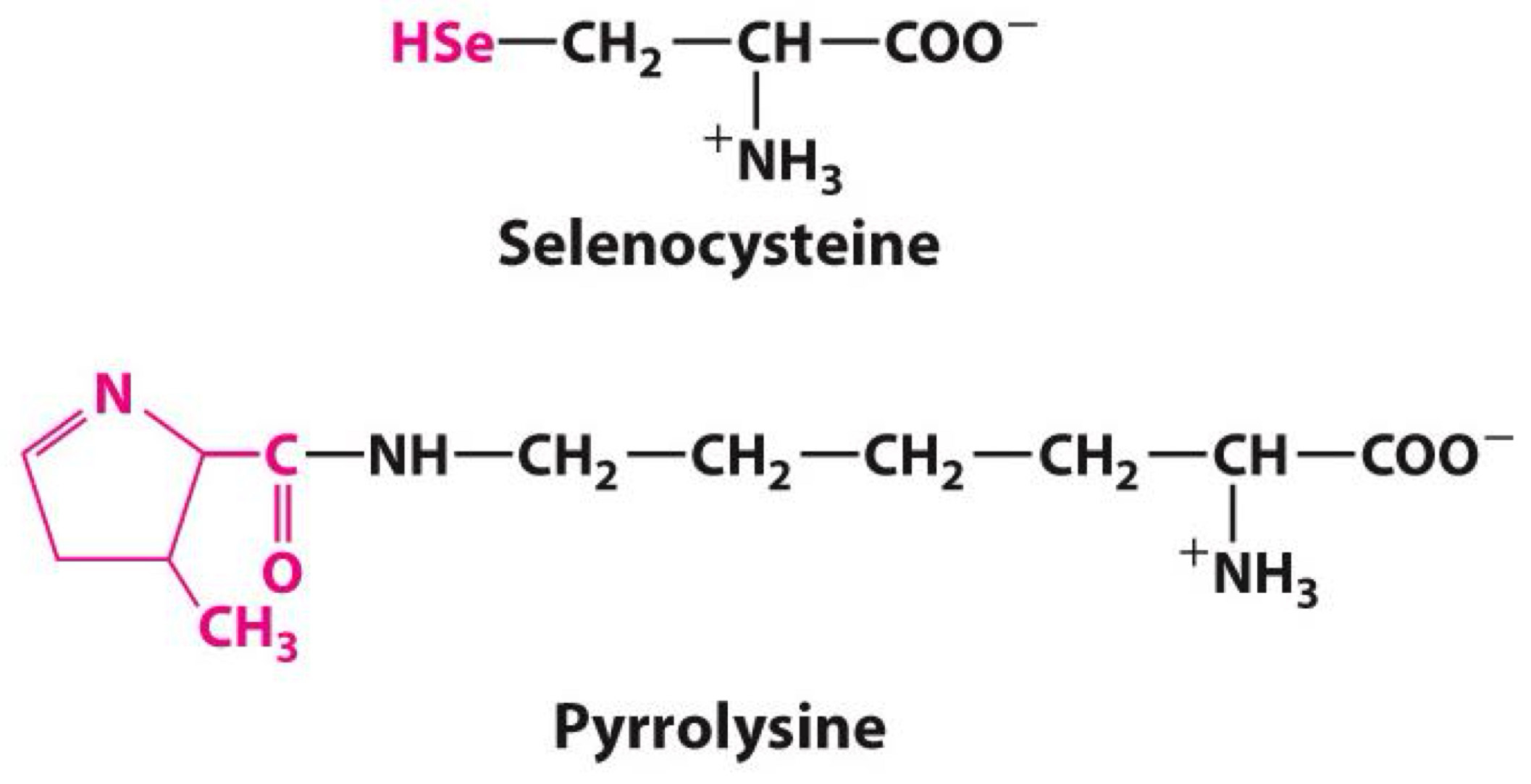
uncommon amino acid 21st and 22nd amino acids
Selenocysteine
Pyrrolysine
◼ Both encoded by DNA
◼ Both part of protein
◼ Not common
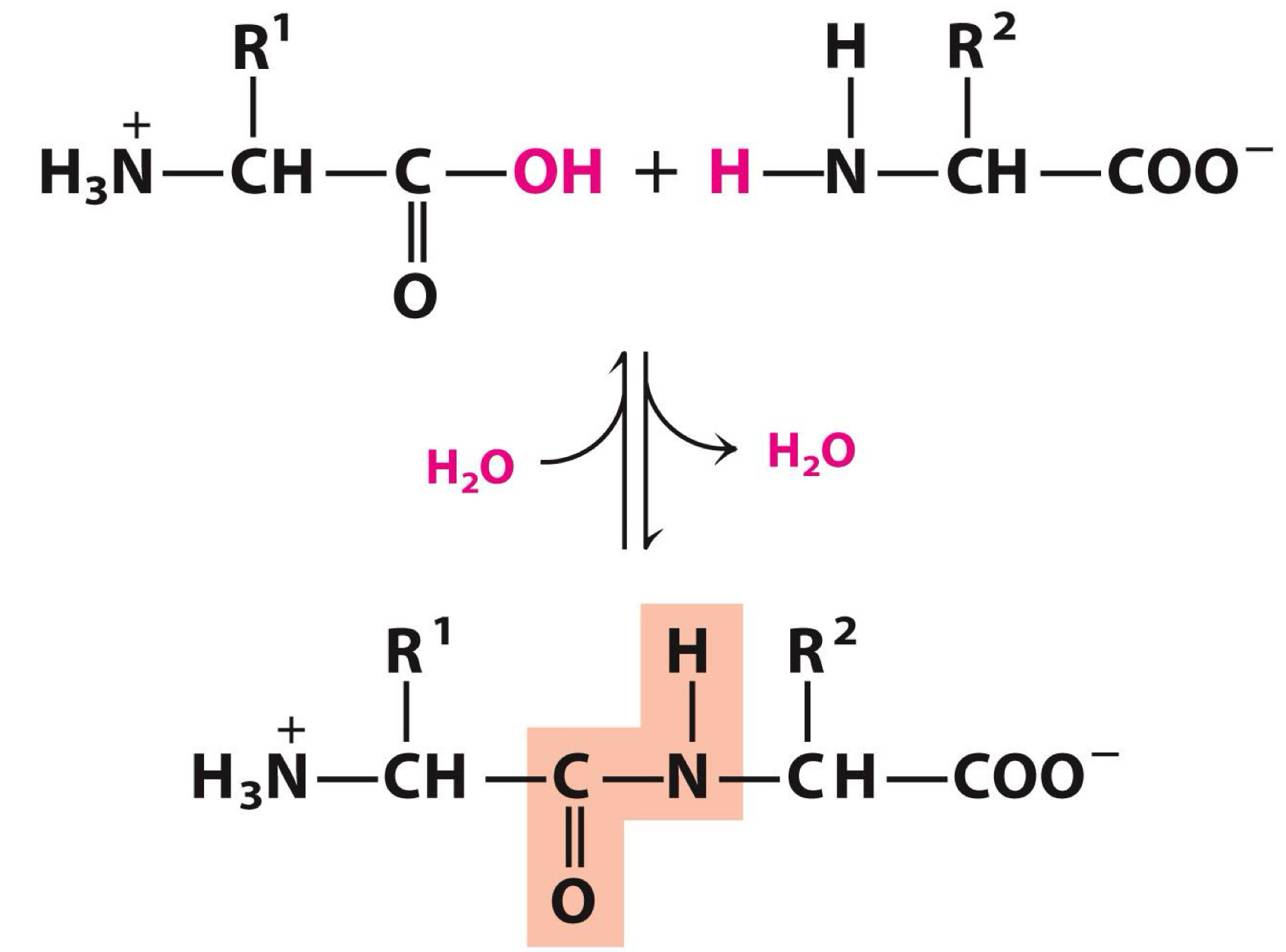
Amino acids polymerize to form peptides
Polymerization involves
Carboxyl group of one amino acid
Amino group another
Peptide bond is formed
An example of a condensation reaction
Peptide ends are not the same
Numbering and naming startfrom the amino terminal end
Dipeptide-Two amino acids
Tripeptide-Three amino acids
Oligopeptide-Up to ~10 amino acids
Polypeptide-Up to ~100 amino acids (~10 kDa)
Protein-More than ~100 amino acids (>10 kDa)
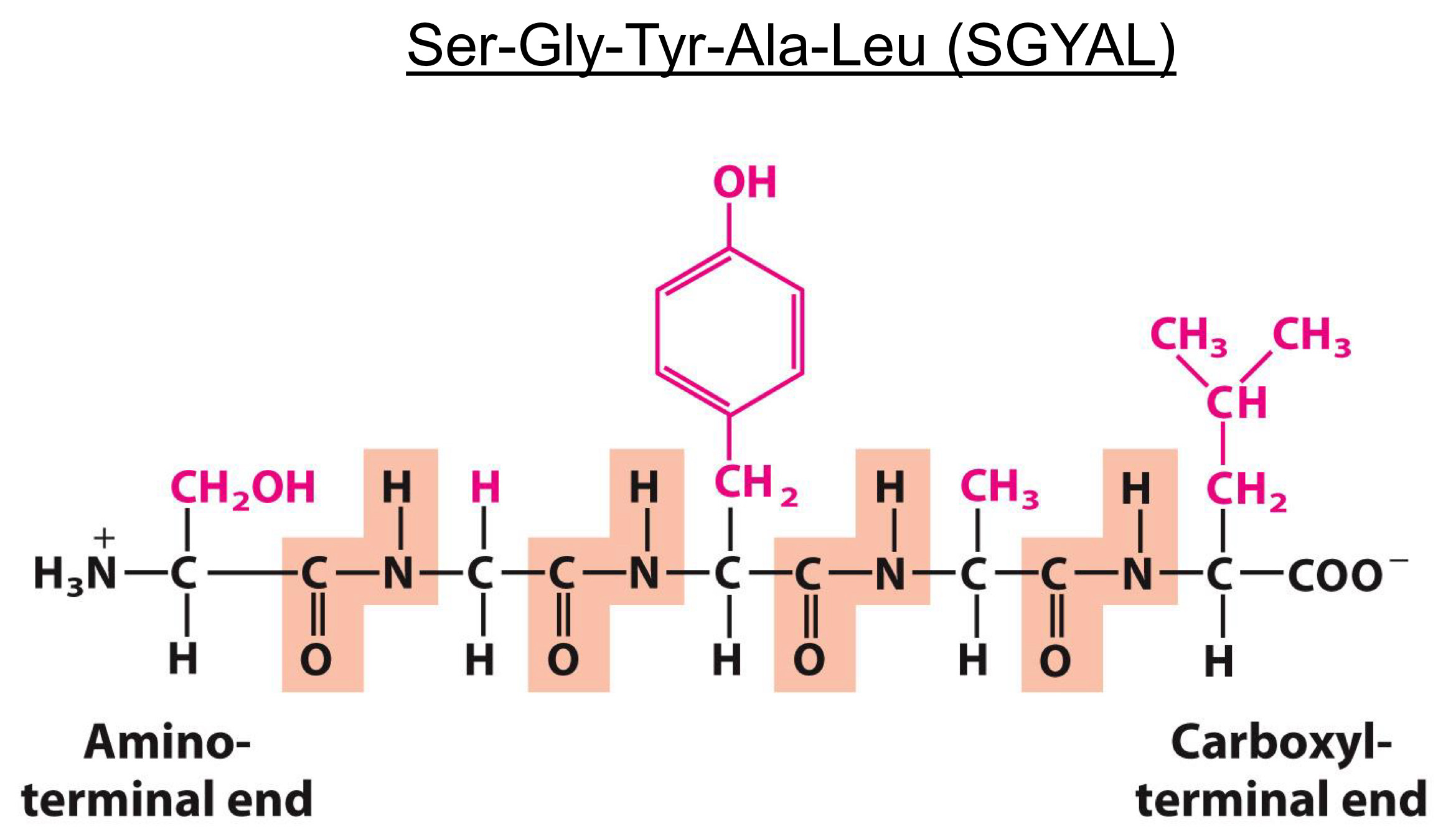
Peptides form proteins
After polymerization, peptide folded into more complex structures
Primary-Amino Acid Residue
Secondary- alpha Helix
Tertiary-Polypeptide chain
Quaternary-Assembled subunit
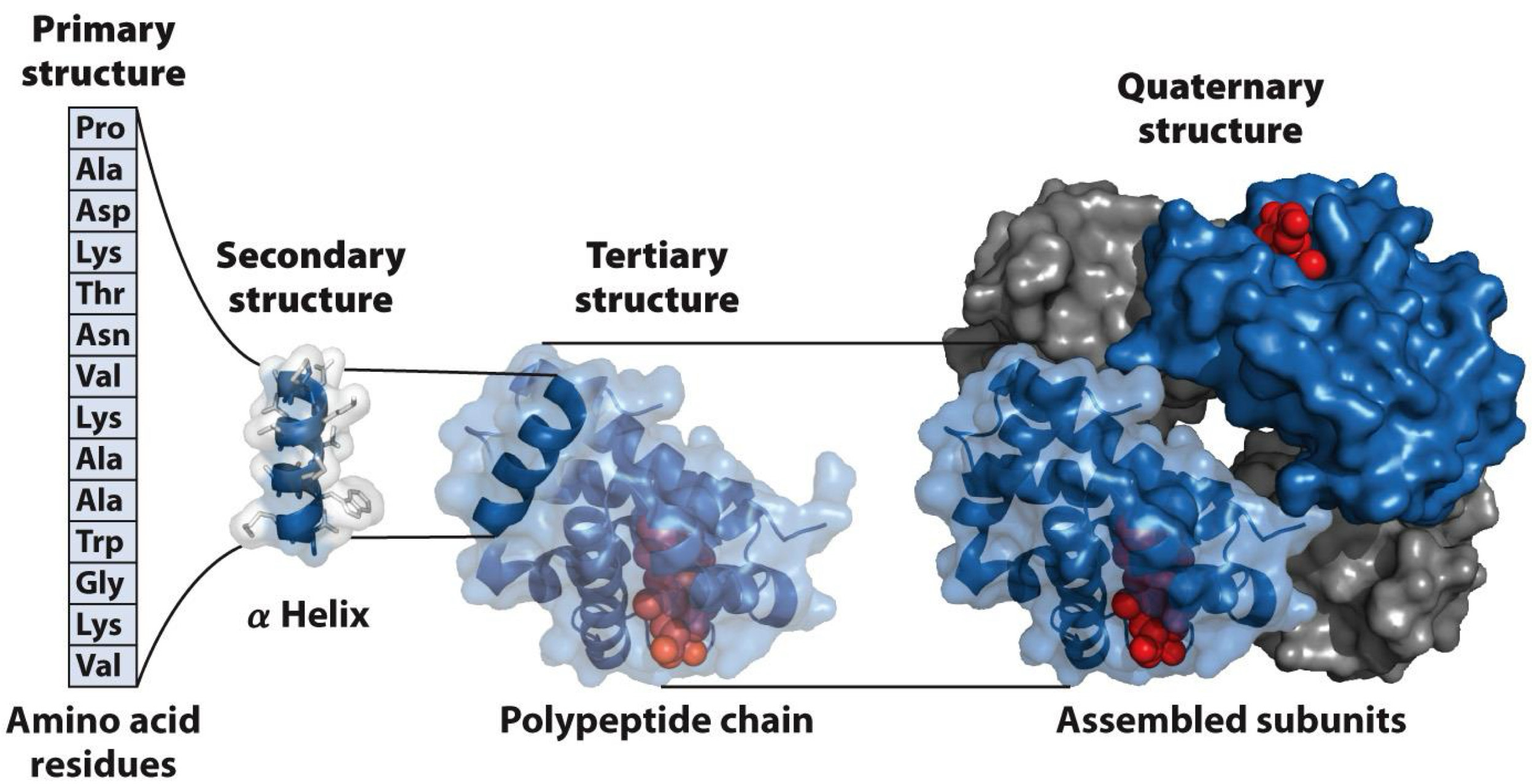
Primary structure-simplest level
sequence of peptide
Determined when amino acids polymerize
Secondary structure
short segments of peptide folded into regular arrangements
Two main types
◼ alpha helix
◼ beta sheet
Other types: Collagen helix(Found only in collagen), Random coil(No regular arrangement)
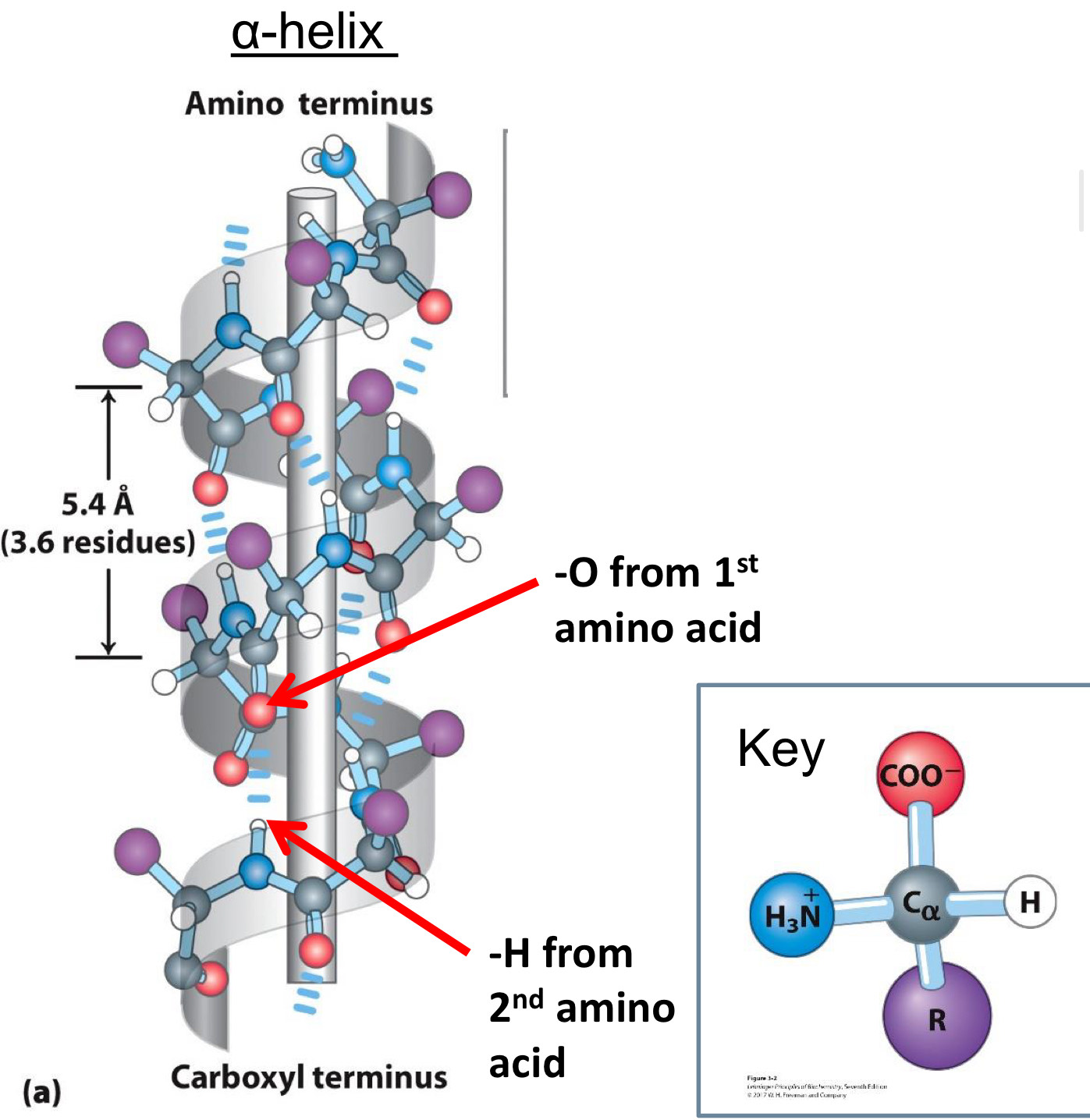
alpha helices are common secondary structures
Have a helical (corkscrew) shape: Held together by hydrogen bonds
◼ Oxygen of carboxyl group of one amino acid
◼ Hydrogen of amino group of second amino acid
◼ First amino acid is four residues away second amino acid
3.6 residues (5.4 Å) per turn
R substituents point out
Some amino acids are common or rare in alpha
helices
Common: Ala & Leu→ Have small, hydrophobic R groups, Pack well into helix
Rare: Proline
- N atom is part of rigid ring, and peptide bond cannot rotate
- Forms kinks in helix when present
Glycine
- Has tiny R group, giving it flexibility to make up other structures
- Tends to make up random coil, not helix
Beta sheets are another common secondary structure
Have pleated sheet-like structure
R groups (side chains) stick out from above and below the sheet
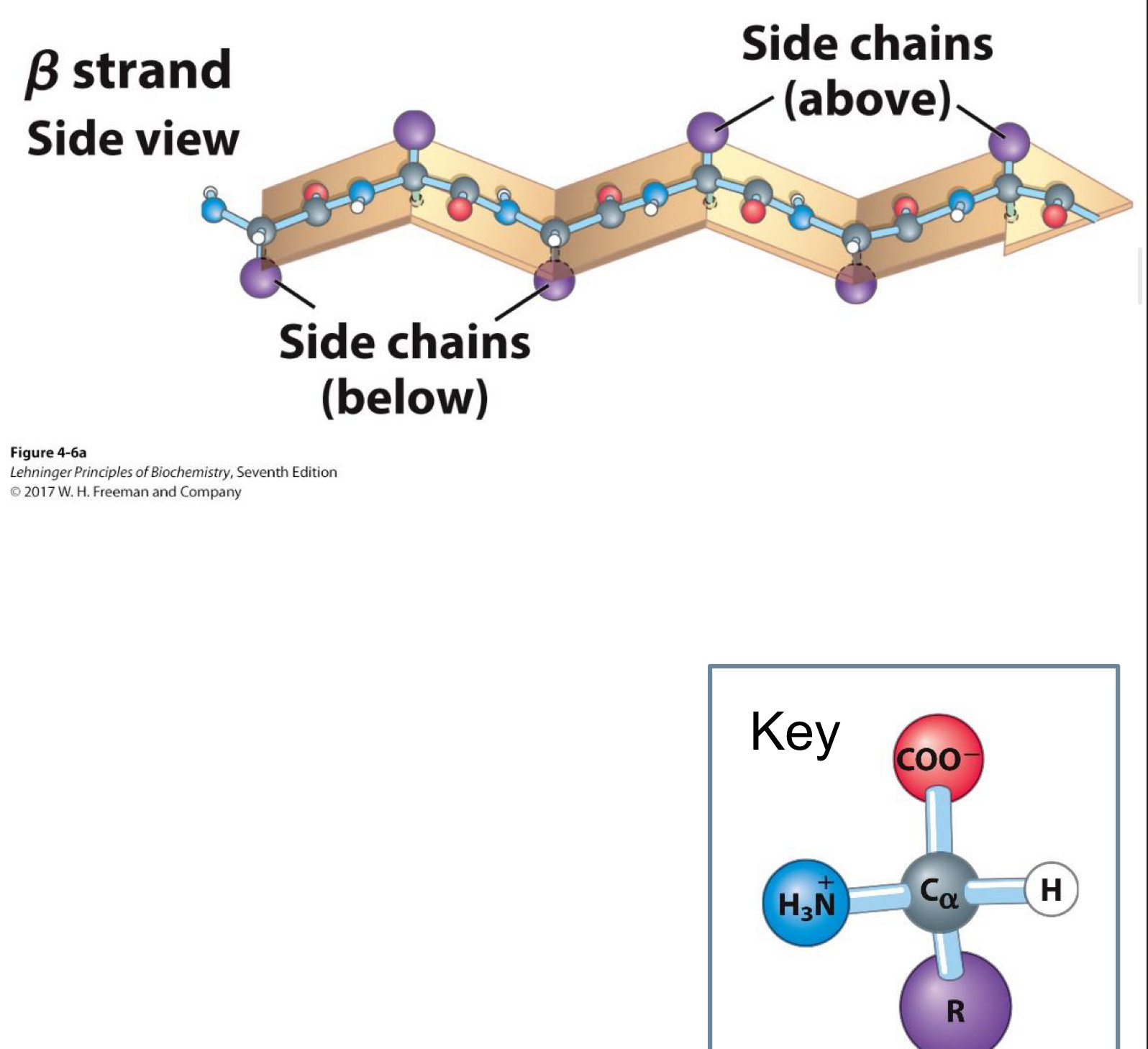
Beta Sheet secondary structure Top view
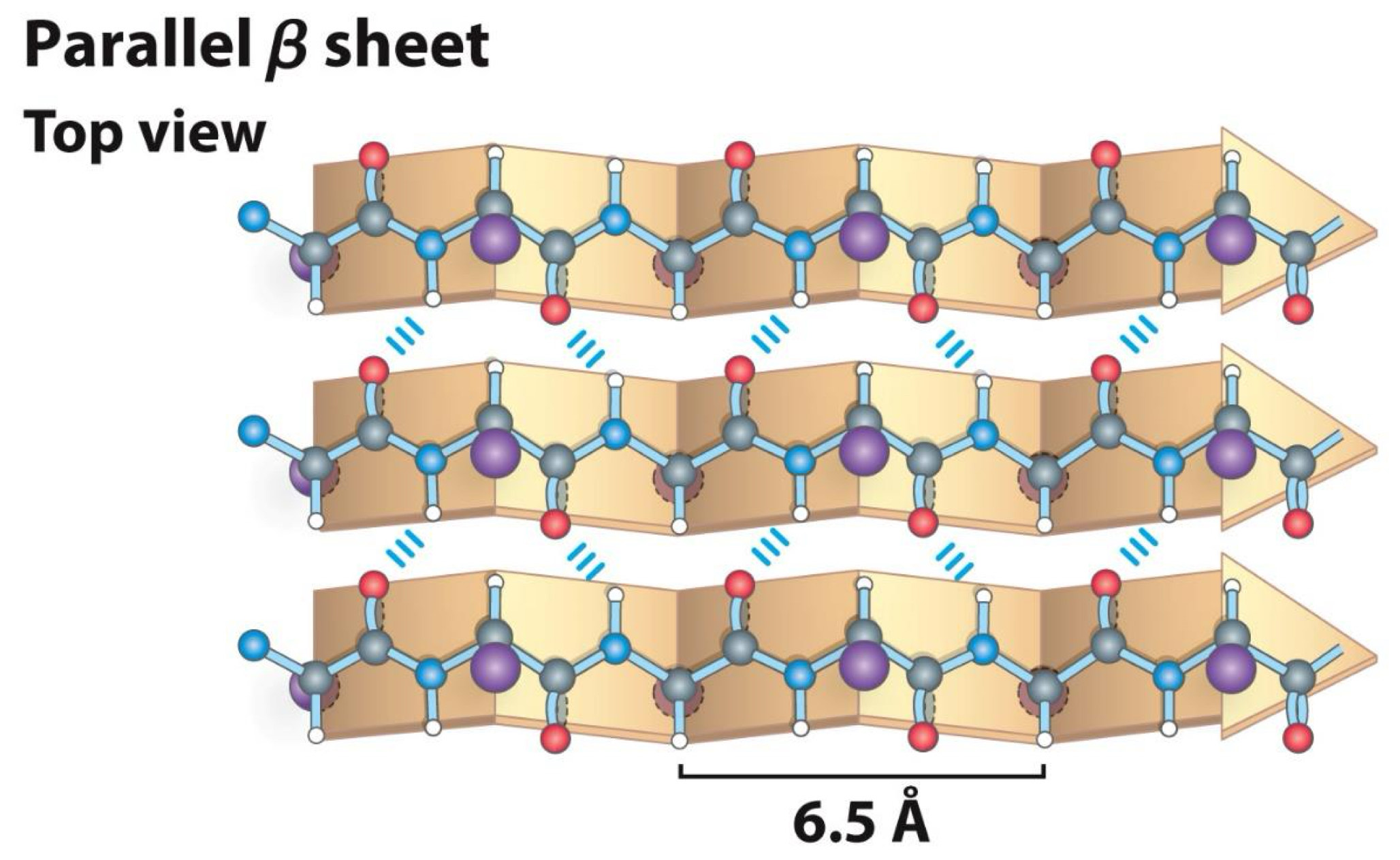
Made up of several strands
-Strands held together by hydrogen bonds between carboxyl group and amino groups
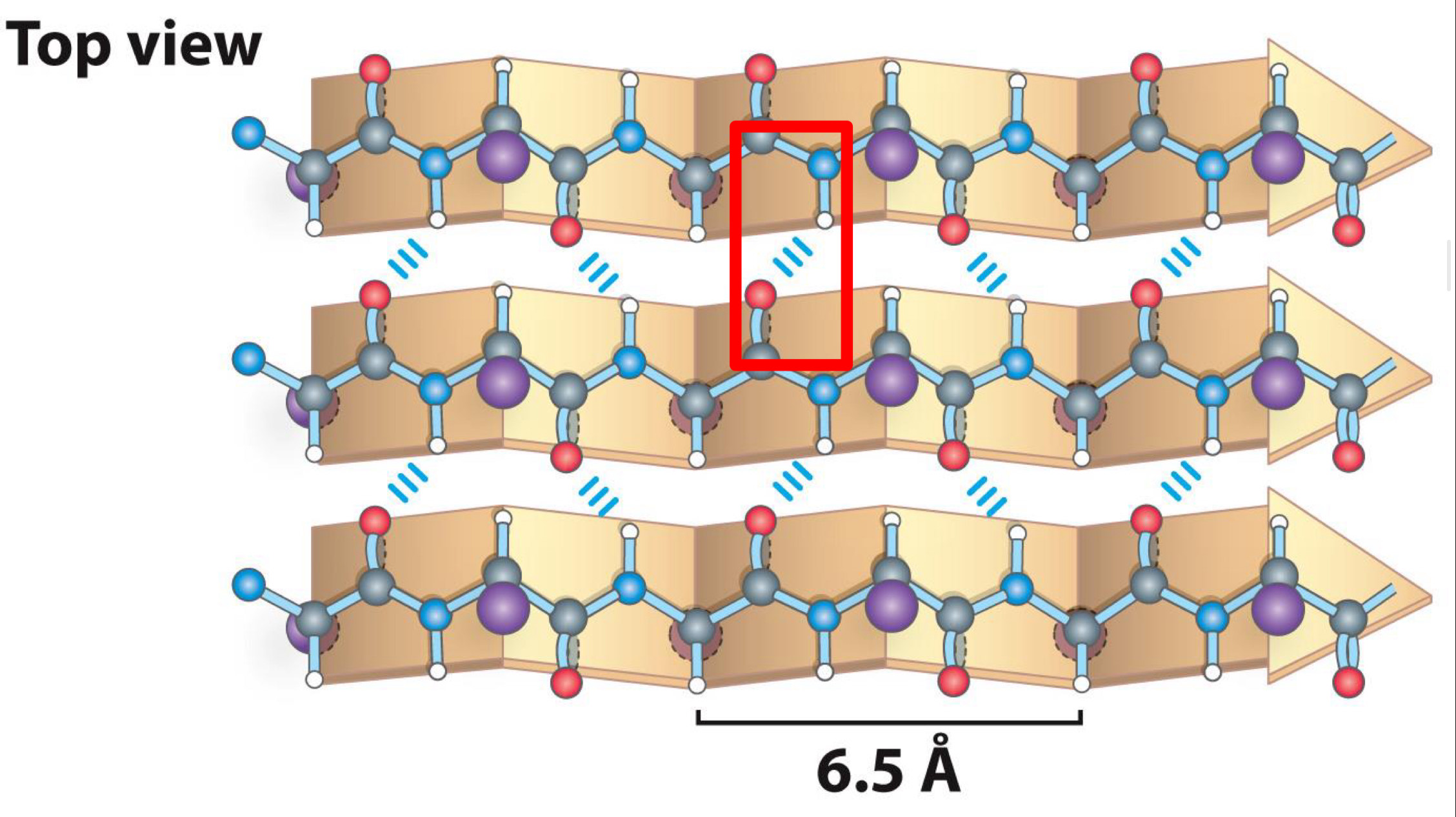
Parallel Beta sheet Top view
6.5 A
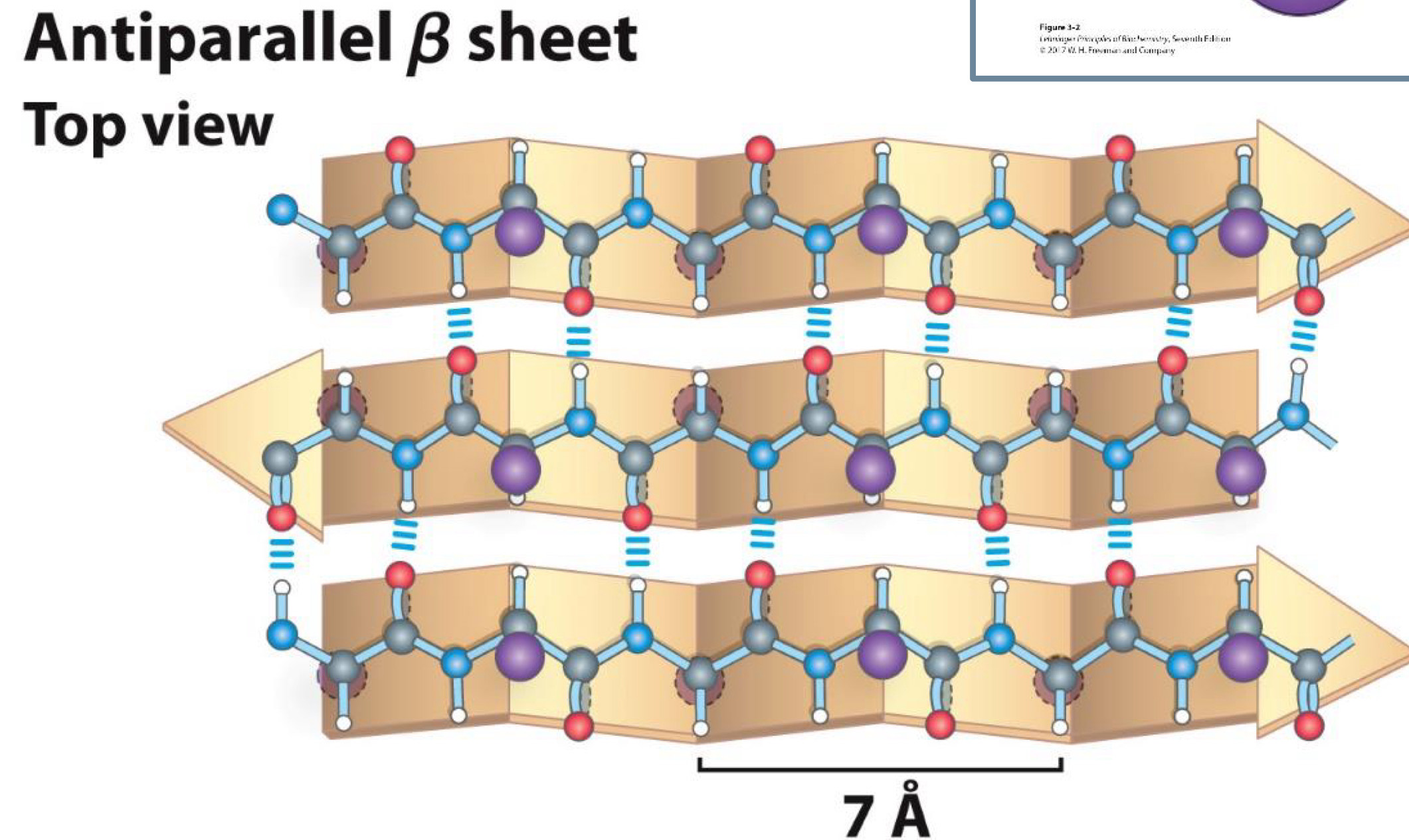
Antiparallel Beta Sheet Top view
7 A
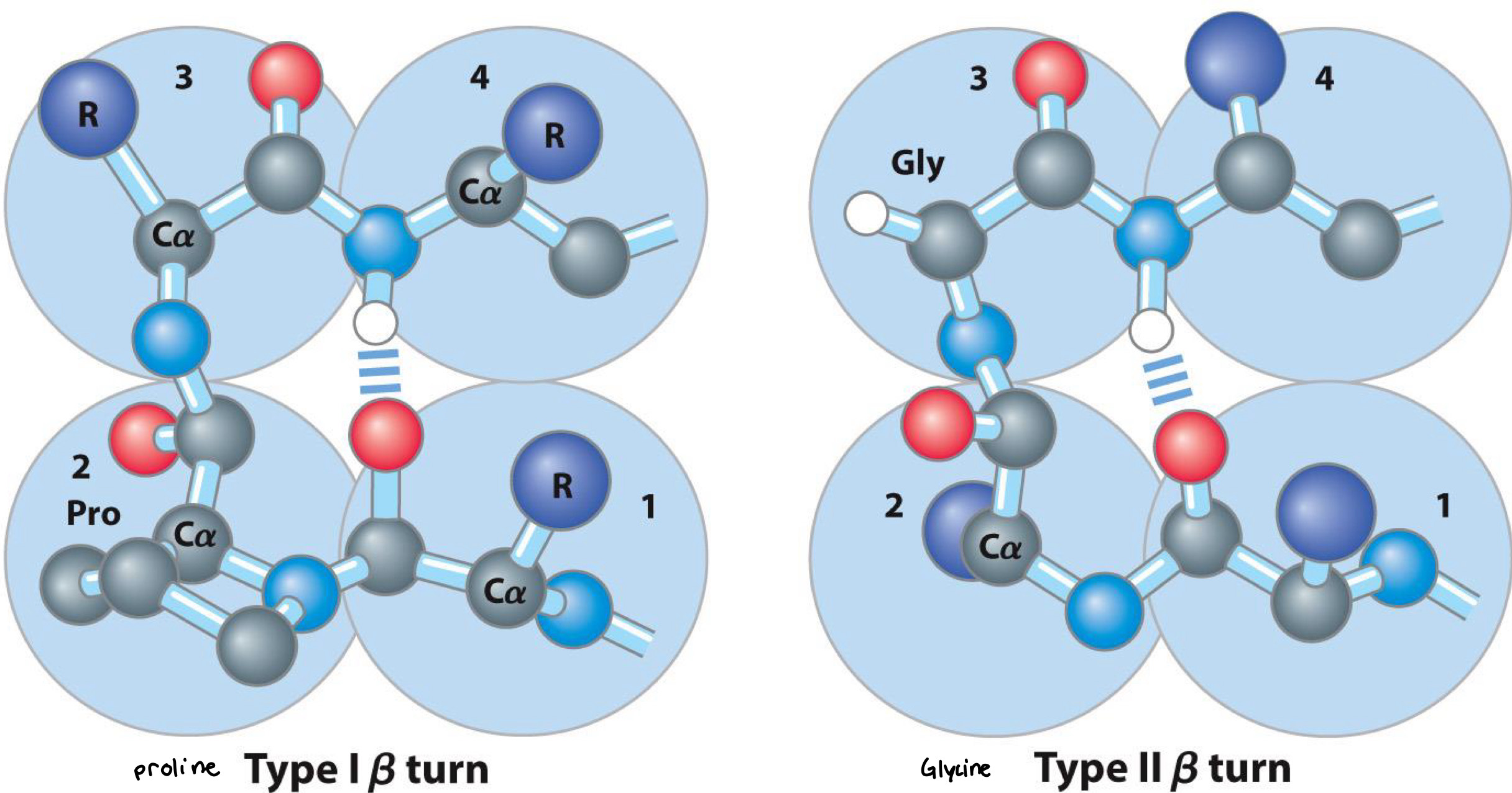
Beta Turn
Part of Beta sheets
Occur whenever strands change the direction
Proline in position 2 or glycine in position 3 are common
Tertiary structures involve folded peptide chains
Involves entire peptide chain, not just short segments
Made up of alpha -helices, Beta sheets, or other
elements of secondary structure
Tertiary structures: 2 types
Fibrous
- Long strands or sheets
- Not soluble (in either water or lipid)
Globular
- Spherical shape
- Soluble (in either water or lipid)
Fibrous- tertiary structure
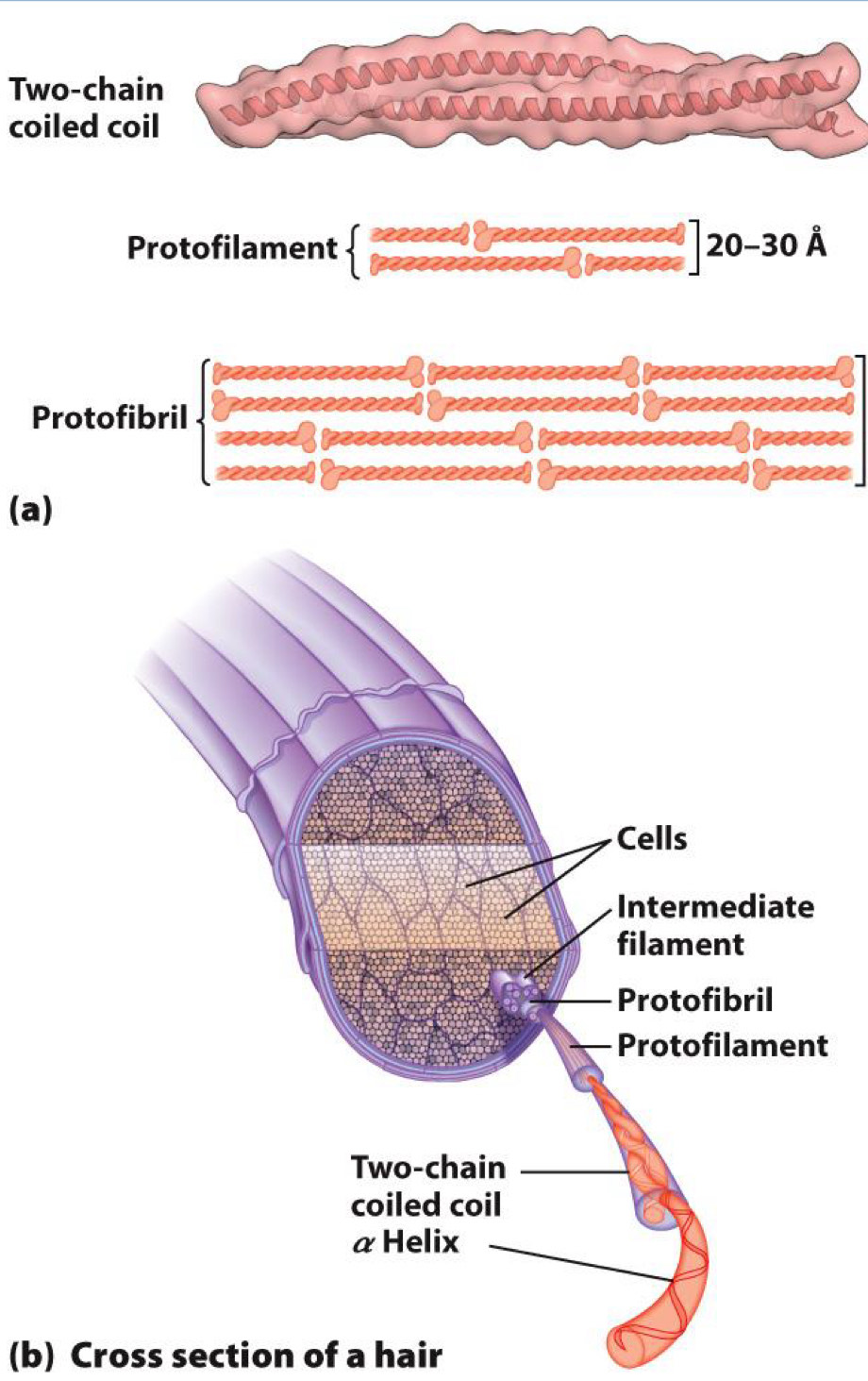
alpha-Keratin
- Found in hair
- Formed from coils of alpha-helix
Silk
- Formed by spiders and moths
- Made up of antiparallel beta sheets
- Small R groups (Ala and Gly) allow the close packing of sheets
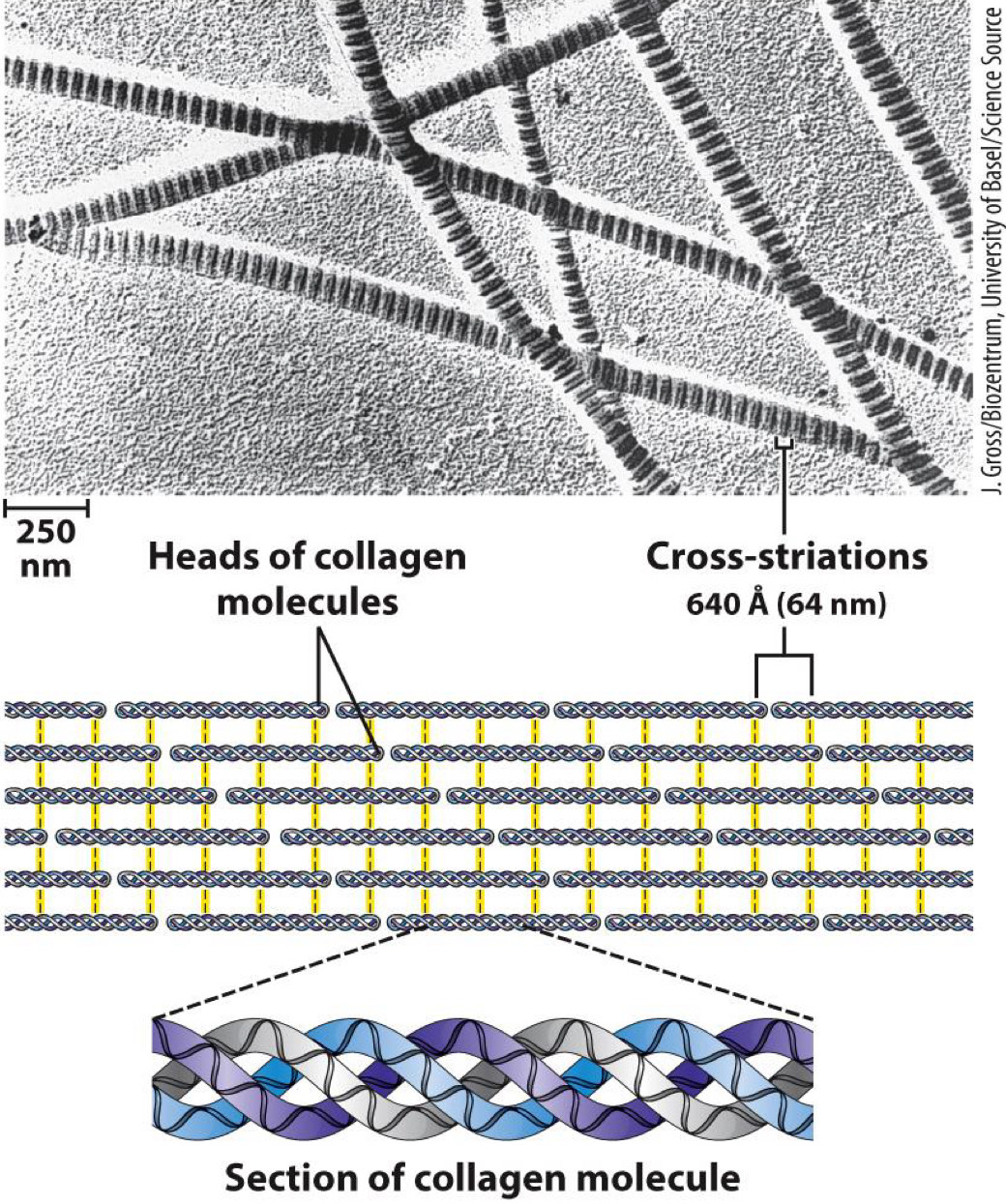
Collagen
- Found in connective tissues
- Made up of collagen helices:
Different from -helix(e.g., 3 amino acid residues per turn)
Helices crosslinked together for strength
Crosslinks are covalent bonds between lysine, hydroxylysine, or histidine residues
α Helix, cross-linked by disulfide bonds
Tough, insoluble protective structures of varying hardness and flexibility
ex. α-Keratin of hair, feathers, nails
β Conformation
Soft, flexible filaments
ex. Silk fibroin
Collagen triple helix
High tensile strength, without stretch
ex. Collagen of tendons, bone matrix
Globular-tertiary structure
not as ordered as fibrous
ex. myoglobin
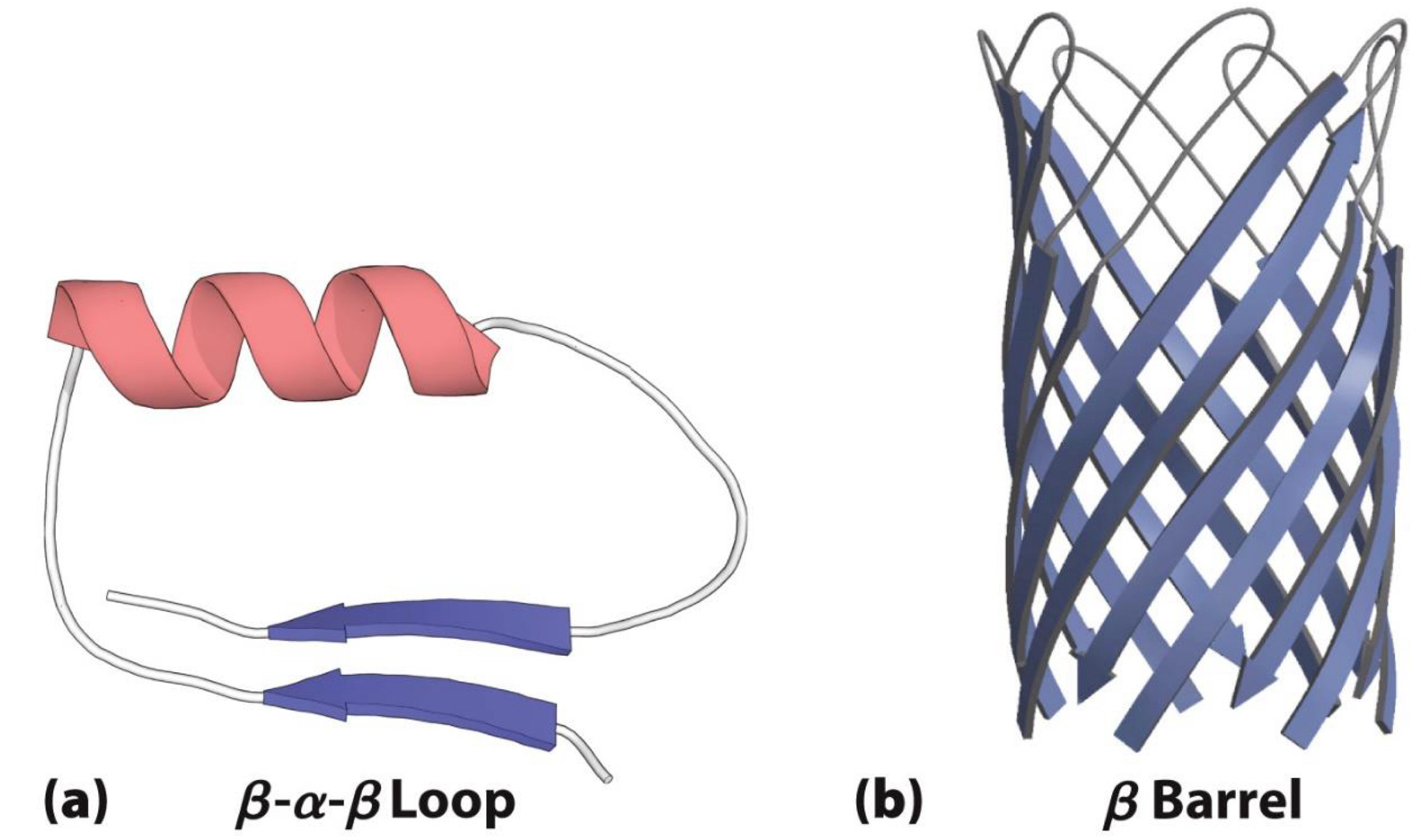
Often made up of motifs
- Contain two or more elements of secondary structure
Beta-Alpha-Beta loop and Beta barrel
Often have multiple domains
- Single entities that are stable when separated from each other
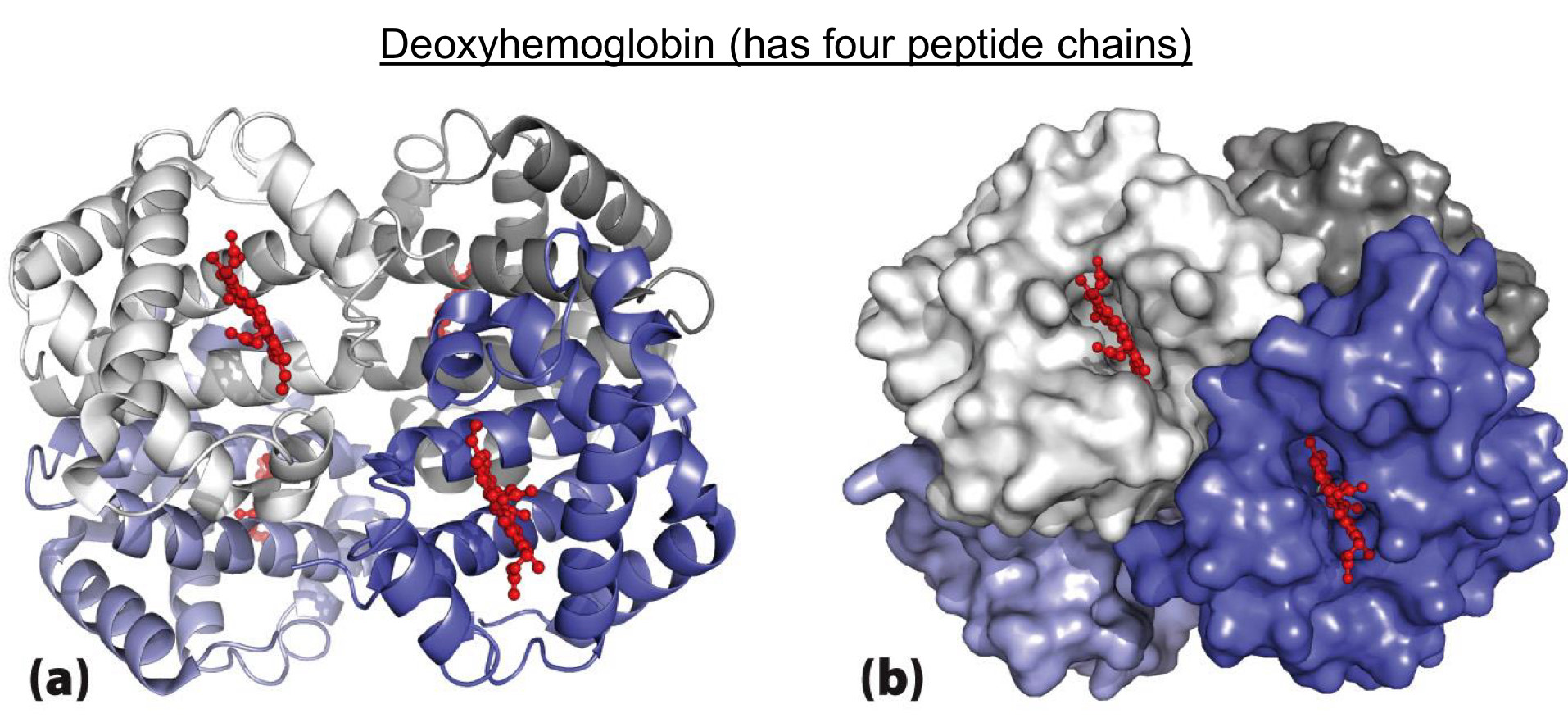
Quaternary structure is the most complex
Formed by multiple peptides (elements of tertiary structure)
ex. Deoxyhemoglobin (has four peptide chains)
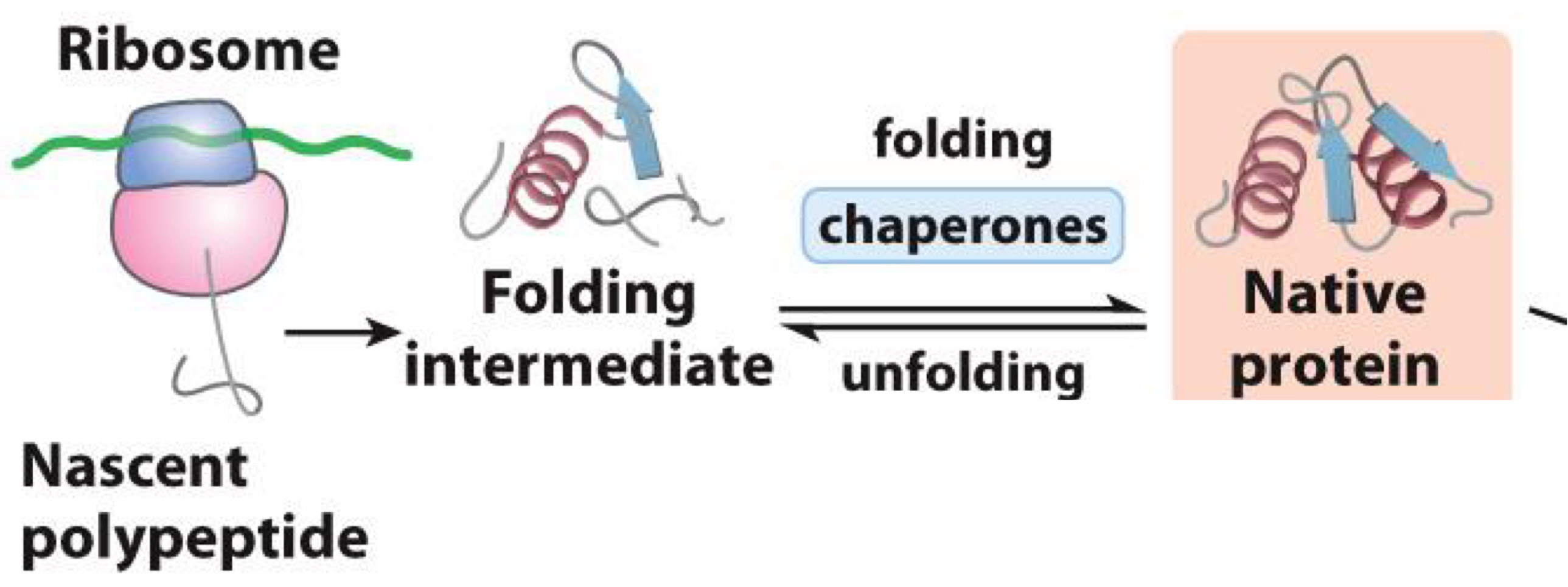
Protien’s folding
Folding is the process by which peptide chains develop tertiary or quaternary structure
- Done by chaperones (other proteins)
- Makes proteins functional
ex. Chaperones in E. coli: Serves as protective bubble for folding
- Protein enters and sealed inside
- Protein folds and is then released
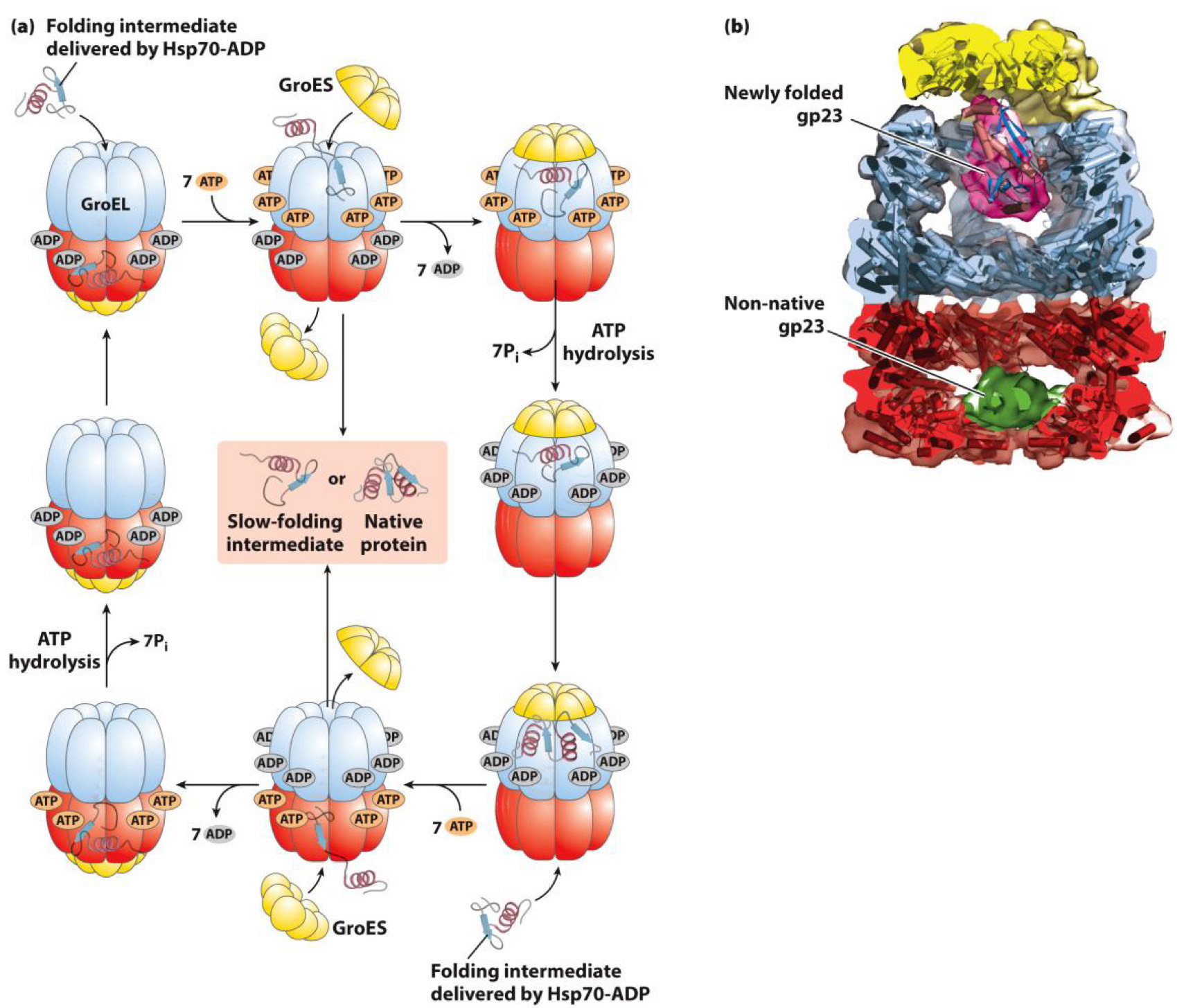
How are proteins unfolded?
Proteins can be unfolded (denatured) by
- Heat
- pH extremes
- Organic solvents
-Chaotropic agents
◼ Disrupt hydrogen bonding
◼ Examples: urea and guanidinium hydrochloride
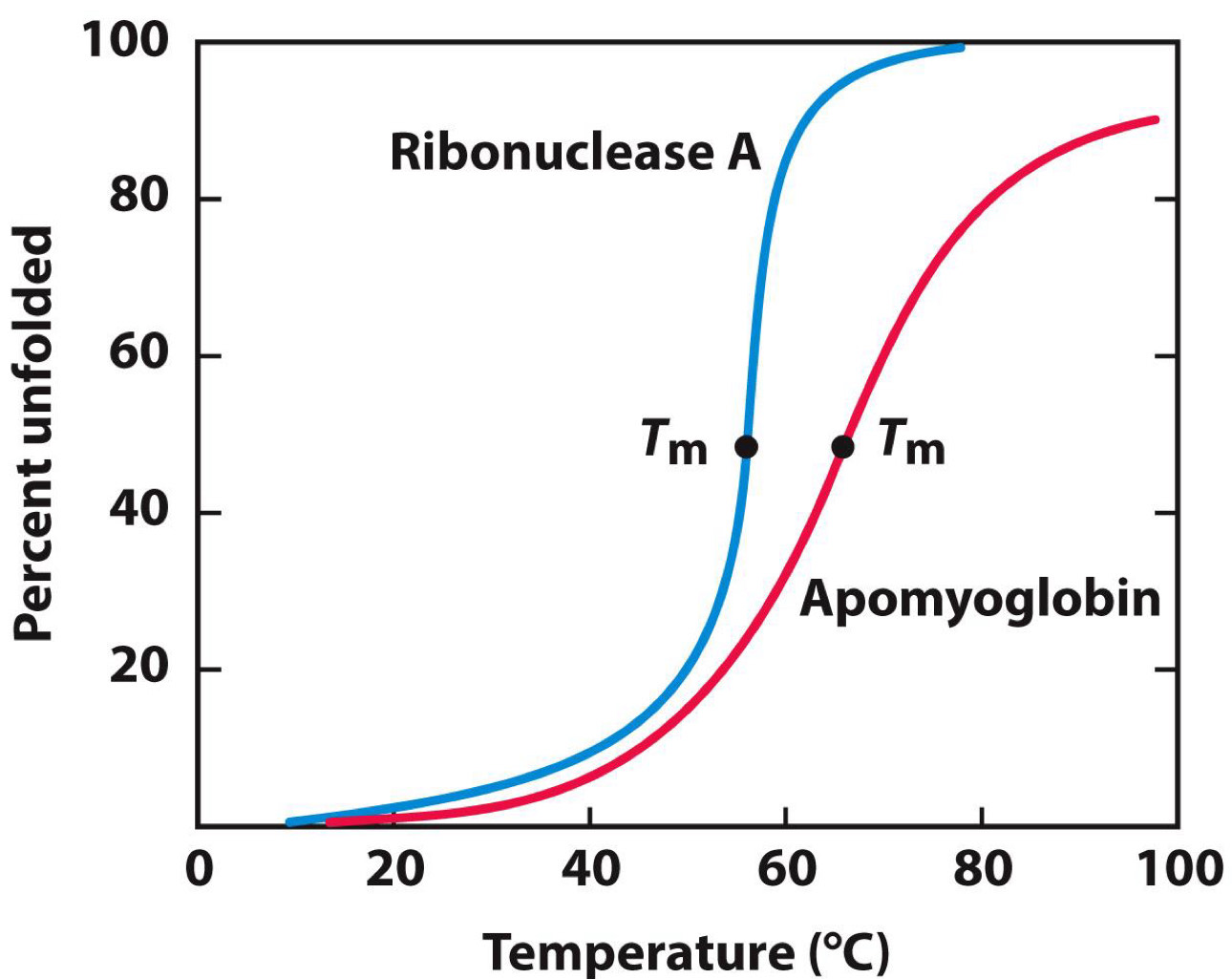
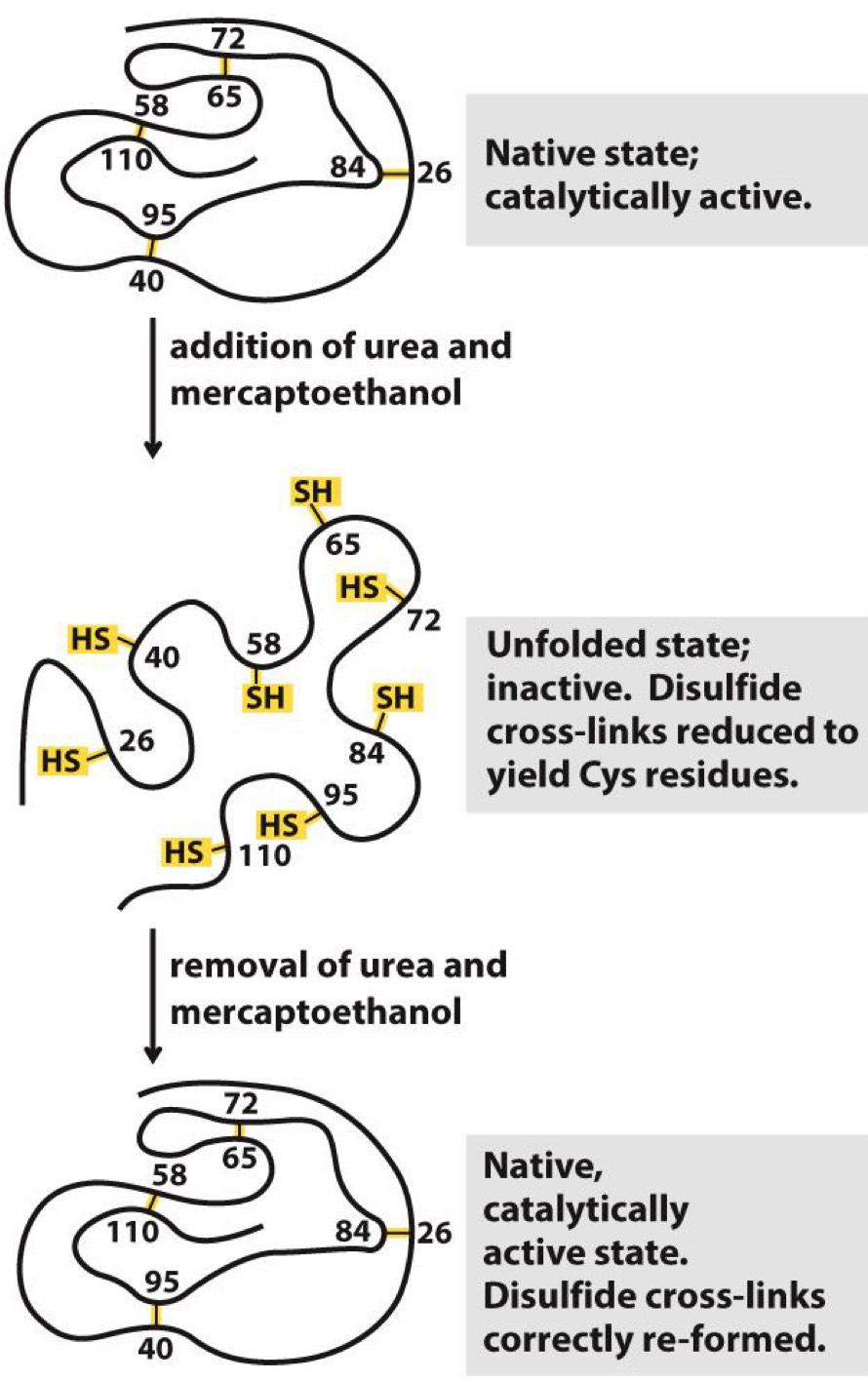
Ribonuclease refolding experiment
Tm= temperature at which half of protein is unfolded
Adding urea and 2-mercaptoethanol causes ribonuclease to unfold
Removing them allows it to spontaneously refold
Function restored when refolded
Misfolding is basis of many diseases
Creutzfeldt-Jakob disease
- One type of protein in brain (human prion protein) becomes misfolded
- One misfolded protein causes more to unfold, leading a domino effect
- Spongiform encephalopathy of brain and death result
Other diseases
- Bovine spongiform encephalopathy in cows
(mad cow disease)
- Scapie in sheep
- Chronic wasting disease in deer and elk
Methods to characterize proteins in the lab
SDS-PAGE-measure the size of proteins
Isoelectric focusing/2D PAGE→ can be done with SDS-PAGE
Chromatography/purification→ purifying large amounts of proteins
Mass spectrometry→ sequence of a proteins
X-ray diffraction→ crystallography reveals the structure
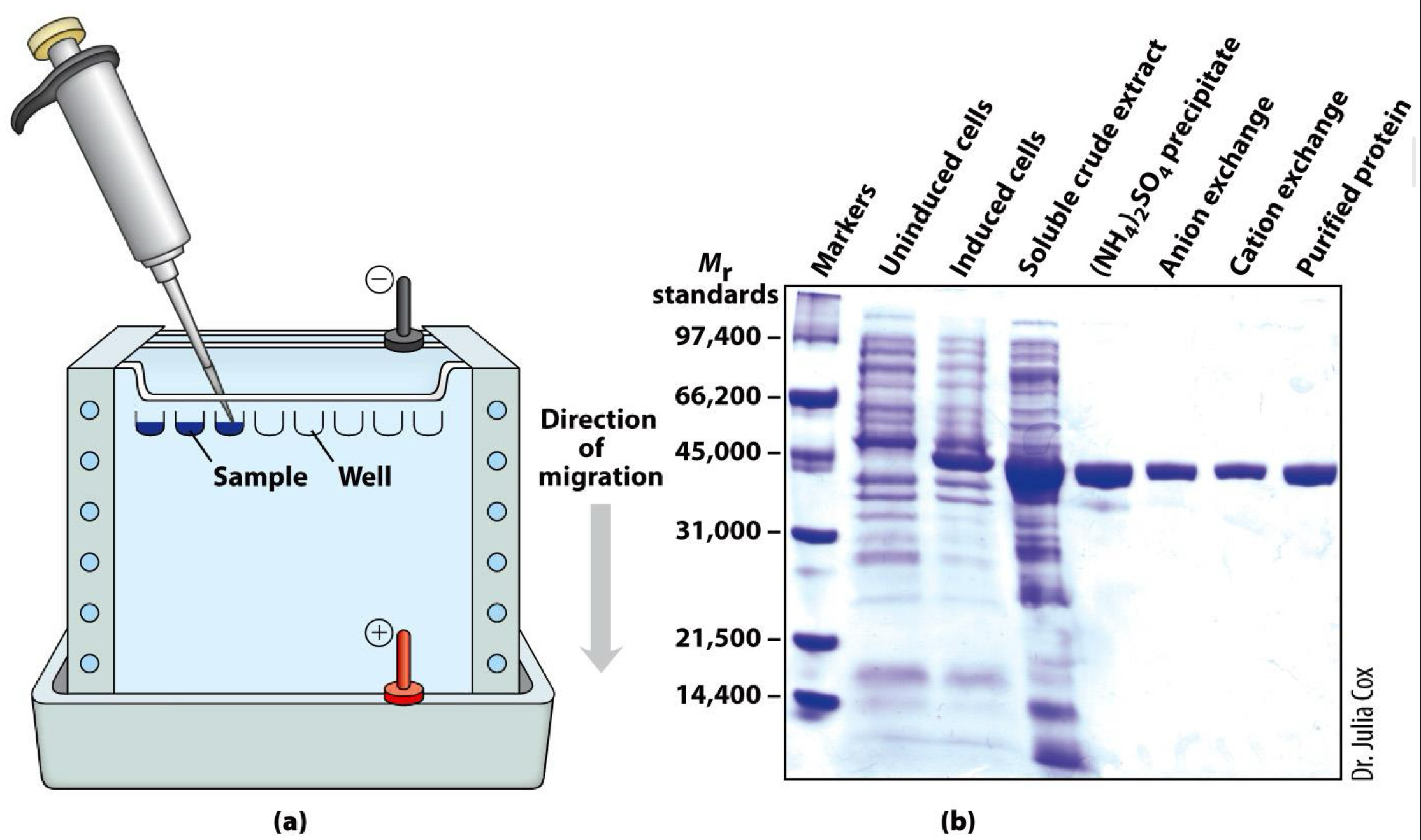
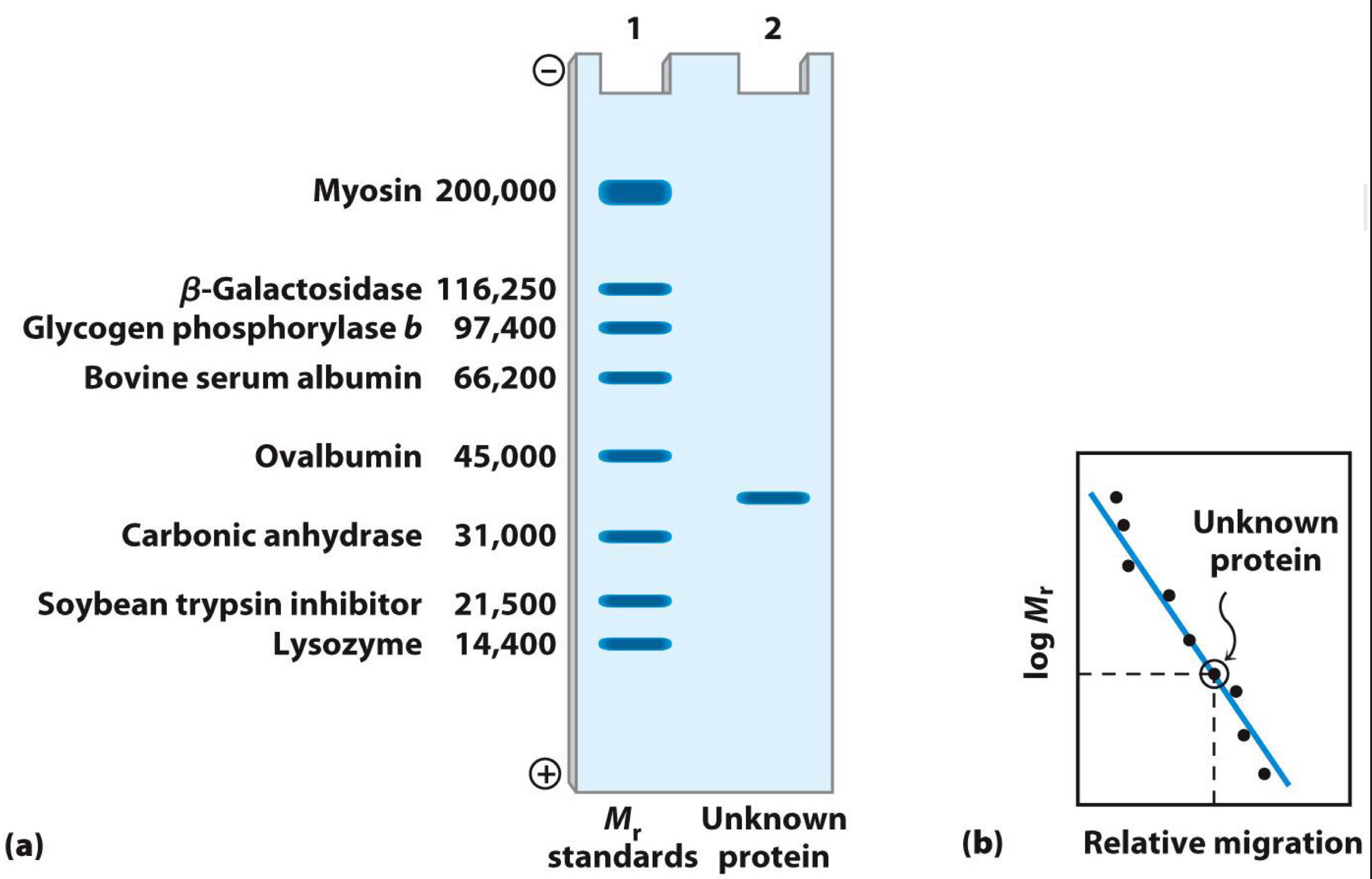
Gel electrophoresis separates proteins by size
Proteins loaded into wells of gel
Electric field applied
Proteins move (migrate) at speed according to size: Small proteins move through gel more easily
Gel is commonly polyacrylamide (or PAGE)
Proteins usually denatured by sodium dodecyl sulfate(SDS)-Technique called SDS-PAGE
Size (molecular weight) measured in comparison to standards
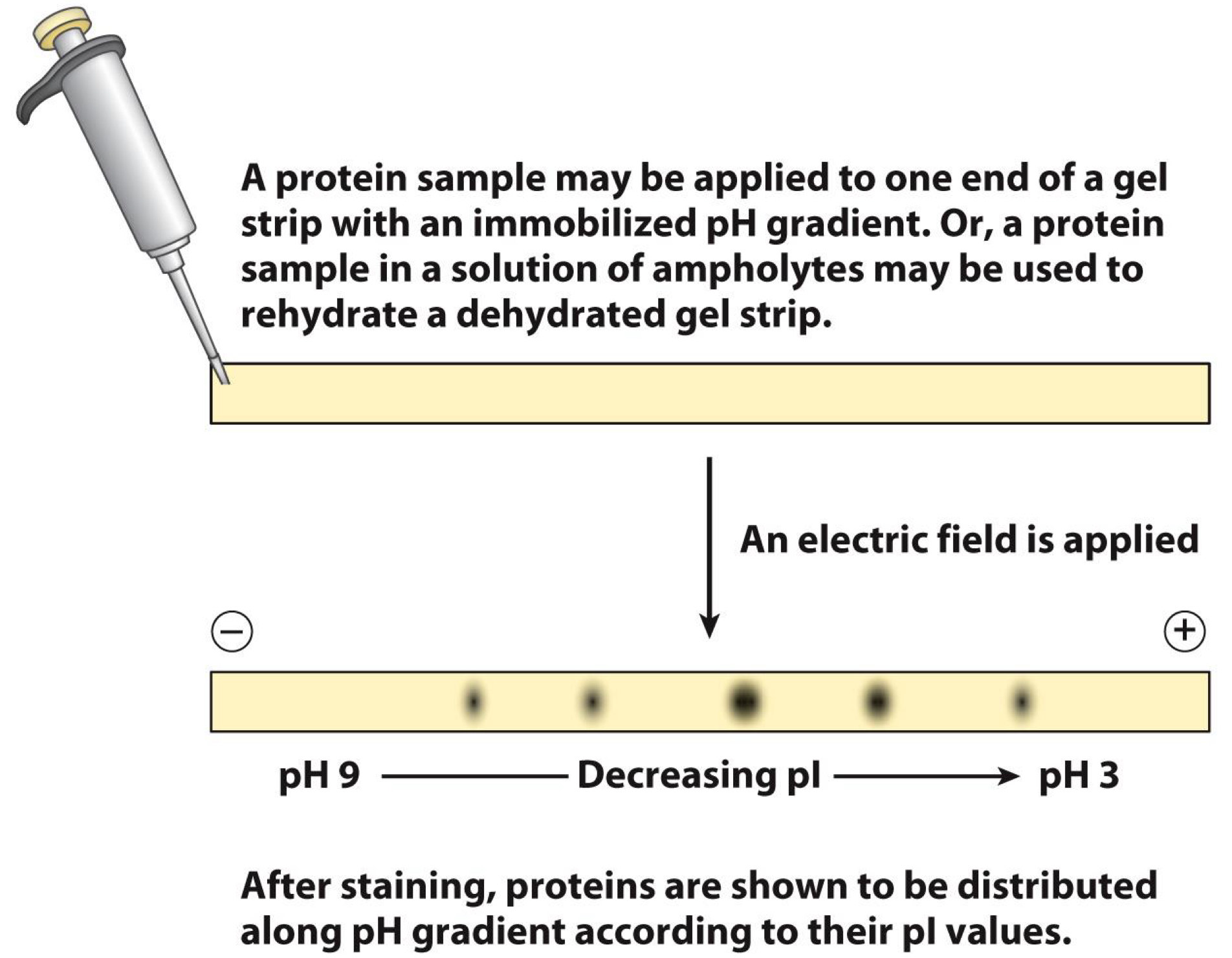
Isoelectric focusing separates proteins by charge
Proteins applied to gel strip
- Strip has pH gradient
Electric field applied
Proteins move until they reach pH= isoelectric point (pI)
- Isoelectric point is point at which proteins have no net charge
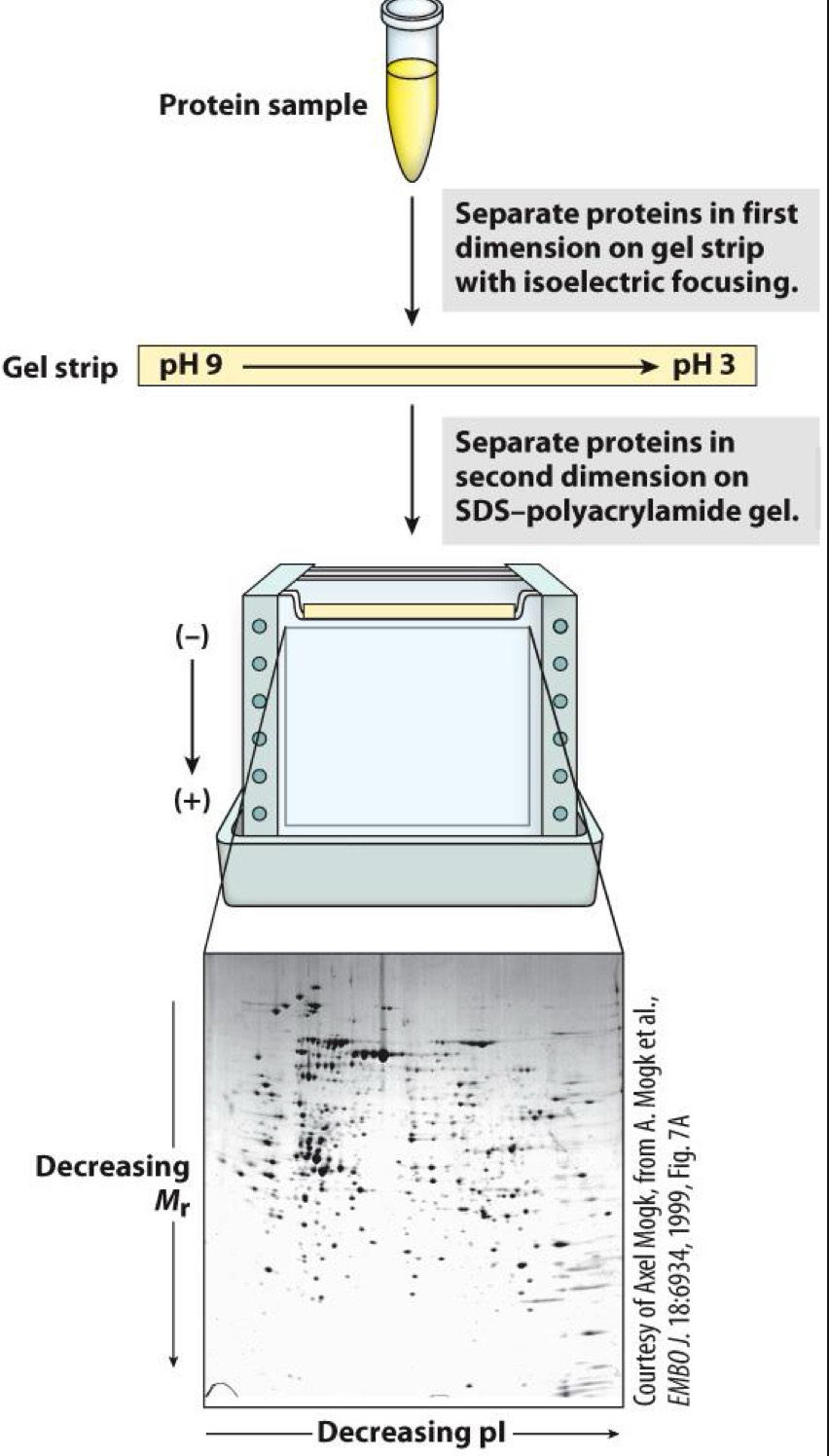
2D gel electrophoresis separates by size and charge
Isoelectric focusing done first (dimension 1)
SDS-PAGE done next (dimension 2)
Separation of complex mixtures possible
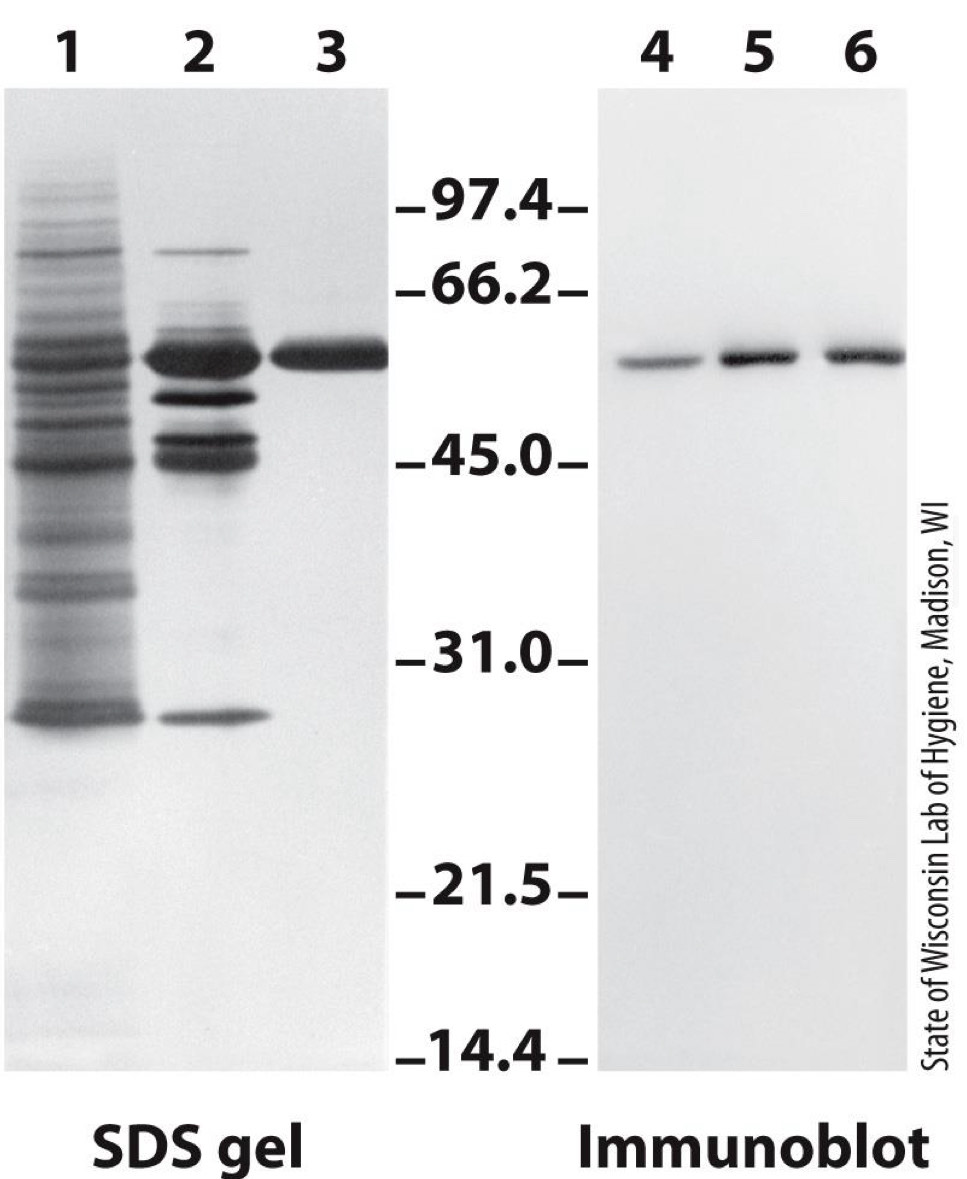
Western blot identifies proteins with antibodies
SDS-PAGE gel is run
Immunoblot is performed
- Proteins are transferred to a membrane
- Proteins in membrane are “probed” with antibody against protein of interest
- Dark color is formed, revealing protein(Series of reaction used to form color)
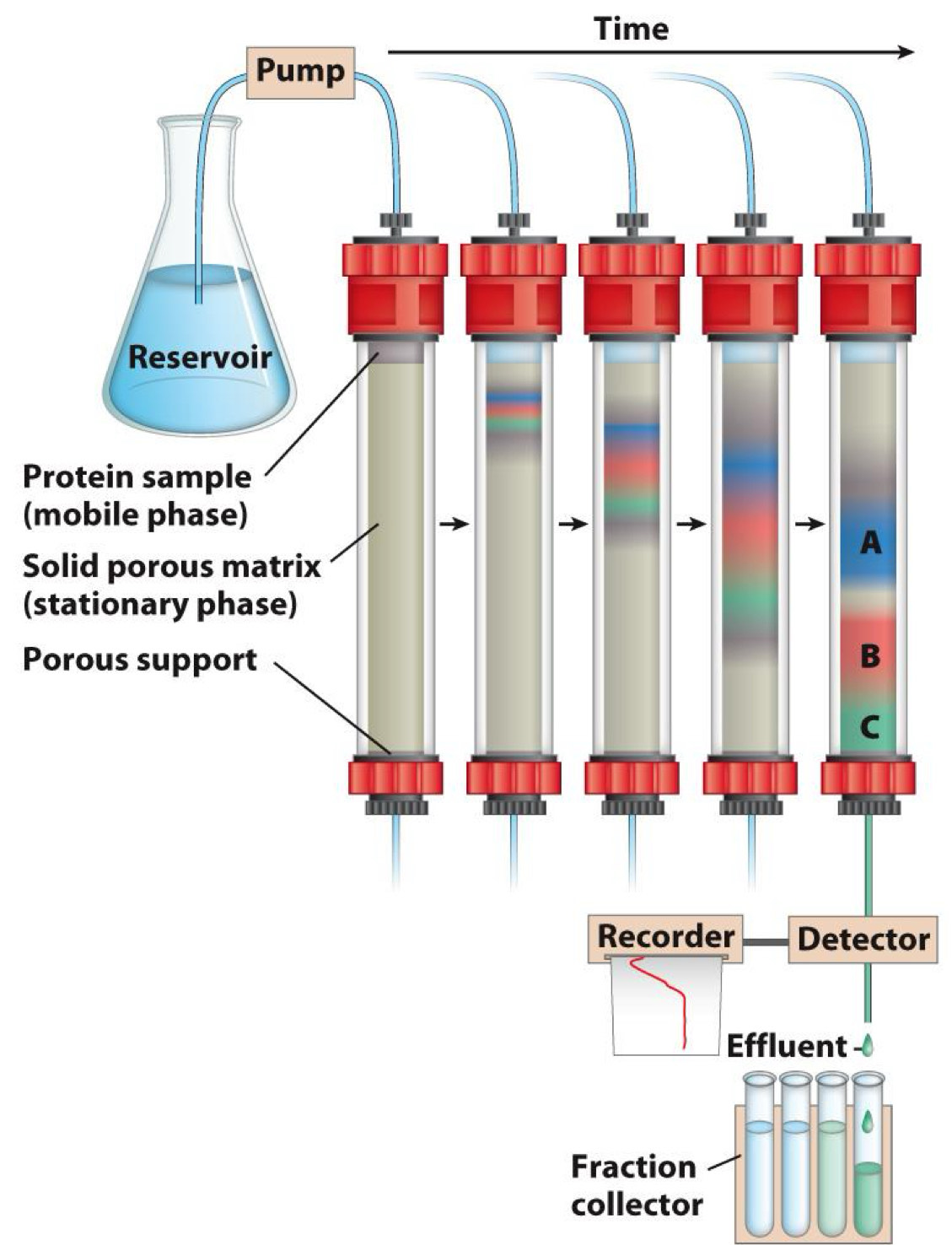
Chromatography is used to purify proteins
Useful for purifying (separating) large amounts of protein
- Protein loaded onto solid phase (porous matrix)
- Eluted from matrix using liquid phase
- Proteins with low affinity for solid phase migrate faster and come off first
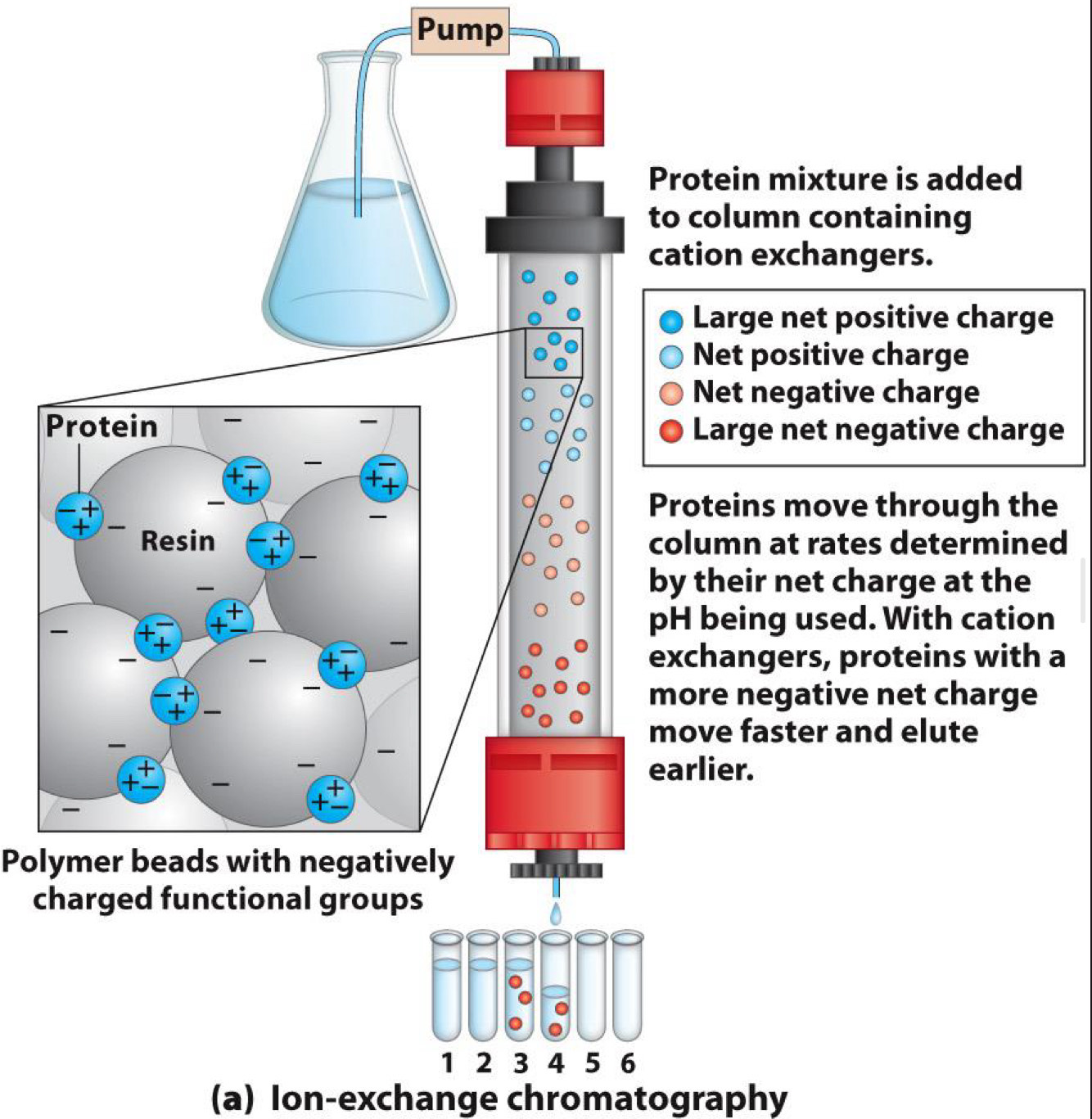
Chromatography-Ion exchange
Ion exchange:
- Separates by charge
- Solid phase has affinity for positive ions (cation exchange)
- Or solid phase has affinity for negative ions (anion exchange)
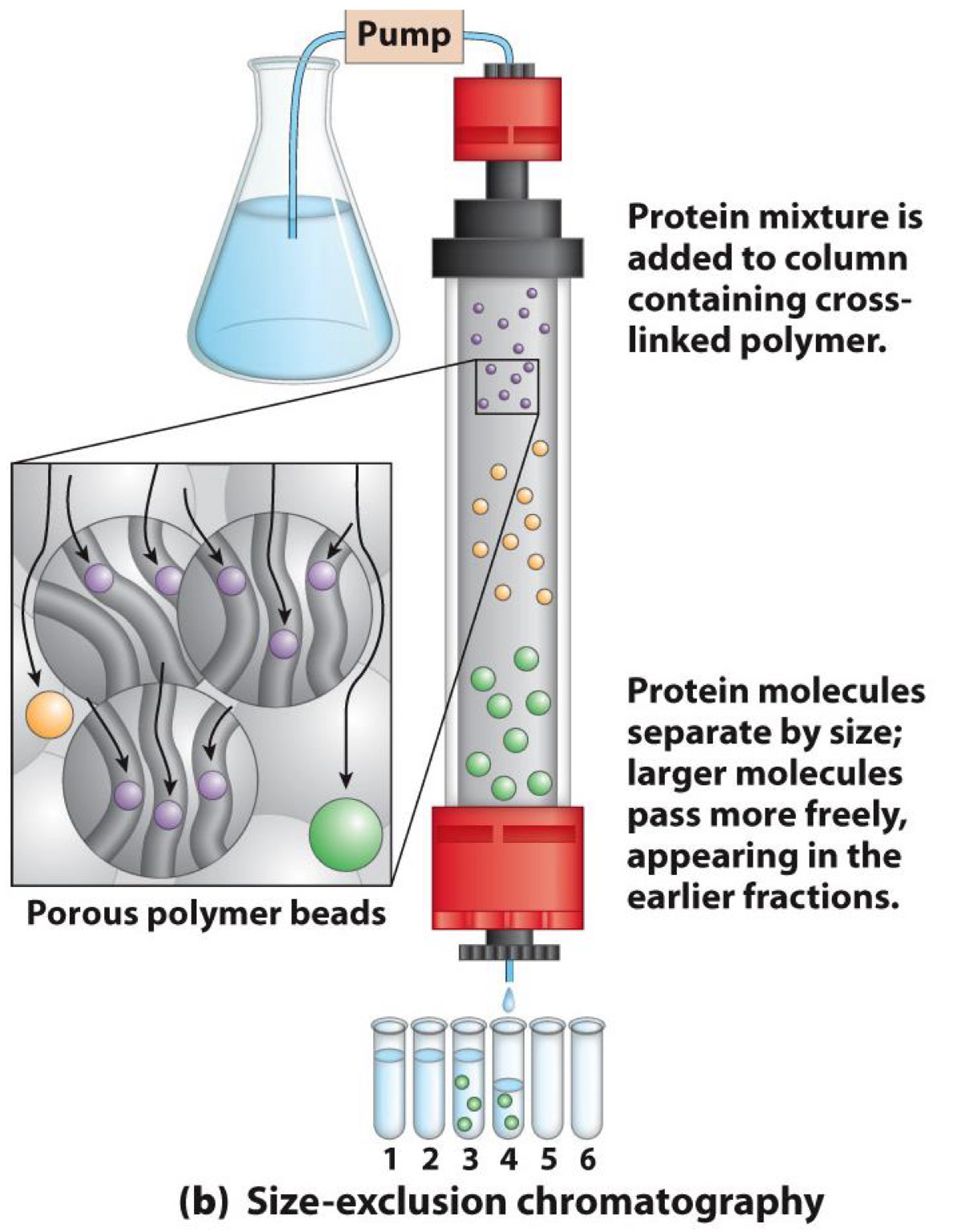
Chromatography-Size exclusion
Separates by size
Solid phase has affinity for small molecules
◼Small molecules trapped in pores of phase
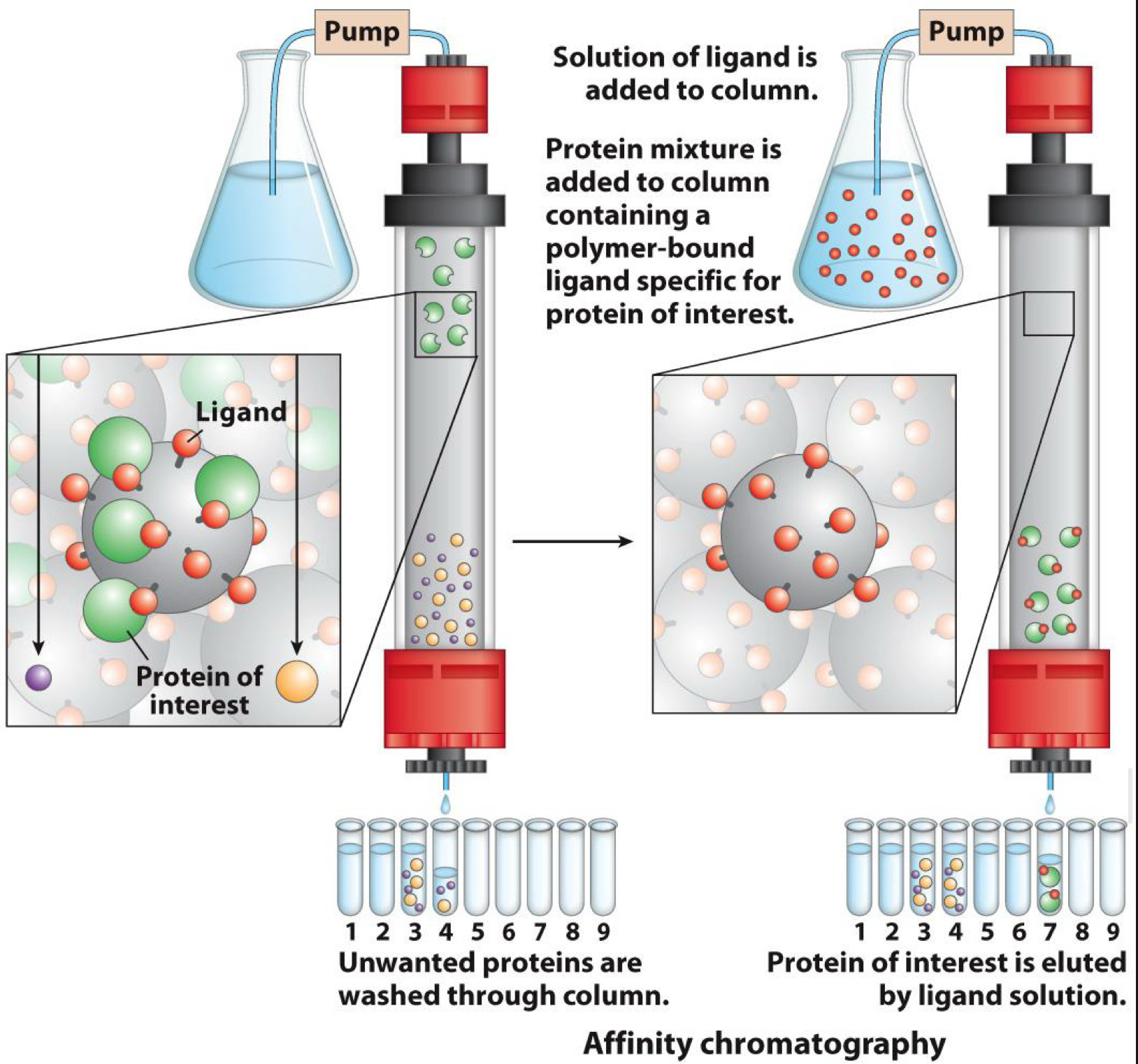
Chromatography-Affinity chromatography
Separates by binding ability
Solid phase has ligand
Ligand binds to (has affinity for) a target on protein
One target is histidine tag added by genetic modification
Several methods are combined to purify proteins
1. Crude cellular extract
2. Precipitation with ammonium sulfate
3. Ion-exchange chromatography
4. Size-exclusion chromatography
5. Affinity chromatography
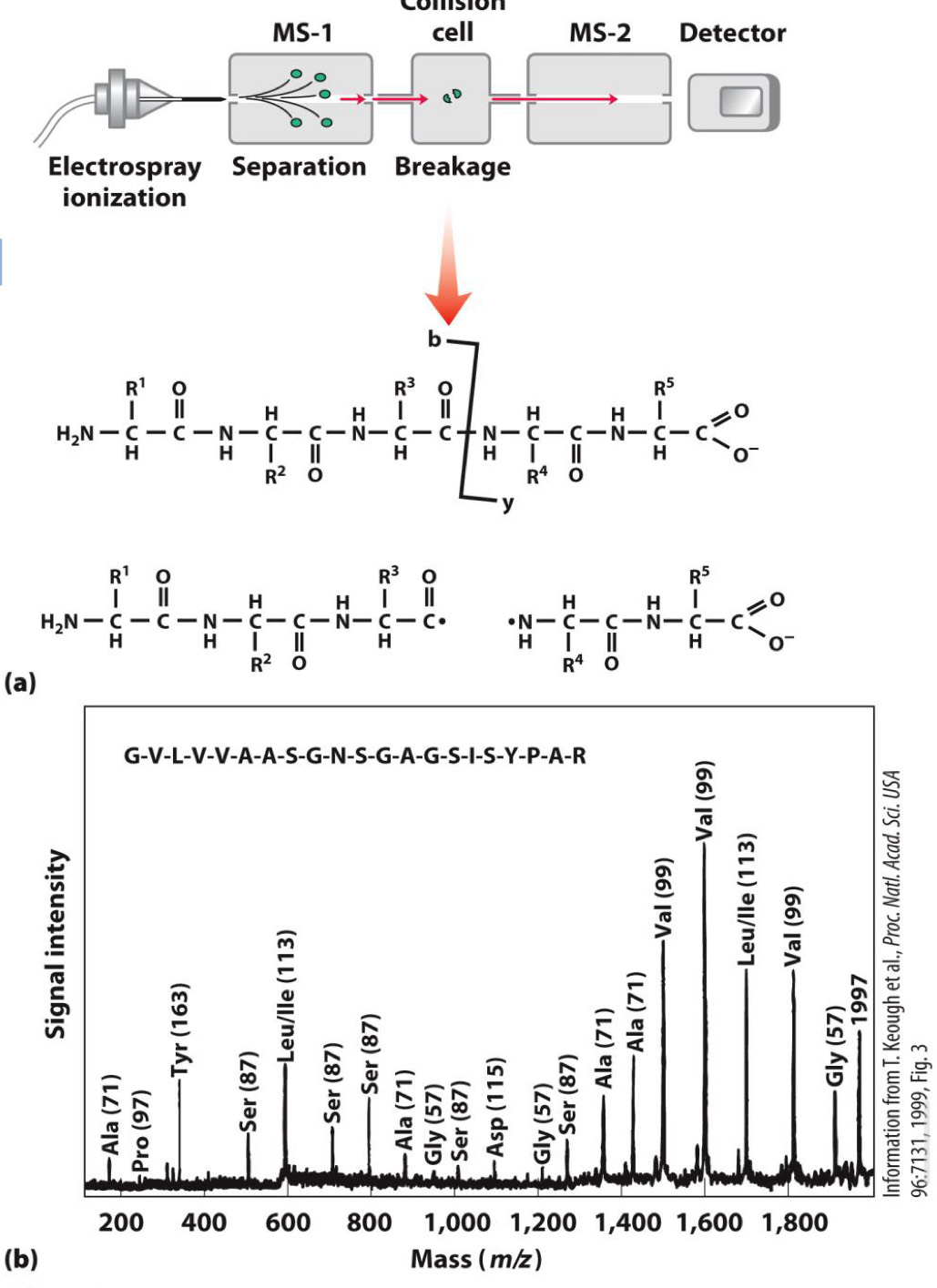
Mass spectrometry reveals protein sequence
Peptide broken into fragments
Mass of fragments measured
Mass of different fragments reveals sequence
X-ray crystallography reveals 3D structure
Crystals of proteins are grown
Crystals put into X-ray beam
Pattern of X-ray diffraction reveals position of atoms (structure)
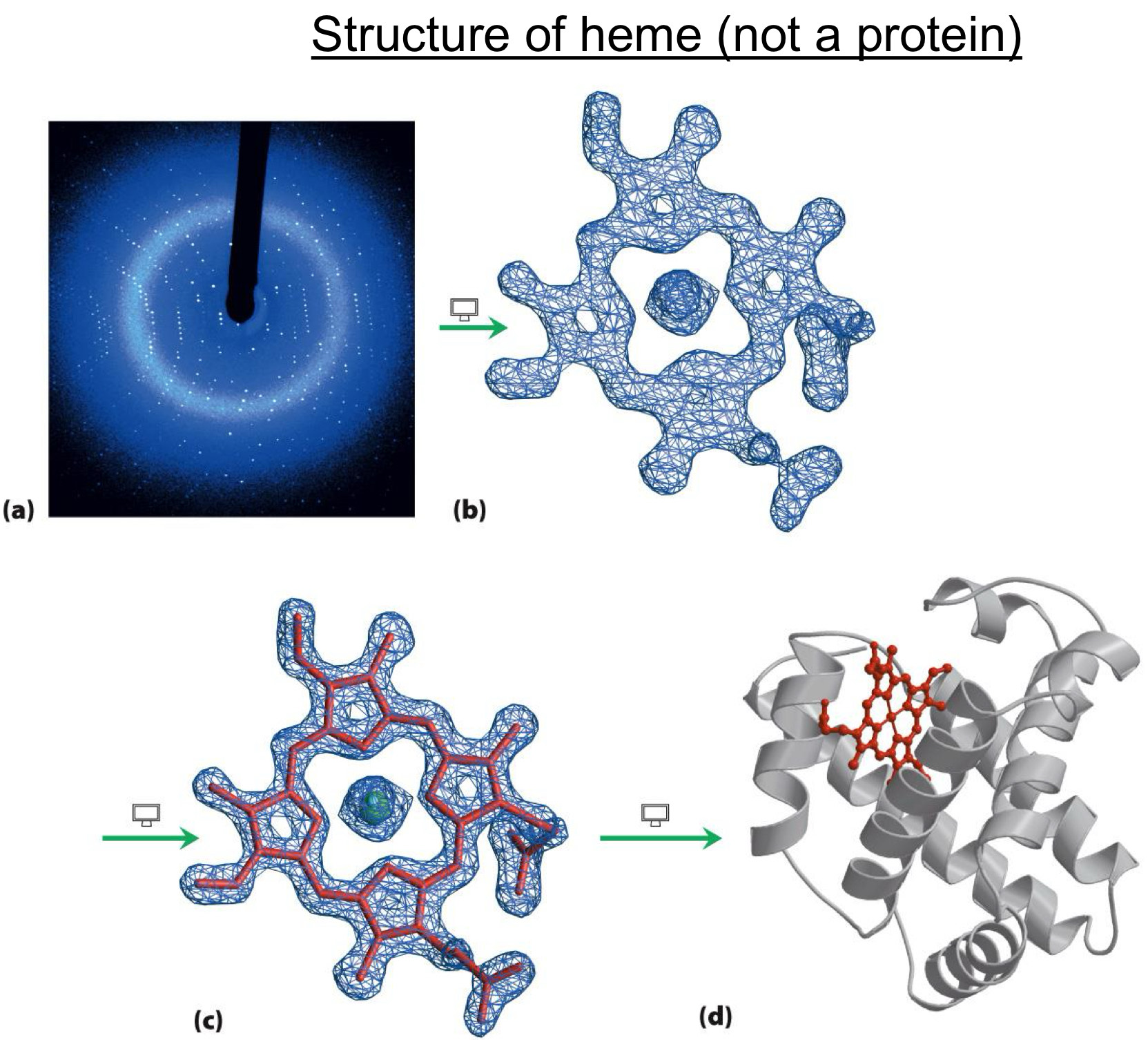
Protein functions
Structure (e.g., collagen)
Catalysis of chemical reactions (enzymes)
Transport of molecules (hemoglobin)
Defense against pathogens (antibodies)
Movement (actin, myosin)
Others
Enzymes bind to:
Antibodies bind to:
Collagen bind to:
all proteins bind to molecules
substrates
pathogens
collagen molecules
without binding, proteins cannot function
Proteins function by binding to ligands
he molecule bound is a ligand
Include ion, small molecule, macromolecules
Best studied example of ligand binding is oxygen binding to
hemoglobin and myoglobin
Hemoglobin binds oxygen as a ligand
Binds to and transports oxygen in blood
Four subunits
Oxygen is bound by heme
Iron (Fe2+) of heme is central
Fe2+ is site of O2 binding
Fe2+ also bound to myglobin (via His residue)
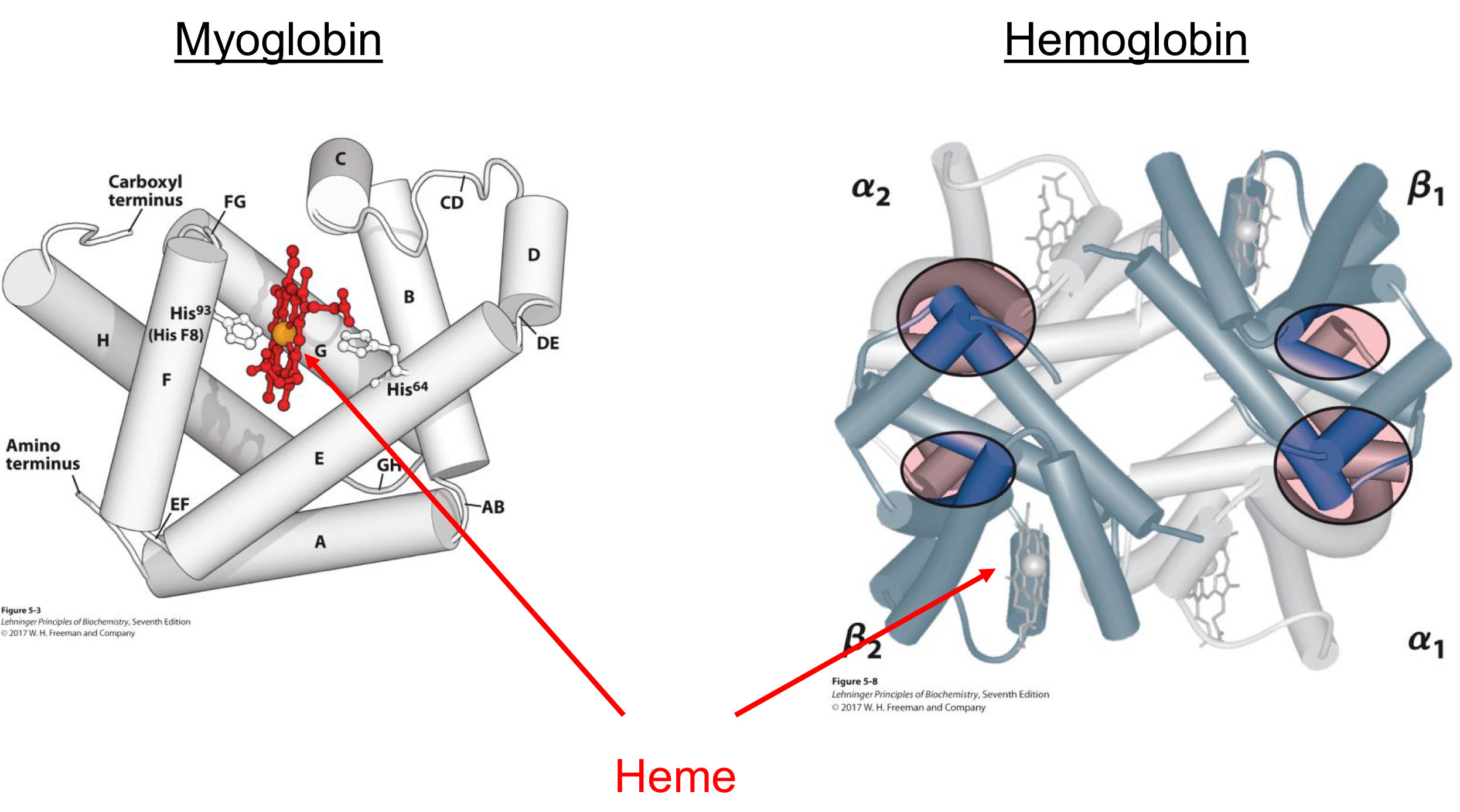
Myoglobin binds oxygen as a ligand
Stores oxygen in muscle cells
One subunit
Oxygen is bound by heme
Iron (Fe2+) of heme is central
Fe2+ is site of O2 binding
Fe2+ also bound to myglobin (via His residue)
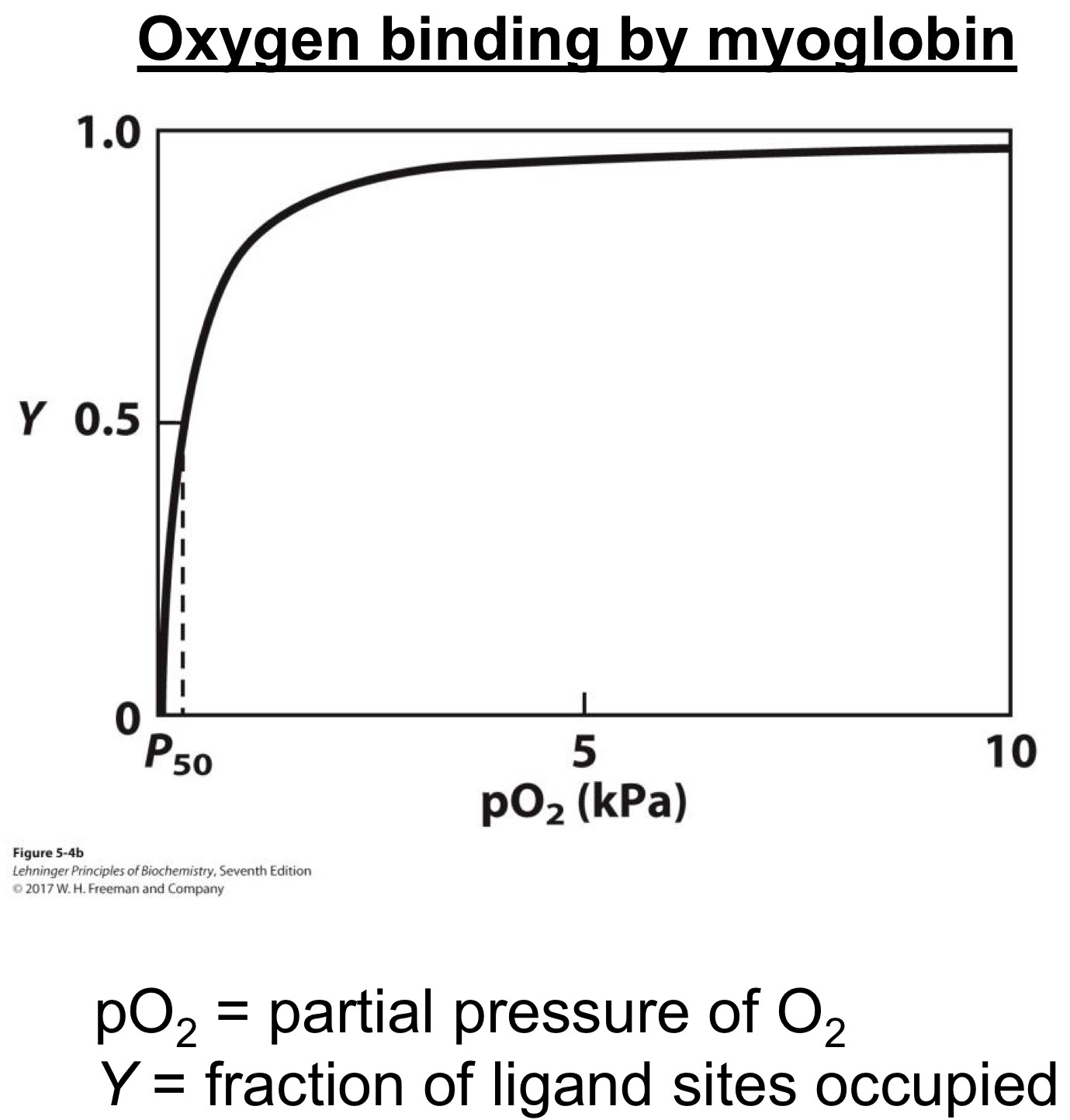
Oxygen binding by myoglobin
The amount of oxygen bound increases with oxygen concentration
In myoglobin, relationship is simple
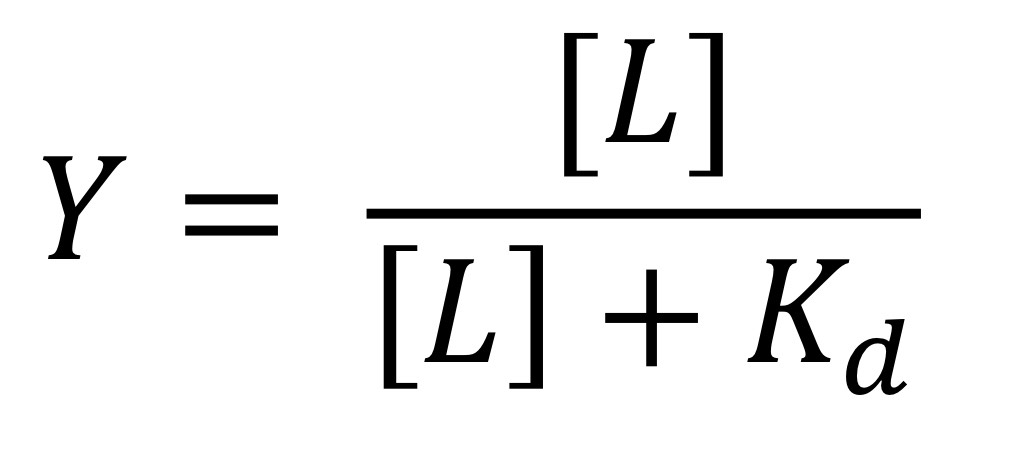
Binding Relationship between oxygen/ligands
Y = fraction of ligand sites occupied
[L] = concentration of ligand (M)
Kd = dissociation constant (M)
Dissociation constant (Kd)
Concentration of ligand where Y =0.5
The higher the value, the lower the binding (affinity)
![<p><span>Y = fraction of ligand sites occupied</span><br><span>[L] = concentration of ligand (M)</span><br><span>Kd = dissociation constant (M)</span></p><p><span>Dissociation constant (Kd)</span><br><span>Concentration of ligand where Y =0.5</span><br><span>The higher the value, the lower the binding (affinity)</span></p>](https://knowt-user-attachments.s3.amazonaws.com/a0c2533d-6108-4152-8770-48931cf7ded8.jpg)
Ligands and proteins have a wide range of dissociation constants
High affinity→ Biotin-Avidin
Antibody-antigen→10^-12-10^-8
Sequence-specific protein-DNA→ 10^-11-10^-7
Typical receptor-ligand interactions→10^-10-10^-5
Low affinity→ Enzyme-Substrate→ 10^-7-10^-3
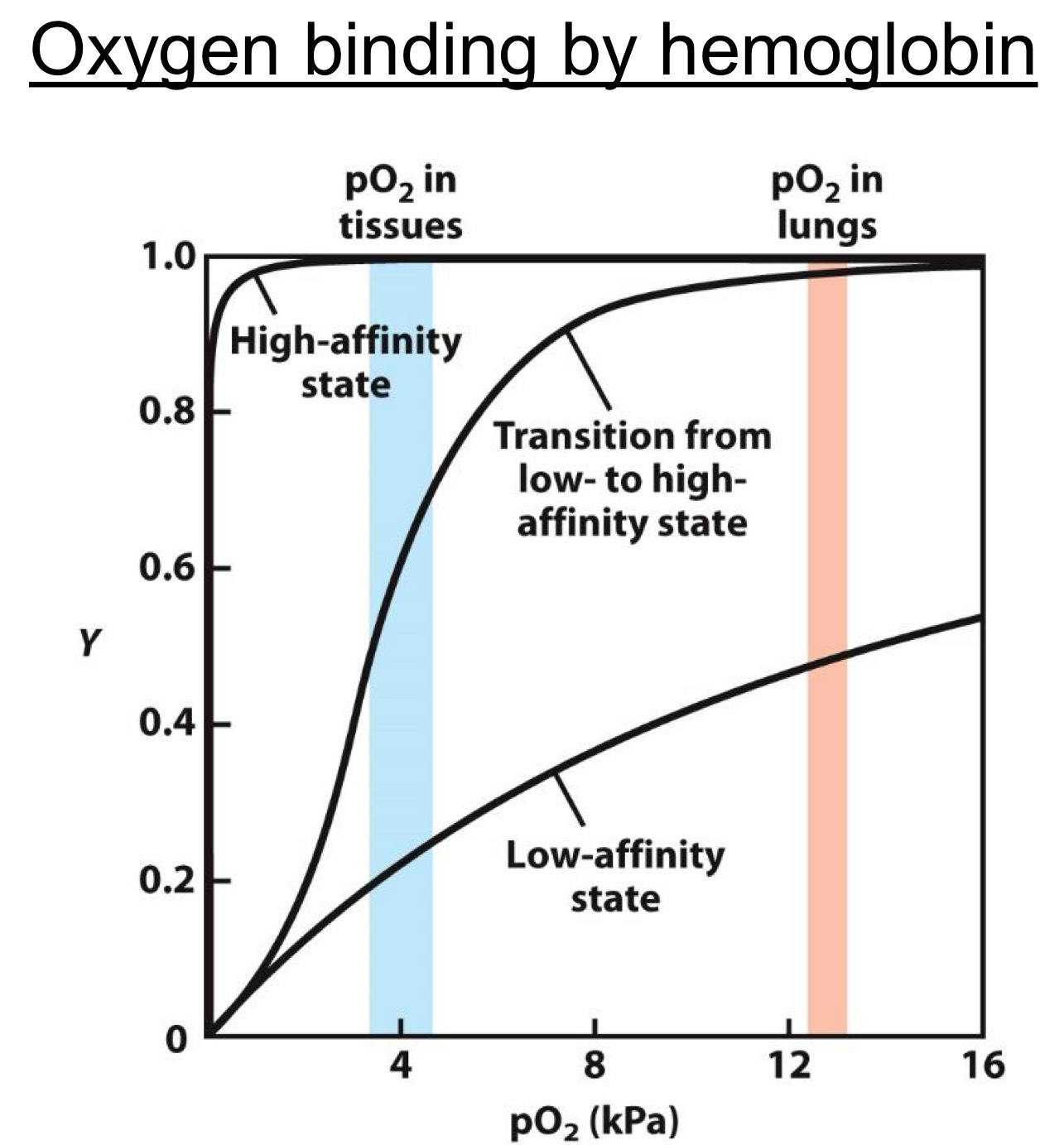
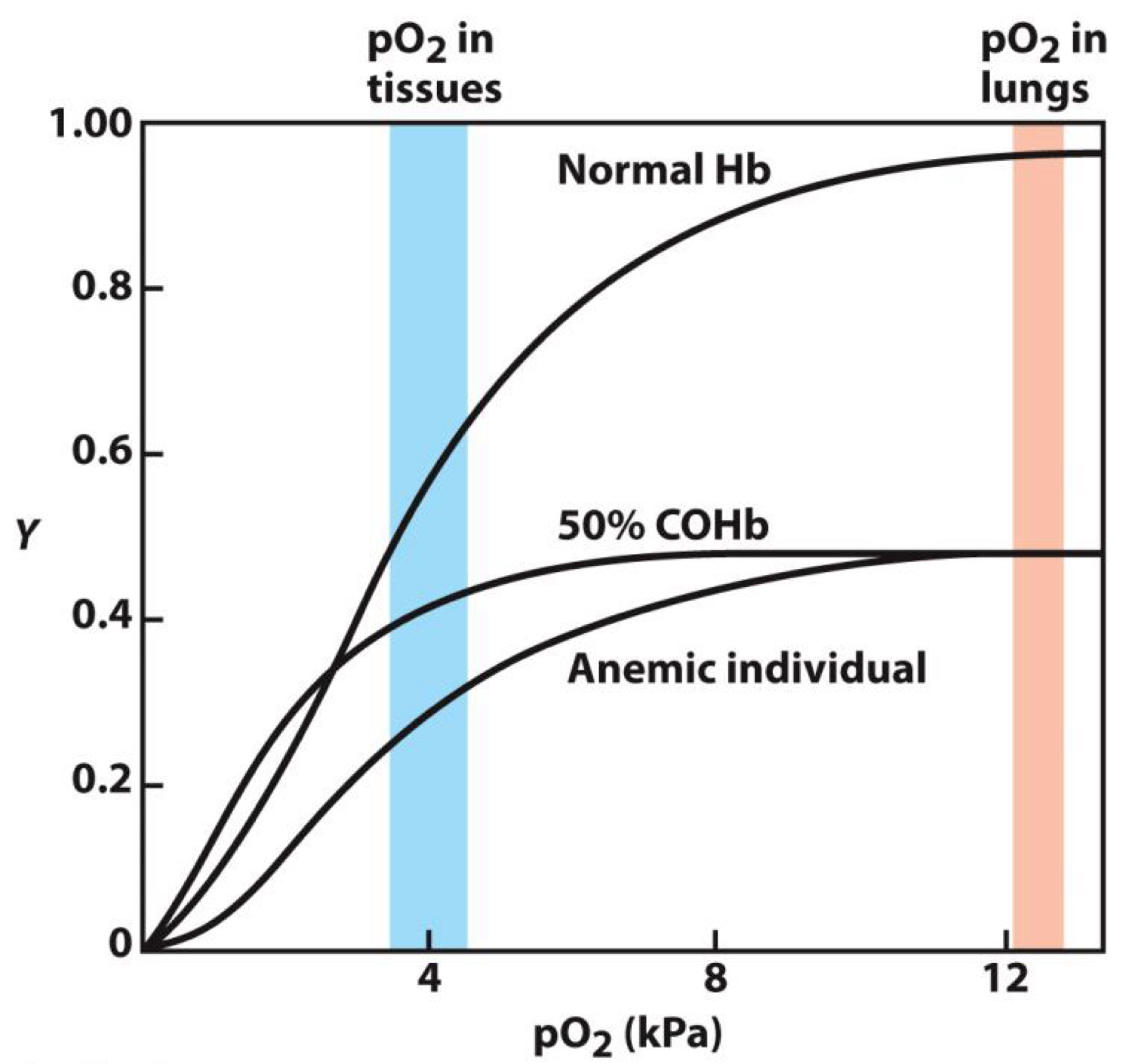
Hemoglobin relationship is more complicated with oxygen
Sigmoid (S) shaped
Due to high and low affinity states(more later)
Allows high uptake of oxygen in lungs and release in tissues
Shape due to two different states (forms) of hemoglobin
Low affinity state(Present at low [O2])
High affinity state(Present at high [O2])
The protein morphs from the low to high affinity state by biding to more O2
![<p><span>Sigmoid (S) shaped</span><br><span>Due to high and low affinity states(more later)</span></p><p><span>Allows high uptake of oxygen in lungs and release in tissues</span></p><p><span>Shape due to two different states (forms) of hemoglobin</span><br><span>Low affinity state(Present at low [O2])</span><br><span>High affinity state(Present at high [O2])</span></p><p><span>The protein morphs from the low to high affinity state by biding to more O2</span></p>](https://knowt-user-attachments.s3.amazonaws.com/1b0c63b7-5279-46f7-b8fb-0aa15d1b7e72.jpg)
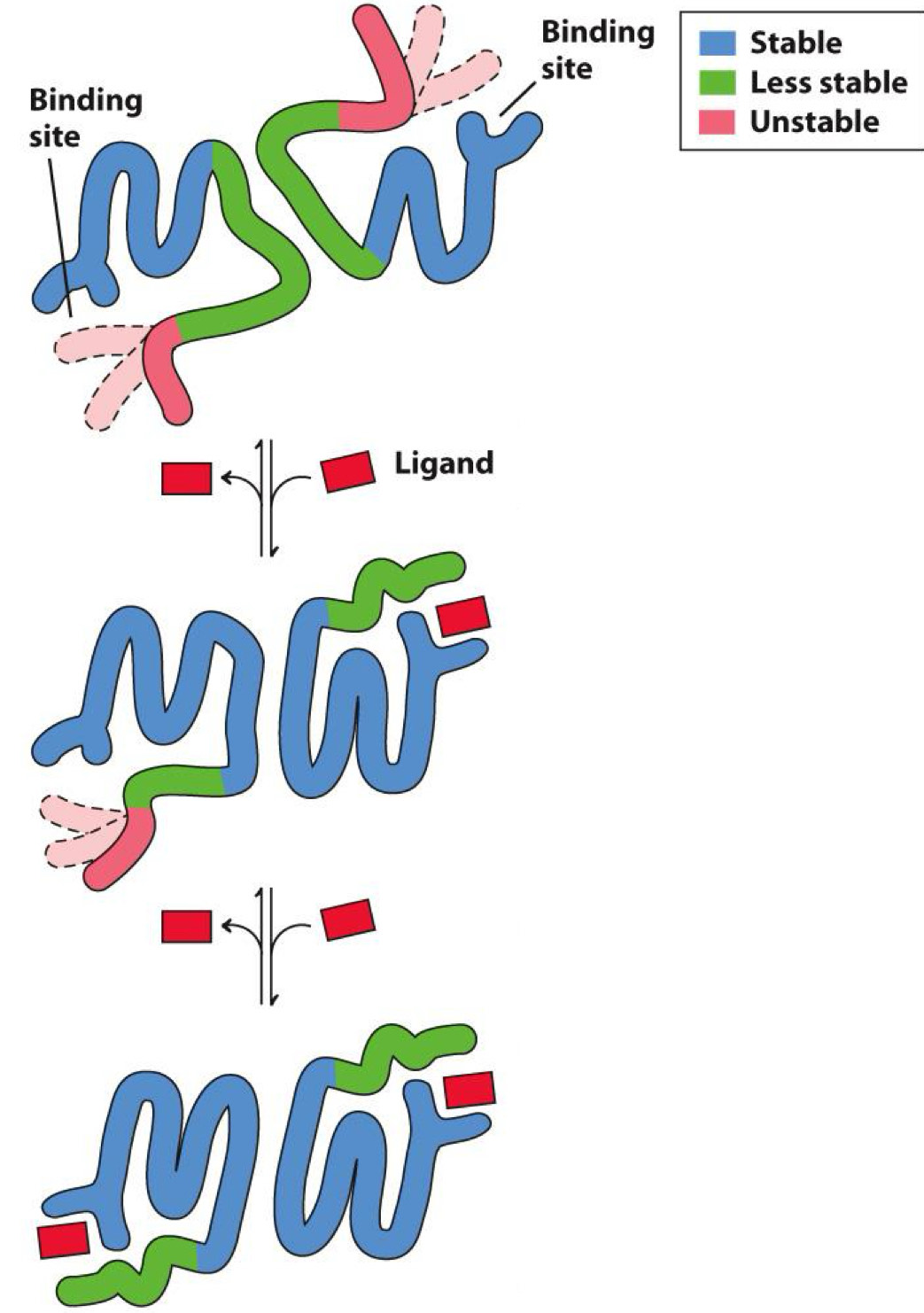
cooperative binding
Happens in hemoglobin
Binding of one ligand causes more to bind
- When ligand binds to one subunit, remaining subunits are stabilized and bind more ligand
How does CO influence O2 with binding to ligands?
Carbon monoxide (CO) prevents O2 from binding to hemoglobin
- CO is in engine exhaust
- Explains why small amounts (ppm) are lethal
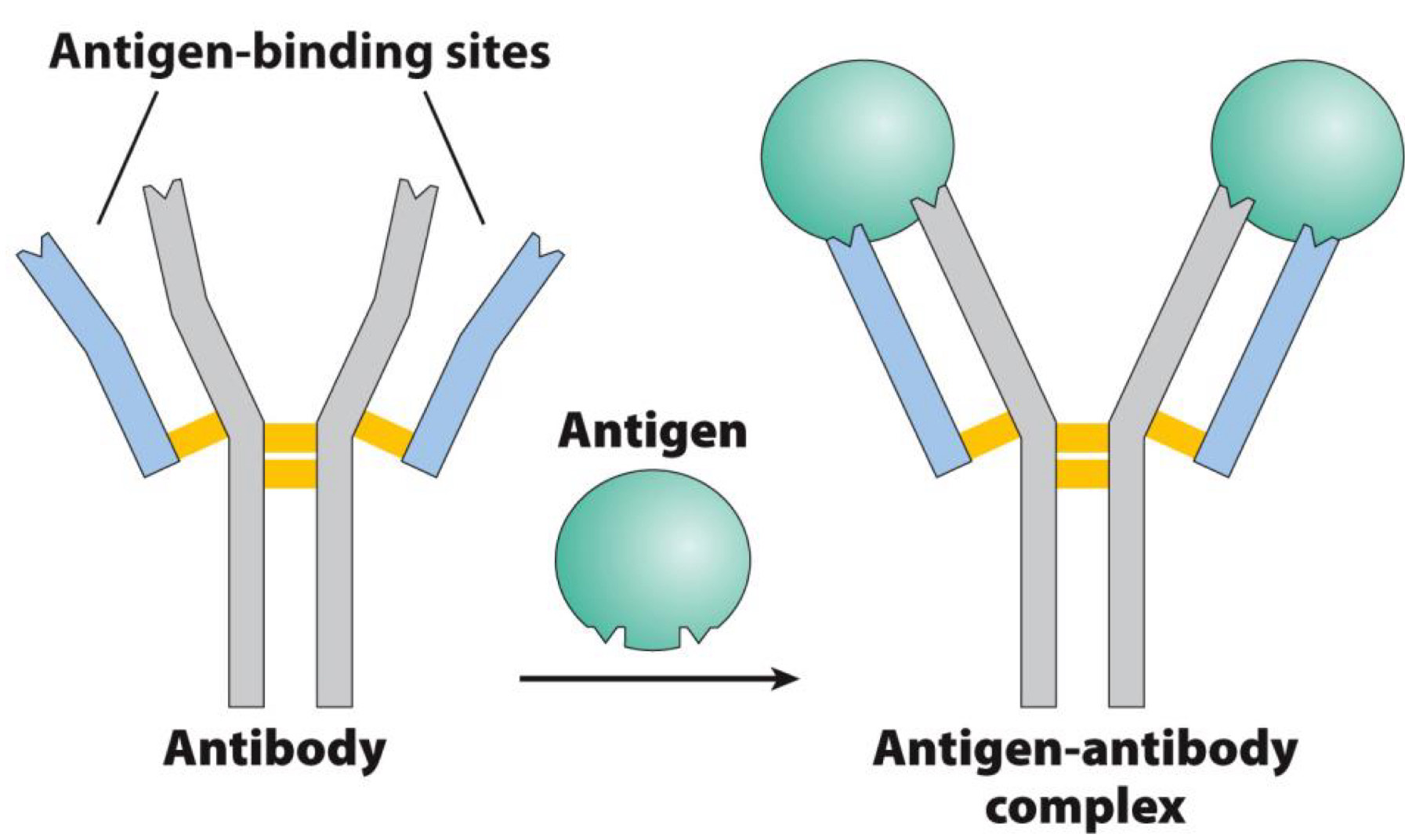
How are antibodies made and work?
Antibodies produced by immune system in response to foreign molecules
Binds target molecule tightly to
- Inactivates it
- Targets it for destruction
Billions of different antibodies can be produced
What do antibodies bind to?
Target molecule is antigen
Antigen is bound by antigen-binding sites
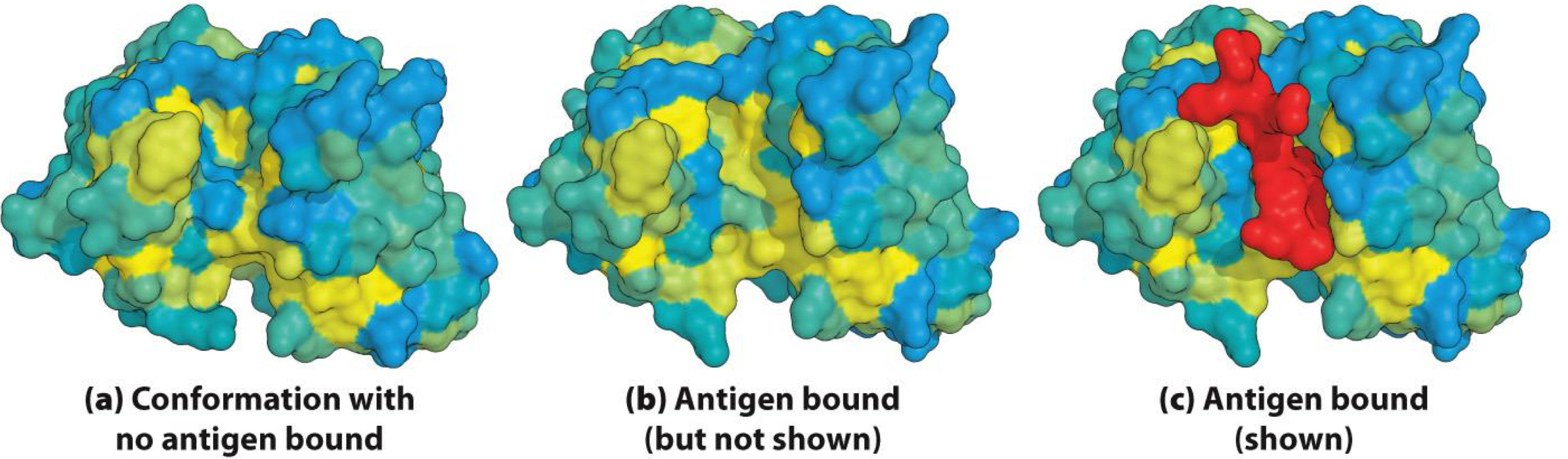
Induced Fit
Antibody changes shape (conformation) when binding to antigen
- Provides tight binding
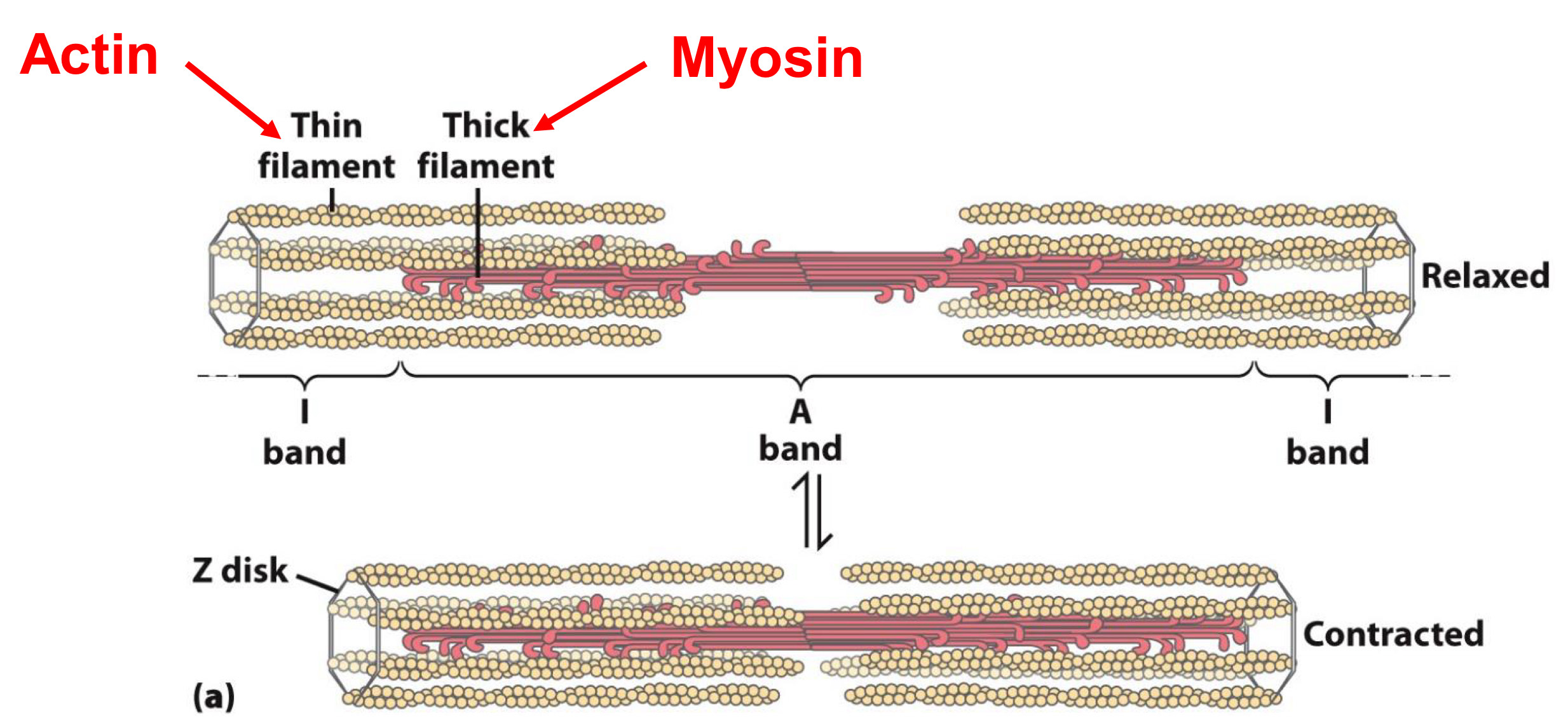
Motor Proteins produce large movements in cell
Motor proteins bind to and move other molecules
Responsible for
- Muscle movement
- Crawling and swimming of cells
Example: myosin and actin in muscle
How is movement caused in muscles?
Muscle contracts when filaments of actin move towards each other
Movement is caused by myosin walking across and pulling actin
thin filament→ actin
thick filament→ myosin(motor proteins)
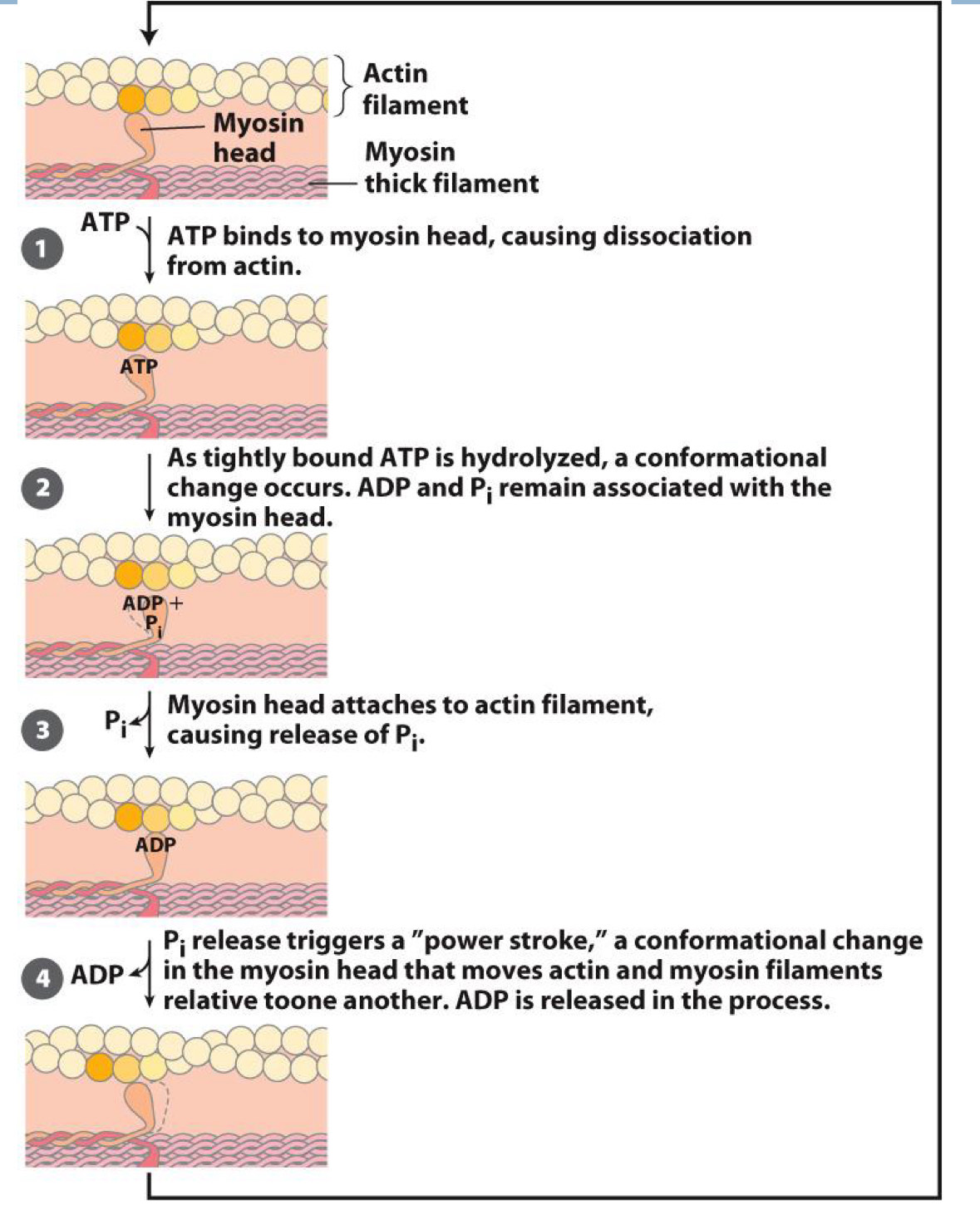
Power Stroke in Muscles
Walking in myosin is catalyzed by ATP
- Binding, hydrolysis, and release of ATP cause conformational changes
ATp binds to myosin head, causing dissociation from actin
As tightly bound ATP is hydrolyzed, a conformational change occurs. ADP and Pi remain associated with the myosin head
Myosin head attaches to actin filament, causing release of Pi
Pi release triggers a “power stroke”, a conformational change in the myosin head that moves actin and myosin filaments relative to one another, ADP is released in the process
Nucleotides and nucleic acids have many functions
Serve as enzymes (ribozymes)
Energy for metabolism (ATP)
Enzyme cofactors (NAD+)
Signal transduction (cAMP)
Gene expression (synthesis of proteins)
Nucleotides contain
Nitrogeneous base
Pentose
Phosphate
Bases are either purines or pyrimidines
Pentoses are either ribose (RNA) or 2’-deoxyribose (DNA)
Nucleoside
Nucleotide without phosphate
Names are different from respective nucleotides (and bases)
Nucleoside Examples
Adenine → base
Adenosine → nucleoside
Adenylate → nucleotide
Ribonucleotides
Components of RNA
Abbreviated as A, G, U, C
Deoxyribonucleotides
Components of DNA
Abbreviated as A, G, T, C
Can also be abbreviated as dA, dG, dT, dC to distinguish from ribonucleotides
Minor nucleotides in DNA
Methylated nucleotides
Example: 5-Methylcytodine- In eukaryotes, marks which genes should be inactive (epigenetic control)
In bacteria, protects DNA from host immune system (restriction enzymes)
Methyl group added after DNA synthesis
Minor nucleotides in RNA
Example: inosine
Found in some tRNAs (in “wobble position” of anticodon)
Made by deaminating adenosine
Polymerize by forming covalent bonds between nucleotides
Covalent bonds are phosphodiester linkages
Linkages create negatively charged backbone
DNA is a double helix- Who proposed the structure?
Structure proposed by James Watson and Francis Crick in 1953- H bonds between bases
A → T
C → G
Explained Chargraff’s rules and consistent with X-ray crystallography
Who showed that DNA was a double helix through X-ray crystallography?
Double helix structure previously shown by X-ray crystallography
Cross = helix
Shown by Rosalind Franklin and Maurice Wilkins
Helix information: major and minor grooves
Bases are 3.4 Å apart
There are 10.5 bases per turn (36 Å)
Two chains are complementary
They run antiparallel
- One runs 5’ to 3’(Numbers are with respect to ribose)
- Other runs opposite
What does the DNA helix suggest?
way for replication
- Strands separate
- New (daughter) strand synthesized(Using old (parent) strand as template), Done by DNA polymerase
- Two new molecules formed(Each with a parent and daughter strand)
Other forms of DNA
Watson-Crick structure is B form
A and Z forms also observed
- Only B and Z forms found in cells
RNA types: 2nd type of nucleic acid
Messenger RNA (mRNA)
Transfer RNA (tRNA)
Ribosomal RNA (rRNA)
Ribozymes (RNA enzymes; includes rRNA)
Others
- Formed as a single strand/helix
Structure of RNA
When it a simple strand (with no internal base pairing), structure is a single helix
Single strand can form internal base pairs
- Leads to complex secondary structures: Hairpins, Loops, Bulges
Single strand can form internal base pairs: Leads to complex 3D structure