Genetic code, protein synthesis, and folding
1/24
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
25 Terms
🔹 Genetic Code Basics
Q: Why is the genetic code redundant?
A: There are 64 codons (4³ combinations) but only 20 amino acids, so most amino acids are encoded by more than one codon. This redundancy (degeneracy) occurs mostly at the third base of the codon.
🔹 Genetic Code Basics
Q: Give an example of a non-standard genetic code.
A: In mitochondria, UGA (normally a stop codon) can code for tryptophan.
🔹 Transcription & Codon Usage
Q: What are the roles of the template and non-template strands in mRNA synthesis?
A:
Template strand (antisense): Used by RNA polymerase to make mRNA.
Non-template strand (sense): Matches the mRNA sequence (except T becomes U).
🔹 Transcription & Codon Usage
Be able to use the genetic code to predict the protein sequence from mRNA and predict how amino acids can be mutated when the base changes.
Q: How do you use the genetic code to predict protein sequence?
A: Translate the mRNA sequence in triplets (codons) using the codon chart.
Q: What’s the effect of a base change on amino acids?
A:
Silent mutation: no amino acid change
Missense mutation: changes amino acid
Nonsense mutation: creates stop codon
Frameshift: alters reading frame (insertion or deletion not in multiples of 3)
🔹 Frameshift Mutations
Q: What is the effect of a frameshift on protein sequence?
A: It alters all downstream codons, usually resulting in a nonfunctional or truncated protein due to premature stop codons.
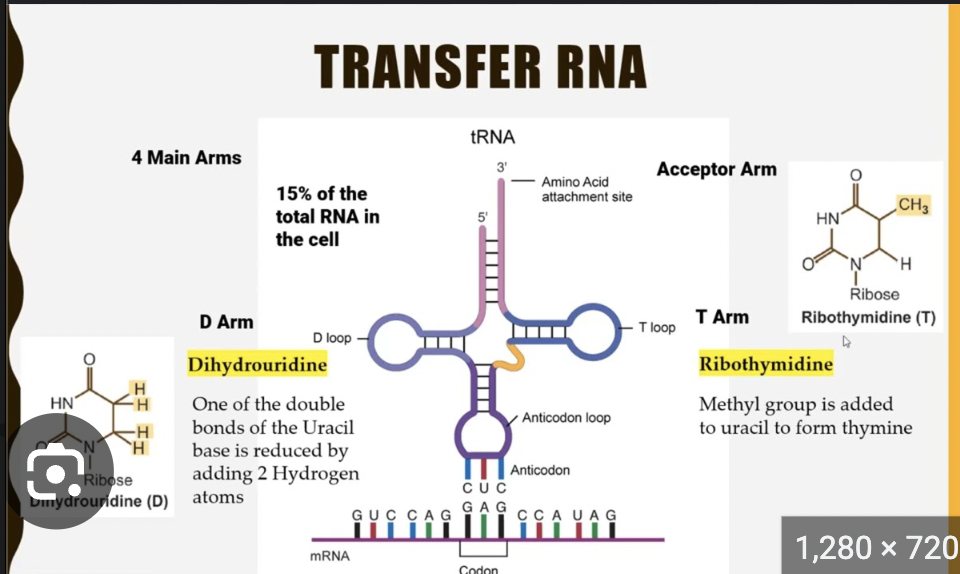
🔹 tRNA and Aminoacylation
Q: What is the structure of tRNA?
A:
Cloverleaf shape
Anticodon loop recognizes mRNA codon
Amino acid is attached at the 3’ CCA tail
🔹 tRNA and Aminoacylation
Q: What enzymes attach amino acids to tRNAs?
A: Aminoacyl-tRNA synthetases (one per amino acid)
🔹 tRNA and Aminoacylation
Q: Which position of the codon allows wobble (promiscuity)?
A: The 3rd position (codon) / 1st position (anticodon) allows wobble base pairing, enabling fewer tRNAs to recognize multiple codons.
🔹 Translation Fidelity
Q: What are the two factors defining translation fidelity?
A:
Accurate aminoacylation of tRNAs by synthetases
Correct codon-anticodon pairing at the ribosome
🔹 Ribosome Assembly & Initiation
Q: How does ribosome assembly begin?
A:
The small ribosomal subunit binds mRNA and initiator Met-tRNA (Met-tRNAi).
Met-tRNA binds the AUG start codon, assisted by initiation factors and GTP.
Q: What is the role of Met-tRNA in starting protein synthesis?
A: The initiator tRNA (Met-tRNAᶦ) carries methionine and pairs with the start codon (AUG), defining the first amino acid in the protein and positioning the ribosome to begin translation.
Q: What is the mechanism of translation initiation in eukaryotes?
A:
Initiation factors bind the small ribosomal subunit and Met-tRNAᶦ.
This complex scans the mRNA 5' → 3' for the AUG start codon.
Once the AUG is found, Met-tRNAᶦ pairs with it.
The large ribosomal subunit joins, forming the complete ribosome and placing Met-tRNAᶦ in the P site, ready for elongation.
🔹 Ribosome Sites & Peptidyl Transferase
Q: What are the three tRNA sites in the ribosome?
A:
A site – aminoacyl tRNA entry
P site – peptidyl tRNA holds growing chain
E site – exit site for empty tRNA
🔹 Ribosome Sites & Peptidyl Transferase
Q: What is the peptidyl transferase reaction?
A: It forms a peptide bond between the amino acids in the P and A sites. This is catalyzed by rRNA (a ribozyme) in the large subunit.
🔹 Termination of Translation
Q: When and how is protein synthesis terminated?
A: When a stop codon (UAA, UAG, UGA) is reached, release factors bind, the peptide is released, and the ribosome disassembles.
🔹 Protein Folding
Q: What is the principle of step-wise protein folding?
A: Proteins fold hierarchically:
Local elements (α-helices, β-sheets) form first
Then domains and full tertiary structure
Final step: native conformation with minimal free energy
🔹 Protein Folding
Q: What are denaturing agents?
A: Chemicals or conditions that disrupt non-covalent interactions, e.g., urea, SDS, heat, low pH.
🔹 Protein Folding
Q: What is the typical last step in protein folding?
A: The final step is the packing of the hydrophobic core, which stabilizes the tertiary structure and determines the protein's functional shape.
🔹 Misfolding and Aggregation
Q: What happens when a protein is overproduced and misfolds?
A: It may form aggregates, be nonfunctional, or toxic to the cell.
🔹 Misfolding and Aggregation
Q: What are typical manifestations of misfolding?
A:
Misfolded proteins can lead to aggregation, loss of function, or toxic gain of function, contributing to diseases like Alzheimer’s, Parkinson’s, Huntington’s, and prion diseases. These conditions often involve amyloid fibrils or inclusion bodies formed from misfolded proteins.
🔹 Chaperones
Q: What do cells use to prevent/correct misfolding?
A: Molecular chaperones:
Hsp70 – binds nascent chains to prevent premature folding
Hsp90 – stabilizes signaling proteins
GroEL/GroES – chaperonin complex that provides a folding chamber
🔹 Ubiquitination & Proteasome
Q: What is protein ubiquitination?
A: A tagging process where ubiquitin molecules are attached to proteins marked for degradation by the proteasome.
🔹 Ubiquitination & Proteasome
Q: What types of proteasomes exist, and when are they active?
A: The 26S proteasome degrades ubiquitinated proteins under normal conditions; the 20S proteasome functions independently to degrade damaged proteins during stress; the immunoproteasome forms in response to immune signals (like interferon-γ) to aid antigen presentation.
🔹 Golgi Sorting
Q: What are the results of apical and basolateral sorting in the Golgi?
A:
Apical sorting: Targets proteins to cell surface/lumen-facing side (e.g., digestive enzymes)
Basolateral sorting: Delivers proteins to interior-facing side or cell junctions
This maintains cell polarity in epithelial cells.