Topic E: Organisation and Control of Euk Genome
1/20
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
21 Terms
Name the non-coding sequences of eukaryotic genomes.
Telomere
Centromere
Promoter
Enhancer
Silencer
What is the role and structure of telomeres?
Structure:
Telomeres are made up of highly repetitive sequences.
Telomeres are non coding DNA.
Telomeres associates with histone proteins.
Role:
Provides a counting mechanism for the number of times a cell has undergone cell division. Once telomere shorten to a critical length, cell will undergo apoptosis, which prevents unlimited cell proliferation.
Prevents the ends of different chromosomes from fusing together.
Protects important genes near the telomeres.
Prevents the ends of chromosomes from being degraded by exonucleases.
What is the role and structure of promoters?
Structure:
Non coding DNA.
Promoters comprise of the TATA Box, which is where the TATA Binding Protein recognizes and binds to, in order to initiate the assembly of the transcription initiation complex.
Located upstream of coding sequences.
Role:
Site at which the transcription factors recruit RNA polymerase, so that the transcription factors and RNA polymerase will bind to the promoter to form the transcription imitation complex.
What is the role and structure of enhancers?
Structure:
Non coding DNA.
Located far away from promoters.
Role:
The activator will recognise and bind to enhancers.
Enhancers increase the rate of transcription, by looping the DNA, increasing interaction between RNA polymerase and promoter which stabilises the transcription initiation complex via increased protein-protein interaction, thus increasing the rate of transcription.
What is the role and structure of silencers?
Structure:
Non coding DNA.
Located far away from promoters.
Role:
The repressor will recognise and binds to the silencers.
This reduces the rate of transcription because it inhibits the assembly of transcription initiation complex.
What is the role and structure of centromeres?
Structure:
Non coding DNA.
Made up of a highly repetitive sequence.
Associates with histone proteins.
Is the constricted region of the chromosome.
Role:
Facilitates chromosomal movement during cell division.
Site at which kinetochore microtubules bind to during cell division.
How can gene expression be regulated in eukaryotes?
Chromatin level (histone modification, DNA acetylation)
Transcriptional level (presence of PES)
Post transcriptional level (5’ cap, 3’ poly-A-tail, splicing)
Pre translational level (assembly of translation initiation complex, stability and half life of mRNA)
Post translational level (protein degradation, biochemical modification)
How does histone acetylation affect gene expression in eukaryotes?
It increases the rate of transcription.
Histone acetyltransferase catalyses the addition of acetyl groups to the positively charged lysine residues of the histone proteins → neutralises the positive charge of lysine residues of histone proteins → reducing electrostatic attraction between histone proteins and DNA → DNA and histone proteins will associate less closely with one another → promoter region of DNA less compact and more accessible to transcription machinery → promotes the assembly of transcription initiation complex → increasing the rate of transcription
How does histone deacetylation affect gene expression in eukaryotes?
It reduces the rate of transcription.
Histone deacetyltransferase catalyses the removal of the acetyl groups from the lysine residues of the histone proteins → restoring the positive charge of the lysine residues of histone proteins → greater electrostatic attraction between histone proteins and DNA → DNA and histone proteins will associate more closely → promoter region of DNA more compact, less accessible to transcription machinery → inhibits the assembly of transcription initiation complex → reducing the rate of transcription
How does DNA methylation affect gene expression in eukaryotes?
It reduces the rate of transcription.
DNA methyltransferase will catalyse the addition of methyl groups to the cytosine residues of the DNA → methylated DNA will recruit histone deacetyl transferase proteins and other transcription repressor proteins → inhibits assembly of transcription initiation complex → reducing rate of transcription.
How do we increase the rate of transcription of DNA in eukaryotes?
Histone acetylation.
Binding of activators to enhancer sequences of the DNA.
How do we decrease the rate of transcription of mRNA in eukaryotes?
Histone deacetylation and DNA methylation.
Binding of silencers to the repressor sequences of the DNA.
How is gene expression regulated in the post-transcriptional level?
5’ Cap:
Guanylyl transferase catalyses the addition of a methyl guanosine nuceloside triphosphate to the 5’ end of the pre-mRNA.
This serves to protect the pre-mRNA from being degraded by 5’ exonucleases.
Facilitates the export of mature mRNA from the nucleus into the cytoplasm for translation.
3’ Poly-A-Tail
Poly-A-Polymerase catalyses the addition of around 200 adenine residues to the 3’ end of the pre-mRNA.
This serves to protect the pre-mRNA from being degraded by 3’ exonucleases.
Facilitates the export of mature mRNA from the nucleus into the cytoplasm for translation.
Splicing
Spliceosomes will excise the introns and join the exons to form a continuous coding sequence.
Alternative splicing allows for many mRNA to form from one pre-mRNA,
How is gene expression regulated in the post-translational level?
Protein Degradation:
Unwanted proteins are tagged by ubiquitin.
Proteasome will recognize and bind to ubiquitin-tagged proteins before injecting them into the proteasome core, where the proteins are degraded.
Biochemical Modification:
Covalent attachment of biochemical groups to polypeptides to transform them into functional proteins.
How is the eukaryotic genome organized?
The double-stranded DNA coils around a core of histone proteins. The negatively charged sugar phosphate backbone of DNA associates closely with the positively charged lysine and arginine residues of histone proteins, thus giving rise to a nucleosome. Nucleosomes are joined by linker DNA which results in a beads on a string structure.
Nucleosome and linker DNA will undergo further folding and coiling to give rise to 30nm chromatin fibres, which appears as a solenoid structure.
The 30nm chromatin fibre will associate closely with scaffold proteins to give rise to 300nm looped domains.
The 300nm chromatin fibre will undergo further coiling and folding, before appearing as 700nm chromatin fibres, which is how metaphase chromosomes appear as.
How is gene expression regulated in the translational level?
Stability of mRNA
The longer the poly-A-Tail, the more stable the mRNA, thus giving rise to a longer half-life of mRNA. This would allow mRNA to be used as a template for translation to occur over a longer period of time.
Translation Initiation Complex:
During translation, the small ribosomal subunit will bind to the 5’ end of the mRNA. Thus, to prevent this, translational repressors can bind to the 5’ untranslated region of mRNA to prevent the assembly of translation initiation complex.
Phosphorylation and dephosphoylation of translation initiation factors can activate or deactivate the translation initiation factors required to initiate ribosomal binding.
Outline the steps of gel electrophoresis.
Mix the DNA samples with restriction enzymes and loading dye.
Using a micropipette, load the mixed DNA samples into the wells located at the negative electrode.
Thereafter, turn on Direct Current and allow the DNA samples to run towards the positive electrode. During the making of the gel, ethidiumum bromide is added to better enable the visualisation of the DNA bands.
Outline the steps of Southern Gel Blotting.
After carrying out gel electrophoresis, place the gel into a solution of NaOH (aq) to denature the dsDNA into ssDNA.
Place the positively charged nitrocellulose membrane above the gel, together with heavy weights and paper towels, in order to generate capillary action that would transfer the ssDNA onto the nitrocellulose membrane.
Remove the weights, and place the membrane under UV to permanently cross link the DNA onto the nitrocellulose membrane.
Incubate the nitrocellulose membrane with DNA probes that will hybridise with desired DNA sequence via complementary base pairing. Thereafter, wash the unhybridised probes away.
Expose the membrane to a piece of X ray film to obtain radiogram.
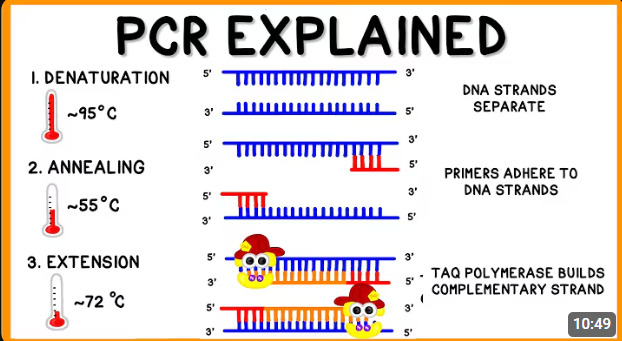
Outline the steps of Polymerase Chain Reaction (PCR)
Heat the sample to 95 degree celsius. This will cause the hydrogen bonds between complementary bases to break, thus denaturing dsDNA into ssDNA.
Cool the mixture to 55 degree celsius, This will allow the primers to anneal via hydrogen bond via complementary base pairing to the flanking sequence of the target DNA sequence.
Heat the mixture back to 72 degree celsius to allow the Taq polymerase to add deoxyribonucleotides to the 3’OH end of primer in the 5’ to 3’ direction using ssDNA as a template.
Outline the limitations of PCR.
Taq polymerase is of bacteria origin, and is unable to perform proofreading, thus incorrect deoxyribonucleotides may be added, and these mistakes may be amplified.
Knowledge on the DNA sequence flanking the target sequence is required in order to design a suitable primer that can anneal to flanking DNA sequence via hydrogen bonds via complemetary base pairing.
Limitations on length of DNA that can be amplified.
Primers are short nucleotide sequences, and there may be non target DNA sequences compleemtary to such primers. Thus, this might result in amplification of unwanted sequences instead.
Outline the advantages of PCR.
Large amount of DNA can be produced from small amount of sample.
Specific, target DNA sequences can be amplified with the use of specific primers.
Large amounts of DNA can be produced in a relatively short time with high accuracy.