Unit 1 DNA and chromosomes (use this one)
1/67
Earn XP
Description and Tags
Questions from Guide, Quiz, and Breakout
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
68 Terms
A gene was originally defined as a unit of heredity that contains instructions that dictate the characteristics of an organism. Can a gene contain instructions for making other types of RNA?
A gene can encode for proteins but not all genes specify for polypeptides. Genes can also contain the instructions for a particular functional RNA molecule that is the end product. The gene is the entire nucleic acid sequence needed to make the functional proteins or RNA molecules. Regulatory RNAs (siRNA, miRNA) or structural RNAs (like tRNAs and rRNAs) or catalytic RNAs (such as those needed for alternative splicing) are examples of the roles that these functional RNAs fill.
Coding regions of DNA can be classified as introns or exons. Define introns and exons. Scientists used to think that “junk” DNA was not important but have observed that some portions of “junk” DNA are highly conserved in its DNA sequence among different species. What does this suggest?
Intron: Noncoding sequence within a eukaryotic gene that is transcribed into an mRNA molecule but is then removed to produce a mature mRNA. The intron is non-coding and will not contribute to the proteins’ amino acid sequence.
Exon: Segment of a eukaryotic gene that is transcribed into mRNA and dictates the amino acid sequence of part of a protein.
The fact that some portions of noncoding DNA sequences have been conserved over the millennia suggests that they play important roles that are not yet understood. For example, some sequences have been identified as serving regulatory functions. It is likely that other noncoding sequences also have important functions.
Origins of replication initiation (ORI) are sites where the replication machinery will initially open the DNA double helix. Is there only a single ORI per chromosome? Is binding of the machinery base sequence specific or are there specific characteristics for ORI?
The DNA replication origin is one of three sequence elements, along with the centromere & telomere, needed to produce a stable eukaryotic chromosome. Eukaryotic chromosomes contain many replication origins to ensure that the long DNA molecules are replicated rapidly. The exact sequence may differ species to species, but attributes such as a high A-T content are common to all ORI’s. Since A-T’s double bond is weaker than C-G’s triple bond, the DNA machinery will be more easily able to break the A-T rich regions apart for the replication machinery to be able to use the DNA strands as templates.
For each of the three biomolecules that form polymers that we discussed, indicate the type of bond that connects the units within the polymer
The three major biomolecules that are polymers are: proteins, carbohydrates, and nucleic acids. Amino acids are held together by peptide bonds within proteins. Monosaccharides are held together by glycosidic bonds within carbohydrates, which includes polysaccharides and disaccharides. Nucleotides are connected by phosphodiester bonds within nucleic acids.
Amino acids have different chemical properties based on their R group. Why is it essential to understand the chemical properties of the R groups?
All amino acids have an amino group, a carboxyl group, and a side chain (R) attached to their alpha-carbon atom. Amino acids are used by cells to build proteins, that are essentially polymers made of amino acids which are joined head-to tail in a long chain. As the chains fold up, they form unique 3-D structures for different proteins. Amino acids in a protein are covalently held together by a peptide bond (bonds formed by condensation reactions), and a chain of amino acids is called polypeptide. The two ends of each amino acid, namely the amino terminus (or N-terminus) and the carboxyl terminus (C-terminus), are chemically different, therefore, giving each polypeptide chain directionality. Projecting from the core atoms, or the polypeptide backbone, are the R groups, that are not involved in forming peptide bonds. It is essential to understand that the chemical properties of the R groups give each amino acid its unique identity. For example, some are non polar and hydrophobic, some are positively or negatively charged, and some are more chemically reactive etc. These R group chemical properties ultimately determine the protein structure and function by dictating how the protein polymer folds into tertiary structures as well as dictating how the mature protein can interact with other proteins in protein complexes (quaternary structure). The distribution of polar and non-polar amino acids govern the folding of any protein as the final folded structure, or conformation, of any polypeptide into a 3-dimensional protein is determined by energetic considerations. Polar amino acid R groups on the outside of a protein can interact with water, whereas non-polar amino acid R groups on the inside of a protein form a highly packed hydrophobic (or water fearing) core of atoms and these can also contribute to the protein folding.
Proteins organization is described by the terms primary, secondary, tertiary and quaternary structure. What do each of these terms mean? What interactions are important for generating this level of organization?
Proteins are one of the four major kinds of biomolecules. Proteins are made up of amino acids. Amino acids can form linear polypeptides via peptide bonds. Due to the properties of the different R groups in each of the 20 amino acids, the polypeptides form bonds between the R regions and can fold upon itself. Every protein has it on own unique way of folding due to its specific amino acid sequence which gives it a specific chemical property. There are 4 levels of protein structures and protein interaction to form 3-D structures of protein. The primary structures are the linear sequence of amino acids in a protein polymer. The secondary structure is a localized interaction of the sequence with a section of the protein. There are two common interactions at this level of folding that include beta sheets and alpha helixes. These interactions occur between the atoms of the backbone. The tertiary structure is the complete 3-D structure of one polypeptide and is considered a mature folded protein. For the tertiary structure, non- linear associations occur and interactions can occur between R groups that are spaced far apart on the linear sequence of the protein. Secondary and tertiary level proteins frequently use hydrogen bonds to fold. At the quaternary structure level, multiple proteins that have previously formed a stable tertiary structure interact. At this level, proteins interact through electrostatic attraction or hydrogen bonds (non- covalent interactions) to form a complex protein molecule. The quaternary interactions are also usually with the R groups.
Both nucleic acid and protein polymers have polarity.
Which chemical structure is on the 3’ end of a DNA strand?
Which structure is normally on the 5’ end of a DNA strand?
Which chemical structure is found on the N-terminus of a protein?
Which chemical structure if found on the C-terminus of a protein?
The chemical structure on the 3’ end is a free Hydroxyl group (OH). The DNA strands are made of 5 carbon sugars and the 3’ name comes from the third carbon on the sugar.
The chemical structure on the 5’ end is a free phosphate group. 5’ end name comes from the group being attached to the 5th carbon of the sugar. This carbon is the one that is outside the sugar ring.
The chemical structure found on the end of an N-terminus protein is a free amine group, which corresponds to the Nitrogen group at the end of an amino acid.
The chemical structure found on the end of a C-terminus protein is a free carboxyl group.
What are three differences between the nucleic acid RNA and DNA?
DNA is double-stranded, made up of two long poly-nucleotide chains; RNA is single nucleic acid polymer.
Both nucleic acids contain nitrogenous bases attached at their C1 carbon of their sugar. Although both DNA and RNA are composed of the nitrogenous bases adenine (A), cytosine (C), and guanine (G) as nucleotide subunits, RNA uses uracil (U) as its fourth base, whereas DNA uses thymine (T).
RNA uses ribose sugar in its sugar-phosphate backbone; DNA uses deoxyribose sugar in its sugar-phosphate backbone. These are both five carbon sugars but differ at the C2 carbon. DNA (deoxyribose nucleic acid) lacks an oxygen at the C2 and only has a hydrogen. RNA (ribose nucleic acid) has a hydroxyl (OH) at the C2 carbon
Describe a double stranded DNA helix. Please make sure to discuss:
the orientation of the two strands of DNA in relation to each other
how nucleotides within a single strand are linked
how the bases between the strands link the two strands
Are the two strands identical? Why?
What direction is the DNA strand read?
The two DNA polymers are antiparallel. This special orientation is the energetically favorable conformation of DNA as it allows the nitrogenous bases to pair while maintaining an equal distance between the sugar-phosphate backbone. It would be energetically unfavorable to distort the backbone using other pairings.
Nucleotides within a polymer are linked by strong/covalent phosphodiester bonds: this covalent bond is formed between the 5' phosphate group of one nucleotide and the 3'-OH group of another. Because of this chemistry, for a polymer, the end with the free hydroxyl group (which is attached to the 3’-carbon position of the sugar) is called the 3’ end, while the end bearing the phosphate group (which is attached via the 5’-hydroxyl group of the sugar) is called the 5’ end.
Hydrogen bonds between the complimentary nitrogen bases help link the two nucleic acid strands together. A-T pairs form two hydrogen bases while G-C pairs form three hydrogen bonds.
No, the two strands of DNA would be considered complementary and antiparallel, not identical. Because of base pairing, every time there is on one strand an Adenine there will always be a Thymine on the other strand at the same location and we call this pairing complimentary pairs. Guanine and Cytosine also form complimentary pairs.
From 5’ to 3’ by convention.
For nucleotides, it is important to understand their structure.
What carbon of the sugar does the nitrogenous base attach to?
The phosphate group is attached to which carbon of the sugar?
DNA and RNA structures are different at which position of the sugar?
How is the nucleotide different than a nucleoside?
The nitrogenous base attaches to the C1 carbon of the sugar.
The phosphate group is attached to the C5 carbon of the sugar
DNA and RNA structures are different at the C2 position of the sugar—in DNA, the C2 carbon is attached to a hydrogen atom; in RNA, the C2 carbon is attached to a hydroxide group (OH).
A nucleotide consists of a pentose sugar, a nitrogenous base, and one or more phosphate groups; a nucleoside consists only of a pentose sugar and a nitrogenous base—it has no phosphate groups attached. This means that nucleosides can never be joined together like nucleotides can because they lack the phosphate groups necessary for this to occur.
What are purines and pyrimidines? In a DNA double-stranded helix, there is always a purine paired with a pyrimidine. Why is this essential for the stability of the helix structure?
Pyrimidines and purines are nitrogen-containing ring compounds. In DNA a purine is always complimentarily paired with a pyrimidine through hydrogen bonds. The two main types of purines that occur in both DNA and RNA are adenine and guanine. The three main types of pyrimidines are cytosine, found in both DNA and RNA, thymine, found only in DNA, and uracil, found only in RNA. This complementary base-pairing between a purine and a pyrimidine enables the base pairs to be packed in the most energetically favorable way that does not disturb the helix’s sugar phosphate backbone. There is a difference in the number of hydrogen bonds between the pairs with a G-C pair having three and an A-T pair having two. Chargaff’s Rule shows that A always pairs with T and that G always pairs with C.
Why did scientists originally lean toward proteins being the hereditary information rather than nucleic acids?
Scientists knew that genes were information containing elements and that they must be formed by biomolecules; and chromosomes are made of nucleic acids and proteins. However, scientists were hesitant to accept that DNA contained the hereditary information because of its chemical simplicity. Nucleic acids are only formed from four nitrogenous bases and amino acids have 20 amino acids. Since proteins are more chemically diverse than DNA molecules, scientists assumed that hereditary information was contained within proteins. It was thought that the larger number of amino acids would provide for more potential outcomes in sequence order to provide genetic information (think if the English alphabet being formed by only four options rather than the 26 letters we are familiar with). It took many experiments (eg. Griffith pneumonia, Avery, MacLeod, and McCarthy bacteria transformation, Hershey and Chase E. coli) to convince the scientific community that DNA, not proteins, contained genetic material.
To determine whether it is DNA or protein that enters the infected bacterial cell, the researchers radioactively labeled the viral proteins with the isotope 35S and the viral DNA with 32P. Which biomolecule would be labeled by each of these radioisotopes?
DNA falls within the biomolecule category of nucleic acids. 32P (Phosphorus-32 is a radioactive isotope of the chemical element phosphorus) and is incorporated in the sugar-phosphate backbone of nucleic acids when cells are grown in the presence of this element. DNA is labeled with the radioisotope 32P in Hershey and Chase’s experiment because phosphorus is found only in DNA, not in proteins. Proteins were labeled with the radioisotope 335S (a radioactive isotope of the chemical element sulfur) in the experiment because sulfur is found only in proteins, not in DNA.
Are all genes on a chromosome encoded on the same DNA strand of the helix?
Genes can be located on either polymer strand of DNA. However, for each individual gene, only one of the two DNA polymer strands actually encodes genetic information, and, in general, coding regions rarely overlap between strands. We will get into more detail with when we discuss transcription. Example: Strand “A” codes for gene “a” and strand “B” codes for gene “b”. Strand “B” is the complementary strand for gene “a” and strand “A” is the coding strand. Strand “A” is the complementary strand for gene “b” and Strand “B” is the coding strand. Either strand can encode genes but for a single gene it will only be coded on one of the two strands.
What are the DNA major and minor groove?
The unique double-helix structure of DNA is maintained through consistent base pairing of adenine to thymine and guanine to cytosine along a phosphodiester backbone. The consistent pairing of a purine to a pyrimidine keeps the antiparallel strands at a uniform width, and the double helix rotates at approximately every 10 base pairs. The coiling from the rotation forms repeating and alternating major and minor grooves, which are binding sites for proteins. The difference in size between the major and minor groove allows for predictable binding patterns based on the surface area available for proteins to interact with the base pairs. The major groove is wider than the minor groove, so it is easier for proteins to bind with those sites and read the exposed base pairs for transcription into RNA.
Is there a correlation between the complexity of an organism and the size of the genome? Is there a correlation between complexity and the number of chromosomes? Do closely related species have to have the same number of chromosomes?
There is no simple relationship between chromosome number, species complexity and total genome size. Over larger differences in genome size, there is a trend between the complexity of an organism and the size of an organism’s genome. BUT the genome size is NOT always related to the complexity of an organism. There is no correlation between complexity of an organism and the chromosome number. It is demonstrated that closely related and phenotypically similar species do not need to have the same number of chromosomes.
Is most of our genome encoding for proteins and RNAs?
Most of our genome is made up of non-coding repetitive sequences with only a small percent making up regions of protein and RNA coding. Our genome is made up of half repeated sequences and half unique sequences. Within the portion of repeated sequences, we find mobile genetic element, simple repeats and segment duplications. Within the portion of unique sequences, we find our genes as well as non- repetitive DNA that make up neither introns nor exons. Non-coding DNA is sometimes referred to as “junk DNA” because its use within the cell has not yet been determined. Sometimes the sequence itself may not be important, but the space it provides may be important for long term evolution and gene activity.
When comparing the homology across multiple organisms, is homology only observed in the exon regions?
Homology across organisms can be observed in both exon and intron regions of a gene. The exon portion of the gene provides coding information and may remain extremely similar across a large array of organisms. The intron regions, which do not provide genetic code, may also remain homologous across organisms, especially if the lineages of those organisms diverged relatively recently. Introns can contain important regulatory information and comparison across homology helps scientists identify important regions.
What are Single nucleotide polymorphisms (SNPs)? Are they harmful?
SNPs, single nucleotide polymorphism, are single nucleotide difference between individuals within the genome at a specific position. Each SNP represents a difference in a single nucleotide. For example, a SNP may replace the nucleotide cytosine (C) with the nucleotide thymine (T) in a certain stretch of DNA. Most commonly, SNPs are found in the DNA between genes. Thus most of them have little or no effect on human fitness. However, when SNPs occur within a gene or in regulatory region near the gene, they could increase one’s susceptibility to disease by affecting the gene’s function.
Is the DNA randomly placed within the nucleus or is there a level of organization in interphase cells? What is the nucleolus?
DNA is not randomly placed within the nucleus but is organized within the nucleus using three different mechanisms. Each chromosome occupies a different region of the nucleus so that different chromosomes don’t become entangled together (we refer to these regions as territories). Some chromosomes attach themselves to the nuclear envelope or to the nuclear lamina. The third mechanism is seen in the nucleolus. The nucleolus is where the parts of the different chromosomes carrying genes that encode ribosomal RNAs cluster together. In the nucleolus ribosomal RNAs are synthesized and combine with proteins to form ribosomes.
Taking advantage of the fact that bacteriophage only have protein and DNA, Hershey and Chase used radiolabeled elements (32P and S35) to identify the biomolecule that carries the heritable information. Which of the following statements is false?
(a) Phage grown in medium containing sulfur S35 will only have labelled proteins.
(b) If S35 was found in the bacteria, Hershey and Chase would have concluded that phage protein enters the host cell.
(c) Phage grown in medium containing 32P will only have labelled proteins.
(d) If 32P was found in the bacteria, Hershey and Chase would have concluded that phage DNA enters the host cell.
c
Name 2 of the major difference between the nucleic acids RNA and DNA
RNA uses the nitrogen base uracil while DNA uses thymine to base pair with Adenine bases
RNA has a hydroxyl group at sugar C2 while DNA has only a hydrogen. (will only receive partial credit if stated that RNA has ribose and DNA has deoxyribose as it doesn’t show you understand the chemical difference)
Name and describe the three regions (DNA sequence elements) needed to produce a stable eukaryotic chromosome.
DNA replication origin - many on a single chromosome, AT rich region to allow less energy to break apart DNA strands
Centromere - holds the two copies of the duplicated chromosome together and attaches to the mitotic spindle, repetitive DNA
Telomere – repeats at the ends of each chromosome, prevents shortening of DNA with each replication.
It has been determined that one strand of a DNA double helix contains 20% adenine and 10% thymine. Given this information, what are the percentage values for each of the following? If it can not be determined from this information alone, please state "not determined":
%T on the other strand of the double helix
%G + C within the entire double helix
%G within the entire double helix
%T within the entire double helix
Given one strand contains 20% A, we know that it will have T nucleotides paired with the A (Chargaff’s rules) so there will be 20% T on the other strand of the helix.
Given one strand has 20%A and 10% T, we know the other strand based on Chargaff’s rules will have 20%T and 10% A. This means the entire DNA helix has 30% A plus T. That leave 70% of the helix to be the amount of G+C.
Because G always pairs with C, we know that half of the 70% or 35% will be the amount of G and the other half (35%) will be the amount of C nucleotides.
In B, we know determined the entire helix had 30% A plus T nucleotides. A and T always pair so we will have the same number. Thus, 15% of the nucleotides within the entire helix will be T nucleotides.
Indicate which of the following statements is true.
(a) Comparing the relative number of chromosome pairs is a good way to determine whether two species are closely related. False, think about the example of the deer species that have very similar phenotypes but very different chromosome numbers
(b) Mitotic cells contain chromosomes that are less densely packed than interphase cells. False, the chromosomes in Mitotic cells are highle packaged into dense chromatin.
(c) More than 40% of the human genome is DNA sequences that do not encode for protein. Our slide showed that more than 50% were non-coding repetitive sequences. This is a TRUE statement.
(d) An organism’s genome size is always directly related to the organism’s genetic complexity. False, there is a loose correlation but this is not always true.
c
What are the two main branches of life and what are key characteristics that are different between these two branches? What are key similarities?
a. Prokaryotic and eukaryotic
b. Prokaryotic; membrane that surrounds, biomolecules floating
c. Eukaryotic is more complex, has nucleus, specialized compartments
d. Both have membrane
e. Both use four biomolecules, protein, carb, lipid, proteins
What is the central dogma? Is this true only for one branch of cells?
a. Central dogma is DNA to RNA to protein and is true for all cells
i. RNA used to selectively make protein that cell type needs
What is the common structure of an individual subunit for a nucleic acid?
A phosphate group – One or more phosphate groups attached to the 5' carbon of the sugar.
A five-carbon sugar
Deoxyribose in DNA (missing an –OH group on the 2' carbon)
Ribose in RNA (has an –OH group on the 2' carbon)
A nitrogenous base, which can be:
Purines: Adenine (A), Guanine (G) – double-ring structures
Pyrimidines: Cytosine (C), Thymine (T in DNA), Uracil (U in RNA) – single-ring structures
How many types of subunits are there? How are they different?
Adenine (A)
Guanine (G)
Cytosine (C)
Thymine (T)
Uracil (U)
Nitrogenous Base – The primary difference. Each nucleotide has a unique base (A, G, C, T, or U).
Sugar Type – DNA has deoxyribose (no –OH on the 2′ carbon), RNA has ribose (has –OH).
Presence in DNA vs. RNA:
DNA uses Thymine (T).
RNA uses Uracil (U) instead of T.
What are the names of the different types of nucleic acids?
Deoxyribonucleic acid (DNA) – Stores genetic information in cells; it is double-stranded and contains the sugar deoxyribose.
Ribonucleic acid (RNA) – Involved in protein synthesis and gene regulation; it is usually single-stranded and contains the sugar ribose.
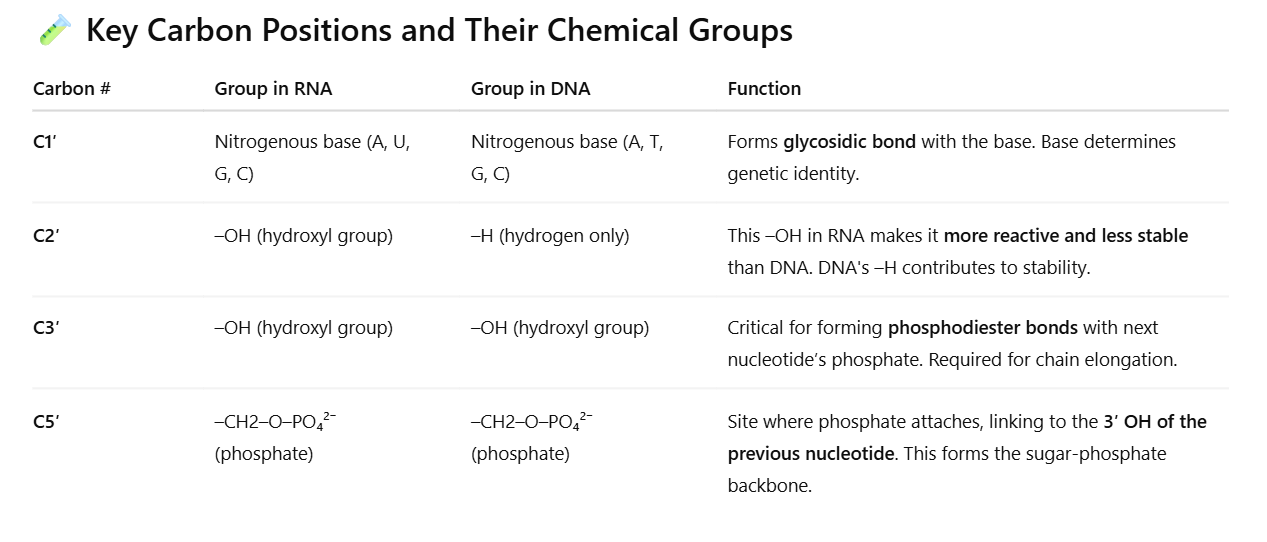
How do the nucleic acid types differ in their structure? Please make sure to discuss the chemical groups on carbon 1, 2, 3, and 5 of the sugar of a nucleotide. In addition, discuss the function of each chemical group
C1′ base attachment: Defines the genetic code (A, T/U, C, G).
C2′ OH group in RNA: Makes RNA suitable for short-term tasks like coding and catalysis (e.g., ribozymes).
C3′ OH group: Enables strand elongation during replication or transcription.
C5′ phosphate: Links to the next nucleotide, forming the backbone and giving the strand directionality (5′ → 3′).
What types of bonds are used to form a nucleic acid polymer?
1. Phosphodiester Bonds
Type: Covalent bond
Where: Between the 3′ hydroxyl (–OH) of one nucleotide’s sugar and the 5′ phosphate group of the next nucleotide.
Function: Forms the sugar-phosphate backbone of a single strand.
Result: Gives the nucleic acid strand directionality (5′ to 3′).
💡 Think of phosphodiester bonds as the links in the chain of a single strand of DNA or RNA.
2. Hydrogen Bonds
Type: Non-covalent bond
Where: Between nitrogenous bases on opposite strands of DNA.
Base Pairing:
A pairs with T (or U in RNA): 2 hydrogen bonds
G pairs with C: 3 hydrogen bonds
Function: Stabilizes the double helix structure of DNA by holding the two strands together.
Does the individual polymer have structural polarity (directionality)?
a. Polarity does not refer to chemical charges; more directionality
b. Yes; 5’ and 3’; naming is associated with what carbon is free
🔁 5′ to 3′ Directionality
Each nucleic acid strand has:
A 5′ (five-prime) end:
Ends with a free phosphate group attached to the 5′ carbon of the sugar.
A 3′ (three-prime) end:
Ends with a free hydroxyl group (–OH) on the 3′ carbon of the sugar.
🧬 Why It Matters
DNA and RNA synthesis always proceeds in the 5′ → 3′ direction, meaning new nucleotides are added to the 3′ OH end.
Enzymes like DNA and RNA polymerases recognize and act based on this polarity.
In double-stranded DNA, the two strands run antiparallel:
One goes 5′ → 3′
The other goes 3′ → 5′
What are the additional levels of organization that occur other than forming a single polymer for some types of nucleic acids? What types of bonds are needed for these additional structures?
After polymer formation, other chemical reactions that lead to structures being made
Read 5’ to 3’ when reading DNA and RNA sequences
Different layers of organization; DNA is double helix; hydrogen bond between bases
Strands are antiparallel
Chargaff’s Rule; A pairs with T, G pairs with C
Pairings allow equidistance; no bulging in backbone; energy efficient
Pairings have different number of hydrogen bonds
We mentioned that phosphodiester bonds are considered strong bonds, but they are not the strongest bonds found in the cell. Why might it be advantageous for phosphodiester bonds to be breakable by the cell?
Strong bonds; so when joined together, they’re stronger than Hydrogen bonds between bases
Strong enough not to arbitrarily break, but are able to repair or edit as needed
Helix rotates as well
Pairing causes both rotation and also necessary spacing
Most energetically favorable
If 21% of the genome contains adenine, what are the percentages of the other nucleotides?
21% adenine is 21% thymine = 42%
Difference is 58%
58%/2 = 29% GC each
If you have 2 separate DNA helices and one helix has 60% GC base pairs and the other helix has 40% GC pairs, which one can withstand more heat before denaturing
More hydrogen bonds can withstand more heat; 60% GC can withstand more because more hydrogen bonds (3)
Heat; trying to break apart bonds
GC has 3, AT has 2; AT breaks first because only 2 Hydrogen bonds
What is the common structure of an individual subunit (proteins)?
Carboxyl, hydrogen, amino acid, R varies
How many types of subunits are there? How do they differ?
20 different subunits, differ by different R groups
What types of bonds are used to form a polymer?
🔗 Peptide Bond
Type: Covalent bond
Where: Between the carboxyl group (–COOH) of one amino acid and the amino group (–NH₂) of the next.
Does the polymer have polarity?
Yes; N and C terminus; free amino group is N terminus, free carboxyl group is C terminus
We read N to C terminus
What additional level of organization may occur after polymer formation? For each level, what types of chemical interactions contribute to the organization?
Primary: Covalent/Peptide
Secondary: Non-Covalent between peptide backbone atoms, localized interactions
Tertiary: Noncovalent; between components of R groups
Quarternary: Non covalent; between different polymers; shape and charges determine the fit
Do all carbohydrate subunits have a common structure?
NO; they don’t have to. We can have branching complexes; much more complexity
Use same type of bond
What type of bond is used to join subunits into a polysaccharide?
Glycosidic
Do lipids have a common structure?
No
Do lipids form polymers?
No
What is a gene?
a. Focusing on molecular biology aspect; contained other information as well; coregulatory information; untranslated regions
b. the entire nucleic acid sequence that is necessary for the synthesis of a functional protein (polypeptide) or RNA molecule
Is a gene only protein coding information?
No it’s also controlling
Is the location of a gene strand specific?
No ; single gene will consistently be found on the same polymer
What is a genome?
a. Sequence of genetic information in DNA
b. the total genetic information of an organism (contains genes and non-coding DNA)
Is there a direct correlation between the complexity of an organism and genome size or chromosome number?
No! Size of genome doesn’t tell us a lot about it
Why is the following statement false? Most of the genome is protein-coding sequences.
1% of genome is protein-coding
What is an intron? What is an exon?
Coding regions called exons – made into protein
Noncoding sequences can be between genes or within genes (introns).
noncoding sequences can function as regulatory sequences (not just “junk DNA”)
messenger RNA
contain sequences that help tell the machinery which of the exon needs to be removed
What is meant if we say that there is sequence homology between organisms? From an evolutionary standpoint, does the presence of homology suggest anything?
a. Homology; compare between organisms we can see similar nucleotide pairings; highly conserved/highly homologous
b. If similar homology; similarly related
c. Regions where its still conserved; something important about those sequences
Are all regions of homology within exons?
No, some are intron
What are SNPs?
Single nucleotide polymorphisms; we can have differences between our population
What are three DNA sequences required for eukaryotic chromosome stability?
Protect ends (telomere)
Replication origin; where its replicated
Centromere; separate DNA after its been copied; one chromosome will go to the daughter we are making (correctly separate copied information)
Are chromosomes organized within the nucleus?
Yes within interphase; we know there are different stages of cell cycle
During interphase, DNA is most extended, not so compacted. There is organization
Example; some type of membrane protein and nucleolus; site of ribosomal RNA genes
A phosphodiester bond forms from a reaction between the_________.
3' phosphate of one nucleotide and the 5' hydroxyl of another nucleotide
5' phosphate of one nucleotide and the 3' hydroxyl of another nucleotide
5' hydroxyl of one nucleotide and the 3' hydroxyl of another nucleotide
5' phosphate of one nucleotide and the 3' phosphate of another nucleotide
5' phosphate of one nucleotide and the 3' hydroxyl of another nucleotide
Which of the following is a TRUE statement about nucleic acids?
DNA polymers will contain Uracil nucleotides
RNA nucleotides are RNA nucleosides with a phosphate group associated with the C3 carbon of the sugar ring
DNA nucleotides will have a hydroxyl group on the C2 carbon of the sugar ring
RNA nucleosides are made of a pentose sugar with a nitrogen base attached to the C1 carbon.
RNA nucleosides are made of a pentose sugar with a nitrogen base attached to the C1 carbon.
All of the following are essential to the stability of a eukaryotic chromosome except ______________.
Origins of Replication
Introns
Telomeres
Centromeres
Introns
All of the following may be used to describe regions of heterochromatin except ___________________.
Gene poor
Transcriptionally active
Highly condensed
Contains repetitive DNA sequences
Transcriptionally active
Which of the following would be considered the basic unit of eukaryotic chromatin structure?
gene
nucleotide
nucleosome
telomere
nucleosome

hydrogen bond
amino group
secondary protein structure
R group
antiparallel
All of the following biomolecules, except _________, are built from smaller organic molecules, or monomers, that link together to form larger polymers.
lipids
proteins
nucleic acids
carbohydrates
lipids

50%
cannot be determined

Answer 1:
TRUE
Answer 2:
FALSE
Answer 3:
TRUE
Answer 4:
FALSE
Answer 5:
TRUE
Answer 6:
FALSE
From each drop down, select the answer the will complete a TRUE statement.
a.["Facultative", "Quiescent", "Constitutive", "Active"] euchromatin refers to regions of the genome that are not currently being transcribed but retain the potential to become active.
b.[Constitutive, Facultative] heterochromatin can become transcriptionally active under certain conditions.
c. The chemical breakdown of a polymer and removing a monomer is called ["condensation", "hydrolysis"] .
d. The sugars in a single carbohydrate polymer are held together by ["peptide", "glycosidic"] bonds.
e. Mechanisms for regulating chromatin states must be ["reversible", "irreversible"] .
Answer 1:
Quiescent
Answer 2:
Facultative
Answer 3:
hydrolysis
Answer 4:
glycosidic
Answer 5:
reversible