Cell Bio Exam 3
1/120
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
121 Terms
What Is the Endomembrane System?
Group of organelles that have membranes associates with eachother and proteins move via vesicles
What organelles are in the endomembrane system? what are not?
nuclear membrane, ER, Golgi, lysosomes, endosomes, and peroxisomes. excluded mitochondria and chloroplasts
What does it mean for protein delivery if an organelle is in the endomembrane system?
If included proteins eneter ER then are moved by vesicles to reach Golgi, lysosome, membrane or are secreted.
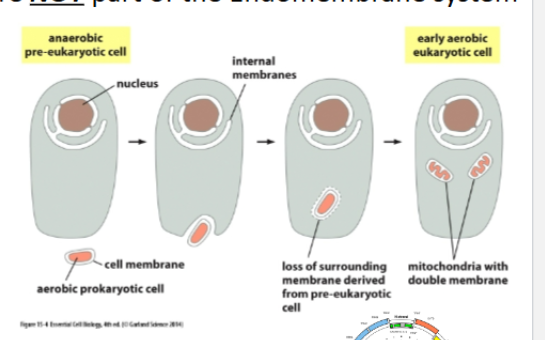
Why aren’t Mitochondria and chloroplasts Part of the Endomembrane System? (Endosymbiont Theory)
Mitochondria and chloroplasts were once prokaryotes that were engulfed by eukaryotes
We know this because they have:
Two membranes (original and plasma)
Circular DNA (seen in prokaryotes)
Divide by binary fission
Prokaryotic-like ribosomes
Cardiolipin in membranes (seen in prokaryotes)
Therefore, they evolved separately and use their own import machinery
What does it mean for protein delivery if an organelle is not in the endomembrane system?
They require a different system to import proteins besides vesicles. Nuclear proteins: enter through nuclear pore, Mitochondrial proteins: enter via TOM/TIM channels
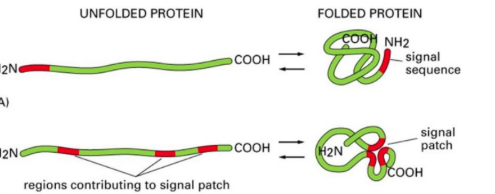
what is a signal sequence?
direct protein to correct location and typically cleaved off once protein reaches destination.
what must a signal sequence be?
nessacary (needed to get to correcy location) and sufficent (correct location)
what is a signal patch?
signal sequence that is only active when folded
what is the nuclear pore complex and why is it needed
gate through which molecules enter and exit the nucleus
needed to tightly regulate what comes in because DNA is in nucleus
What is the nuclear localization signal (NLS)
A signal that proteins bound for the nucleus have
formed by lysine/arginine positive charge
forms a signal patch (created in a folded protein)
Is the NLS removed?
No, needed repeatly during cells life
are nuclear import proteins folded or unfolded?
folded because of the signal patch
helper proteins of nuclear import? (4)
Importin (Nuclear Transport Receptor)
Binds NLS in the cytoplasm for import
moves cargo through nuclear pore
Nuclear Pore Complex (NPC)
Allows import of large proteins only when bound to importin
Ran-GTP (inside nucleus)
Binds importin → forces cargo release
Ran-GDP (in cytosol)
Form created after GTP hydrolysis
Cannot remain bound to importin
what is the energy soucre of nuclear import
GTP hydrolysis by Ran drives directionality.
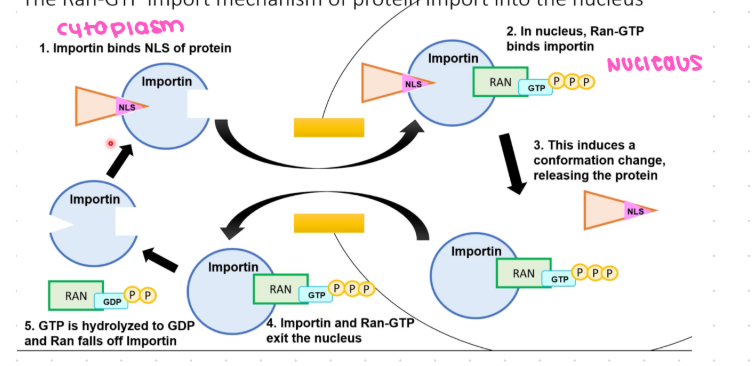
steps of nuclear import
importin binds to NLS on folded cargo protein in cytosol
Importin undergoes conformational change, so it is recognized by fibrils
Receptors direct cargo protein to the nuclear pore by interacting with fibrils
compound goes into the pore complex
Inside the nucleus, Ran GTP binds to importin
importing releases cargo protein back to the cytoplasm
Importin and Ran GTP exit the nucleus
Ran hydrolases GTP to GDP in cytosolthe
Ran falls off and releases importin and repeats processthe
what id during nuclear import
nls is mutated
defective importin
Ran GTP/GDP cycle fails
cargo stays in cytosol
no transport
import stops because cargo cant unload and recpetor cant recyle
Where are nuclear proteins made?
On free cytosolic ribosomes.
Where are mitochondrial proteins made?
On cytosolic ribosomes.
Signal Sequence for mitochondria
needed
positive charges (argiine/lysine)
N terminus
when protein enter mitochrondia are they folded or unfolded?
unfolded (linear) because pores are narrow, chaperones help keep unfolded
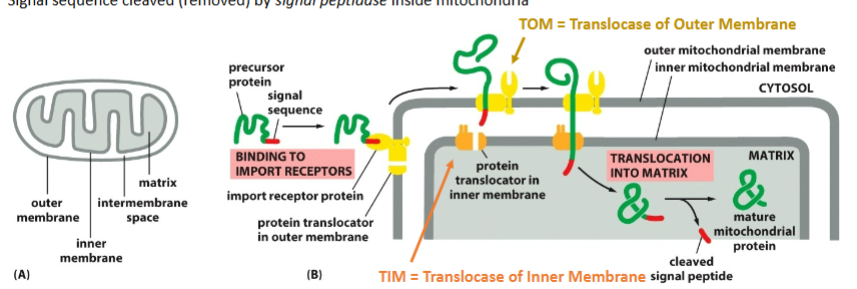
Helper Proteins for mitchondrial import? (5)
Receptor protein on outer membrane — recognizes the signal.
TOM complex (Translocase of Outer Membrane)
First channel protein encounters.
TIM complex (Translocase of Inner Membrane)
Pulls protein into matrix.
Chaperones
Keep the protein unfolded
Pull the protein inside
Signal Peptidase
Removes the N-terminal signal once inside
Energy Source of mitchondrial import
ATP for TIM
Is the signal cleaved in mitochondrial import?
Yes removed by signal peptidase
steps for mitchondrial import?
signal recognized by the repressor on the outer membrane
Translocases of the outer membrane (TOM) get protein out of the outer membrane
TIM (translocase of the inner membrane) gets protein out of the inner membrane to the matrix, which needs ATP
When Tom finds Tim, the protein goes through the channel
Once the matrix protein is folded and the signal sequence is cleaved
effects of possible defects
1- Mutated signal sequence
2- Defective TOM/TIM
3- Chaperone failure
1- can’t bind the receptor, so it stays in the cytosol
2- protein stuck between membranes
3- protein folds and can’t enter
What is the significane of ER import
entry into endomembrane system, ER → Golgi → lysosome → endosomes → plasma membrane → secretion.
where do ER bound proteins start?
On free cytosolic ribosomes
When signal appears, ribosome is moved to the ER to finsih translation
ER signal sequence properties
nonpolar hydrophic amino acis
near N terminus
are proteins folded or unfolded during er import?
Imported as a linear chain during translation
- Folding happens INSIDE the ER lumen
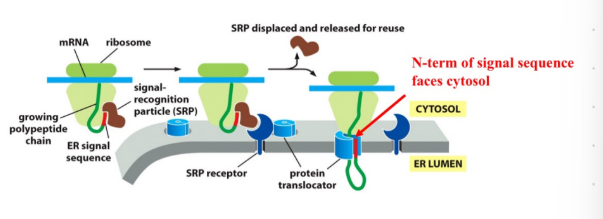
helper proteins for ER import (5)
SRP (Signal Recognition Particle)
Binds ER signal
Pauses translation
SRP Receptor
Located in ER membrane
Accepts SRP+ribosome complex
Translocator Channel (Sec61)
Opens to allow polypeptide to enter the lumen
Signal Peptidase
Cuts signal off for soluble proteins
ER Chaperones
Help folding inside ER
Prevent misfolding/unfolded protein response
energy source for ER import
GTP
is signal cleaved for er import
YES → for soluble ER proteins
NO → for many transmembrane proteins (internal start-transfer sequences stay embedded)
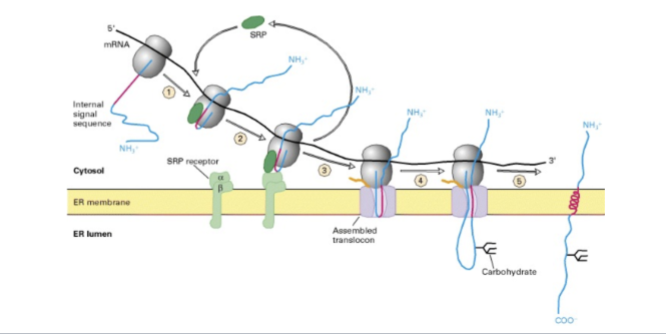
er import steps
Ribosomes begin translation in the cytosol.
ER signal emerges, the SRP binds and pauses translation
SRP brings the ribosome to the SRP receptor on the ER
ribsomeones transferred to the tranlocator
SRP leaves and the polypeptide chain is threaded into the lumen via the translocation channel, and translation continues in the ER lumen
Signal may be cleaved by signal peptidase.
Protein folds with the help of ER chaperones.
Post-translational modifications occur:
N-linked glycosylation (on Asn)
Disulfide bond formation (unique to ER lumen)
What happens if these are defective inthe ER import
No SRP
Translocator broken
Signal peptidase missing
Chaperone failure
The protein continues in the cytosol, reaching the wrong location
Proteins cant enter ER
signal not removed, protein misfolded
unfolded protein response ER stress
single pass (N terminus in lumen)
N-terminal signal starts transfer
Stop-transfer anchors protein
Signal is cleaved
Result:
N inside lumen
C in cytosol
transmembrane domain embedded
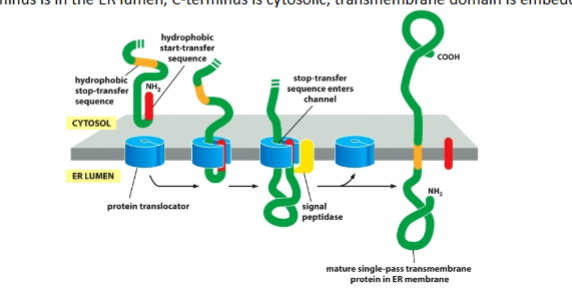
single pass (cytoplasmic N terminus)
Internal start-transfer
Not cleaved
Result:
N in cytosol
C in lumen
transmembrane domain embedded
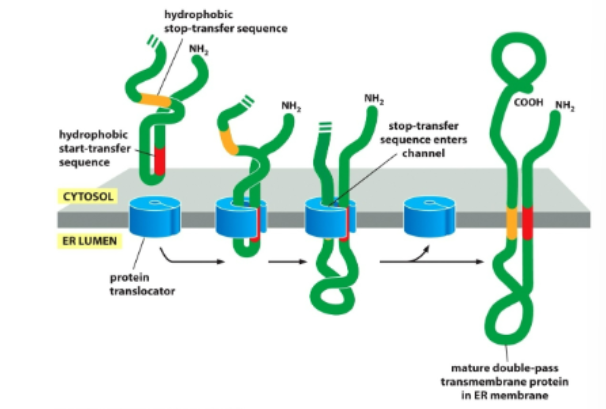
double pass/ more
N and C terminus in cytosol
Repeated start-transfer and stop-transfer sequences create multiple membrane spans.
Orientation depends on the first start-transfer.
Intenral start-transfer signal sequence never removed
Where does glycosylation occur?
ER lumen (N-linked - lumen side) → modified in Golgi
sugar covalenly bonded = glycoprtiens
occurs as protein is being translated and translocated
Disulfided bond formation
occurs in ER lumen (oxidizing) between cysteine amino acid
protects teritrary structure
what is direction of secretory (exocytic) pathway?
out of cell: goes outward from cell interior :ER → cis-Golgi → trans-Golgi → plasma membrane → secretion
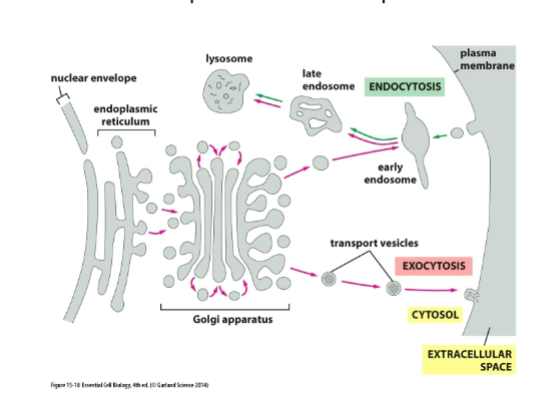
purpose of exocytosis
bring proteins to the cell membrane
let’s specialized cells respond to the environment
allows proteins to communicate with proteins outside of cell
cell replaces membrane every 30 mins
purpose of endocytic pathway?
Bring nutrients inside
Remove old receptors
Balance membrane added by exocytosis: fulid balance
defend against pathogens
how do vesicles form?
budding
three basic steps of budding process
picking the correct cargo
building a coat around the budding vesicle
pinch off the vesicle
how does the vesicle pick the correct cargo
Cargo doesn’t randomly fall into vesicles — it’s selected.
Cargo receptors grab specific soluble proteins
Adaptins connect these receptors to coat proteins
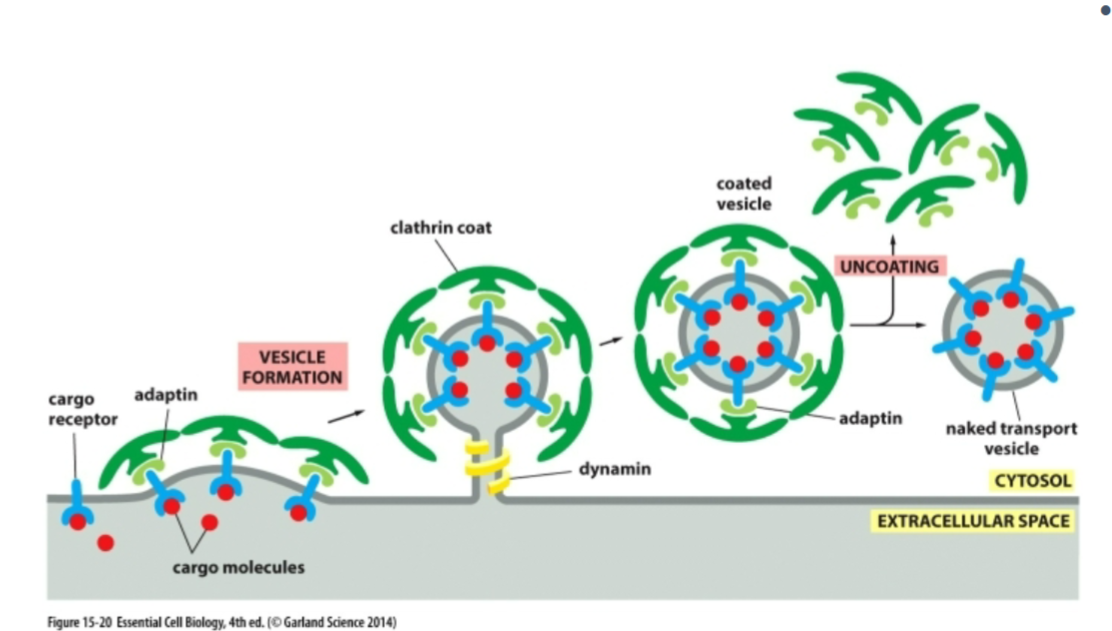
why does the cell build a coat around the budding vesicle? what are the three main types?
Coat proteins give the vesicle curvature and identity.
clathrin, COPII, COPI
what do cargo recpetors do
bind specific proteins via signal
what does adaptins do
connect cargo recpetors to coat proteins
players or clathrin-coated vesicles?
cargo receptors (bind soluble cargo)
Adaptin (links receptors to clathrin)
Clathrin (forms outer coat)
Dynamin (GTPase that pinches off vesicle)
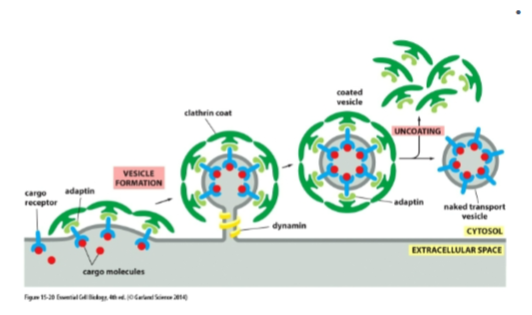
pathway of clathrin coated vesicle
Golgi → endosome; PM → endosome)
pathway of COP coated vesicle (1 and 2)
1: Golgi → ER or Golgi → Golgi
2: ER → cis-Golgi
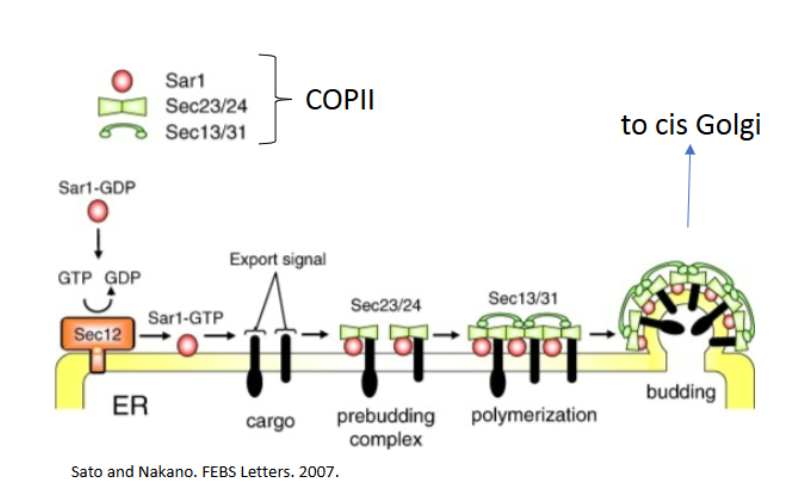
what does COPII coated vesicles do?
transport from ER to cis golgi
COPII coated vesicles players
Sar1 (GTPase)
GDP form = inactive in cytosol
GTP form = inserts into ER membrane
Sec23/24 = inner coat + selects cargo
Sec13/31 = outer coat
No dynamin required
how do vesicles know where to go (identity)?
every vesicle has a molecular ID code so it fuses with the correct membrane.
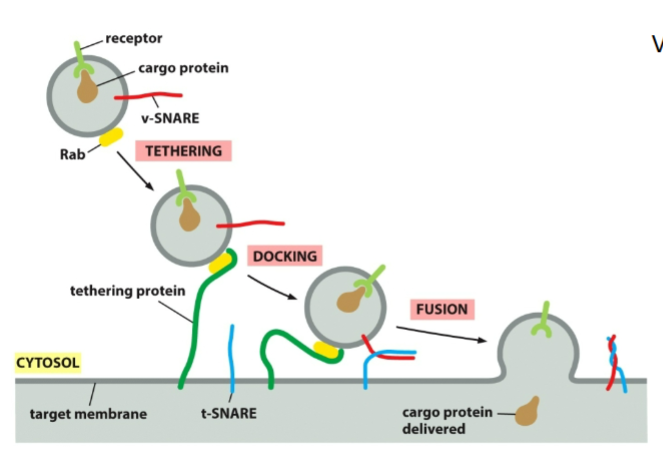
Purpose of Vesicle Budding
Move proteins between endomembrane organelles
Deliver membrane components
Select cargo with high specificity
Maintain flow between ER → Golgi → PM
Feed lysosomes during endocytosis
Rab proteins
Small GTP-binding proteins
Bound to vesicle membranes
Each membrane type has a unique Rab
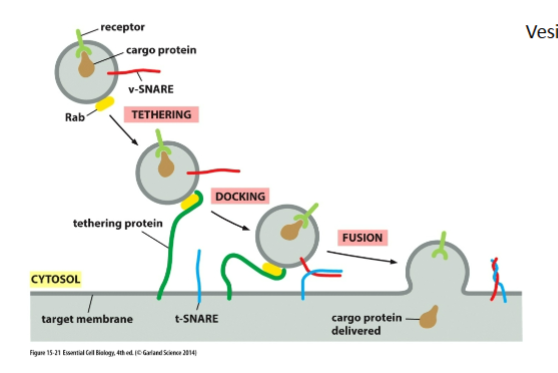
Tethering proteins
On target membrane
Recognize correct Rab
Bring vesicle VERY close (~1.5 nm)
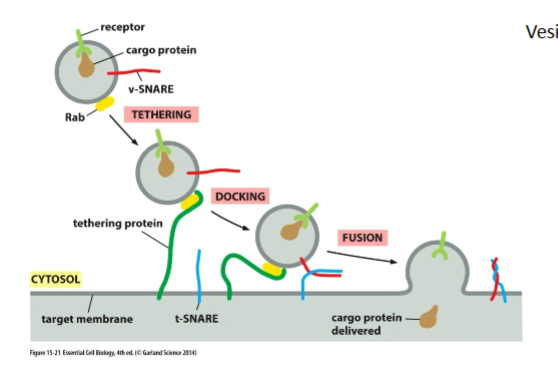
SNAREs
v-SNAREs on vesicle
t-SNAREs on target
Form a 4-helix bundle to “zipper” membranes together
Squeeze out water → allow membrane fusion
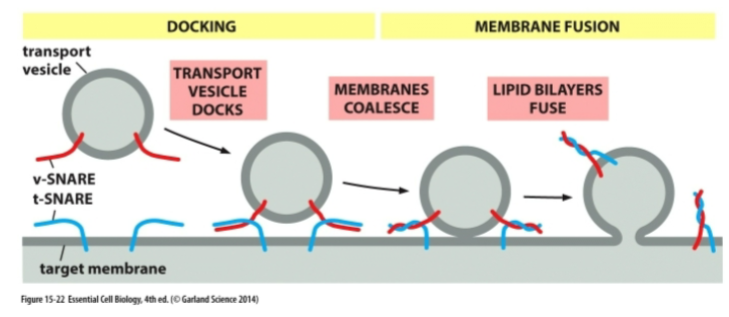
mechanism for vesicles fusing with target membrane
Vesicle arrives at destination using microtubules + motor proteins
Rab–tether interaction docks vesicle
v-SNARE + t-SNARE wrap tightly, pulling membranes very close
Water is forced out → membranes fuse
Vesicle collapses into target membrane
NSF + SNAP use ATP to separate SNAREs for reuse
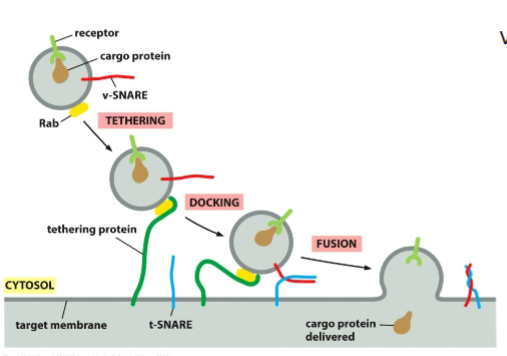
is vesicle fusion engertically favorable?
no. Fusion is energetically UNFAVORABLE because two membranes must come extremely close.
whta if Rab mismatch with tether
vesicle docks at wrong organelle or fails to dock → NO fusion.
what if v snare is mutated
Docking happens, fusion cannot occur → vesicle accumulates.
what if clathrin is missing
Receptor-mediated endocytosis fails
what if sar1 cant bind to GTP?
COPII coat cannot form → proteins trapped in ER.
what is Unfolded Protein Response (UPR)
UPR is a protective mechanism activated when misfolded proteins build up in the ER and there are not enough chaperones. Er is clogged bc chaperones are overwhelmed, and quality control can’t keep up.
what happens during UPR
Cell makes more chpaerone to fix misfolded proteins, protein synthesis slows down, er grows to give more space for protein folding.
why do cells need UPR
To prevent the ER from being damaged by misfolded proteins.
Without UPR, misfolded proteins would:
clog the ER
create toxic aggregates
disrupt the entire endomembrane system
Constitutive exocytosis
occurs in all cells all the time
unregulated
supplies newly made lipids and proteins to the plasma membrane
carries secreted proteins which are released to the outside of cell
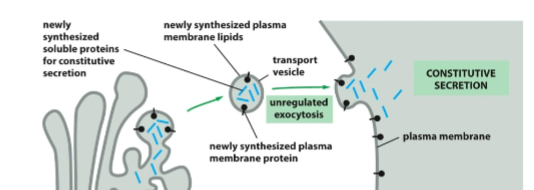
what does Exocytosis deliver
transmembrane proteins to the plasma membrane in correct orentations
realses solu proteins to the extracellular environment
new phospolipids
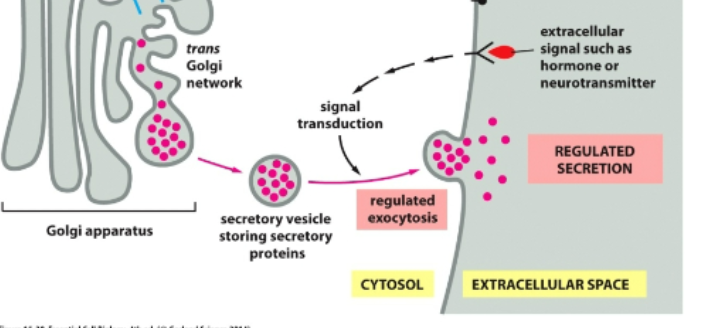
regulated secretion
Specialized secretory cells
Stored in vesicles wating for signal
Released only when triggered
Example: insulin release
endocytosis pathway
PM → vesicle → early endosome → late endosome → lysosome
3 types of endocytosis
pinocytosis, phagocytosis, Receptor-Mediated Endocytosis
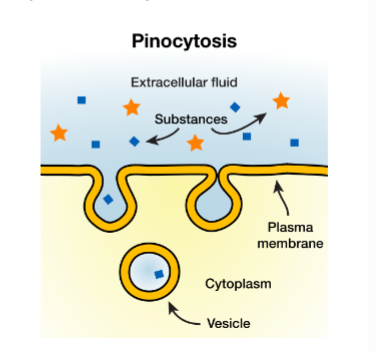
Pinocytosis
“cellular drinking”
constituve (always on)
ingest parts of membrane w fluid that has small particles in it
balances fluid loss in exocytosis
Feeds lysosomes
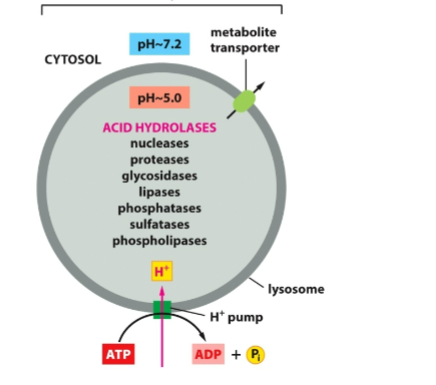
why is lysosome unique?
enzymes only function inside lysosme due to low pH
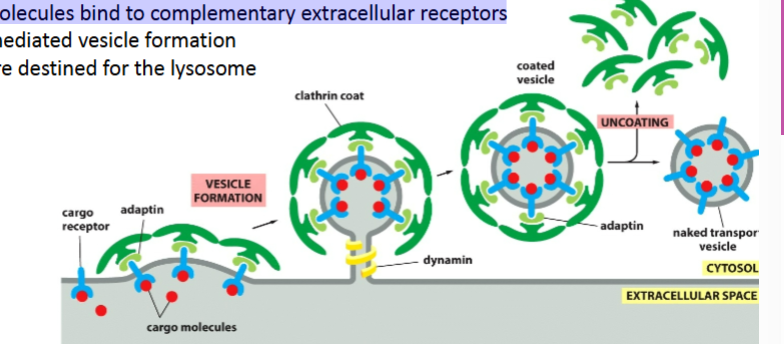
receptor mediated endocytosis
Specific cargo injesyed
Requires clathrin (receptor selects specific cargo)
Vesicles are destined for the lysosome
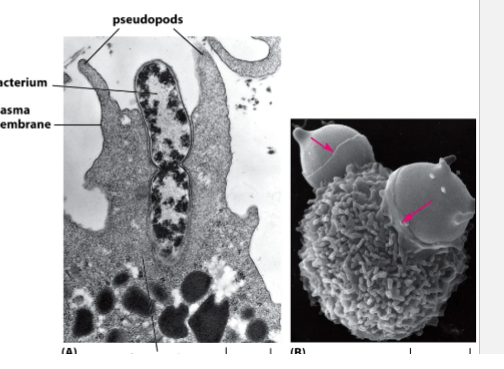
phagocytsois
Large particles (bacteria, debris)
Performed by specialized cells (immune system)
Defense: neutrophils/macrophages kill pathogens
where does core-oligosaccharide modification occur
er
where is Mannose-6-phosphate (M6P) tag added, what does it do
cis golgi. Tells the cell: “This protein belongs in the lysosome”
how is mgp recongized
M6P receptor (M6PR) binds M6P-tagged hydrolases
M6PR packages them into clathrin-coated vesicles headed to endosome → lysosome
what pathway do lysosomal hydrolases follow?
ER → cis-Golgi → trans-Golgi → endosome → lysosome
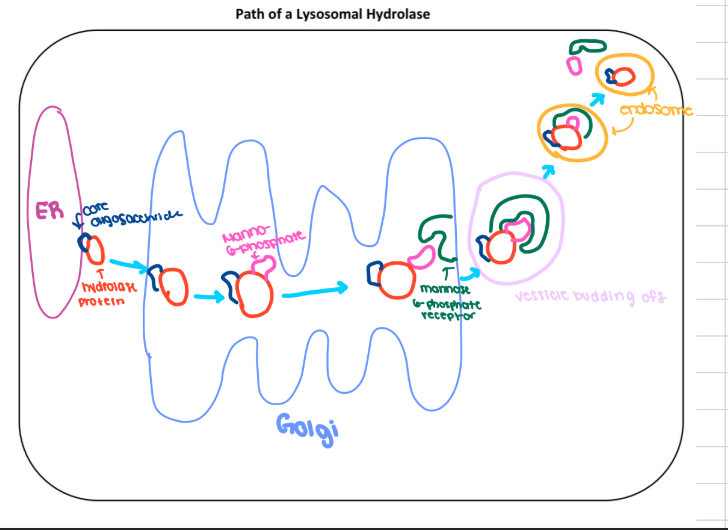
steps of lysosome pathway
core oligosacchride added in er to hydrolase
enters cis golgi and m6p adds
m6p receptor recongized it in transgolgi
vesicle buds off and enters endosome
then enetrs lysomes and m6p and m6p receptor recyled

what is chondrocyte and purpose
Only cell type in cartilage
Produce & maintain extracellular cartilaginous matrix: allows joins to move
Cartilage cushions joints, shapes bones & facial features
what is Fibroblasts and purpose
Connective tissue cells sampled in Ana’s test
Used in lab assays because they contain lysosomes
Why do LSDs affect cartilage?
Chondrocytes get swollen, stop making matrix
Cartilage becomes thin, weak, underdeveloped
explains Ana’s:
Hip/knee stiffness
Club feet
Facial abnormalities
Reduced mobility
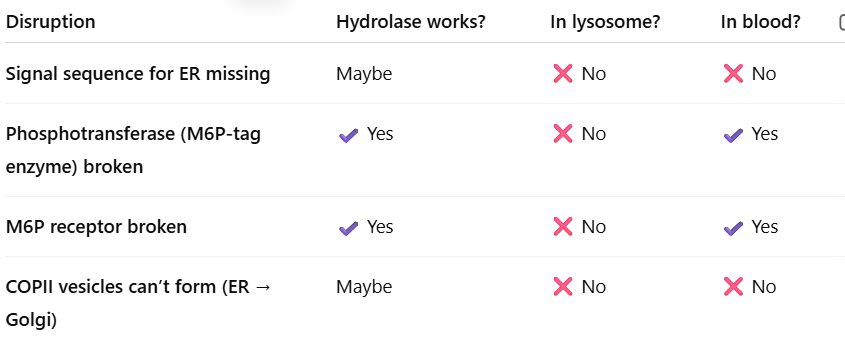
Why do Ana’s lysosomal hydrolases show up in her BLOOD?
Her blood tests showed extra hydrolase activity, and her fibroblasts had low activity (lysosomes empty
This means:
Enzymes never reached the lysosome
Enzymes were secreted outside the cell instead
This only happens when the M6P tag is missing OR M6P receptor can’t bind (only one enzyme tags them so if it doesnt work none are tagged)
what happens if M6P tag is missing
the Golgi thinks hydrolases are just normal secreted proteins → sends them out of the cell → they end up in the bloodstream.
What went wrong in Ana?
mutation in the enzyme(GNPTAB) that adds M6P tag on hydrolase.
Without M6P → NOTHING gets to the lysosome
EVERYTHING gets secreted
be familar w this
What are Intermediate Filaments?
One of the three cytoskeletal filaments (MT, Actin, IF)
Most stable filament: tp take on stress
Rope-like → stretchy + strong
Found mostly in animal cells under mechanical stress:
epithelial cells
neurons
muscle cells
IF monomer Structure
One polypeptide
Has head (N) and tail (C)
Middle = α-helical rod

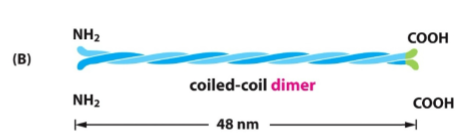
IF dimer Structure
Two monomers twist together
“Coiled-coil”
Held by hydrophobic interactions
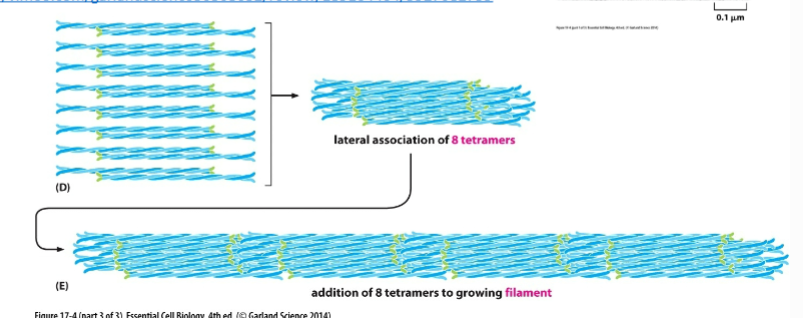
IF Tetramer structure
Two dimers anti-parallel (opposite directions)
Ends look the same → NO polarity

Final IF strucute
8 tetramers pack together
Twist into a rope-like cable
Rope can stretch → gives strength
Won’t break easily
Why can IFs withstand mechanical stress?
Rope-like structure distributes tension
Can shift from α-helix to β-sheet when stretched
Accessory proteins help cross-link them
Cytoplasmic IFs (3 types)
Keratin Filaments (epithelial cells)
Vimentin Filaments (connective tissue)
Neurofilaments (neurons)
Keratin Filaments (epithelial cells)
Most diverse IF family (54 genes)
Found in skin, hair, nails
Give tensile strength
Link to other cells through desmosomes
Disease: epidermolysis bullosa simplex
Mutated keratin → skin blisters from minor friction
Vimentin Filaments (connective tissue)
Found in bone, muscle cell
Job = anchor organelles (nucleus, mitochondria, ER)
Neurofilaments (neurons)
Found in axon cytoplasm
Provide space-filling properties → increase axon diameter
Nuclear IFs (Lamins)
Form nuclear lamina under nuclear envelope
Provide nuclear shape + stability
Regulate cell division:
Phosphorylate → disassemble in early mitosis
Dephosphorylate → reassemble in late mitosis
Which cells contain which IFs?
Epithelial cells → Keratin
Muscle cells → Vimentin-like IFs
Neurons → Neurofilaments
All animal cells → Nuclear Lamins