PHSI3009: Module 2 - Cell Signalling
1/59
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
60 Terms
List the 2 types of input into a cell
Contact (e.g. gap junctions, eph/ephrin, integrin)
Diffusible (e.g. peptide hormones, lipid hormones, gas - NO)
more controversial examples: ions and osmotic gradients, molecules (e.g. glucose)
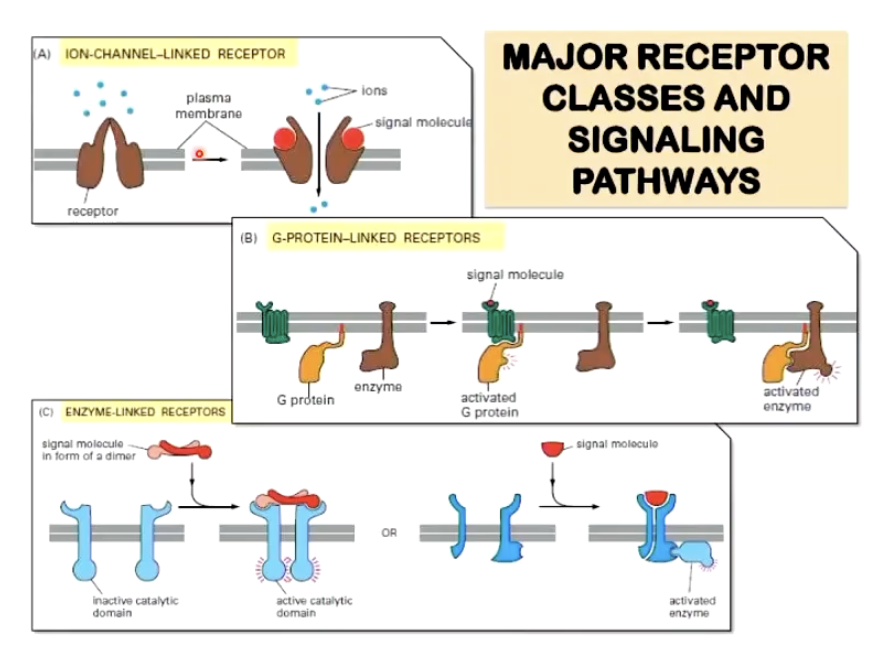
List the 3 major receptor classes and signalling pathways
Ion-channel-linked receptor: moves ions between cells
E.g. GABAa
G-protein-linked receptor: modulating something that diffuses inside a cell (tends to be immediate response)
E.g. GPCR - control of exocrine secretion
Enzyme-linked receptor: triggers reactions (tends to be longer-term response)
E.g. tyrosine kinase receptor - insulin receptor
What is an example of clinical relevance of contact signalling?
connexin expression regulates tumour progression (e.g. 46, 43)
What is an example of clinical relevance of receptor signalling?
Insulin resistance in diabetes resulting from receptor dysfunction
What are intracellular membrane compartments and their signifiicance?
Intracellular membrane compartments are membrane-bound organelles that allow for the spatial and temporal separation of incompatible biochemical processes.
each membrane bound organelle has a unique composition of proteins and unique function → separates eukaryotic cells from prokaryotic cells
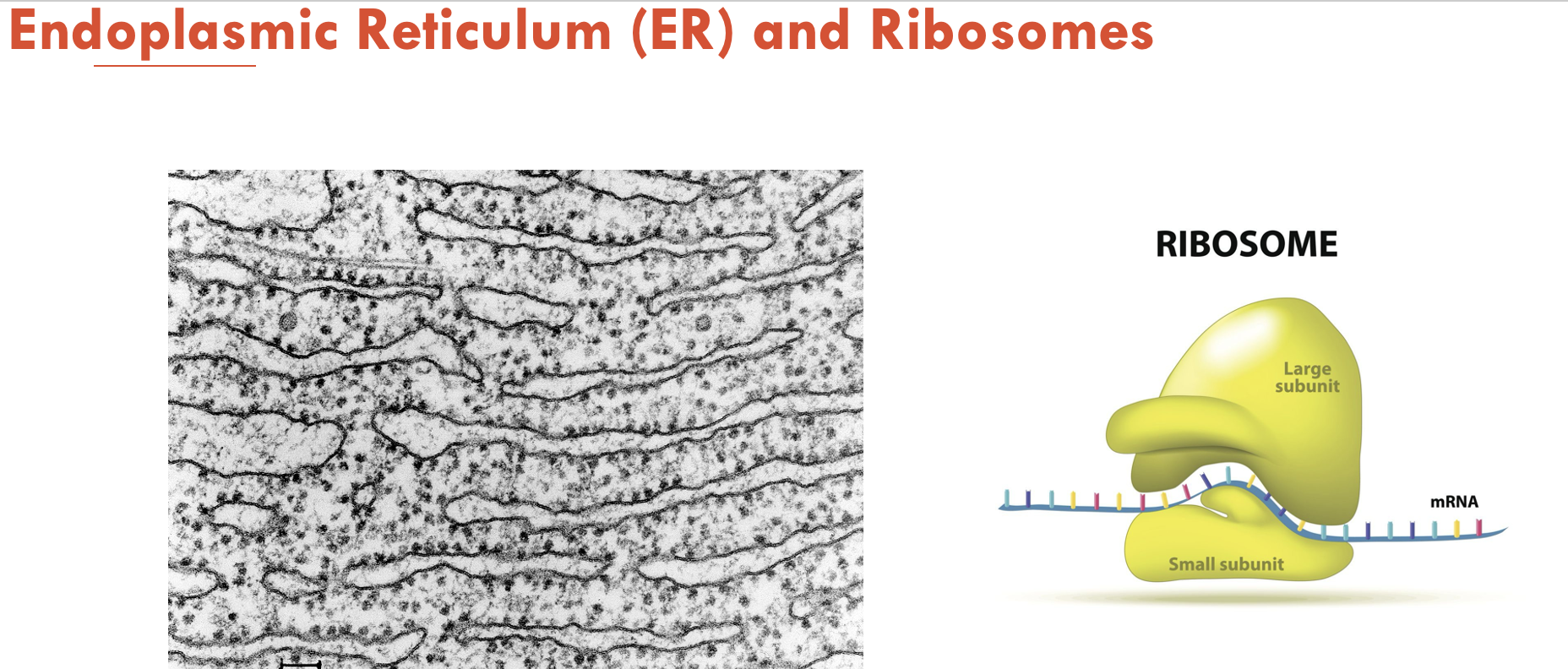
Protein synthesis is initiated on (A - part of cell) which may be found (B - 2 locations). Smooth ER is different to rough ER in that (C)
A - ribosomes (each made of one large and one small subunit)
B - free floating in the cytoplasm or bound to form a complex in rough ER
C - it is not associated with ribosomes
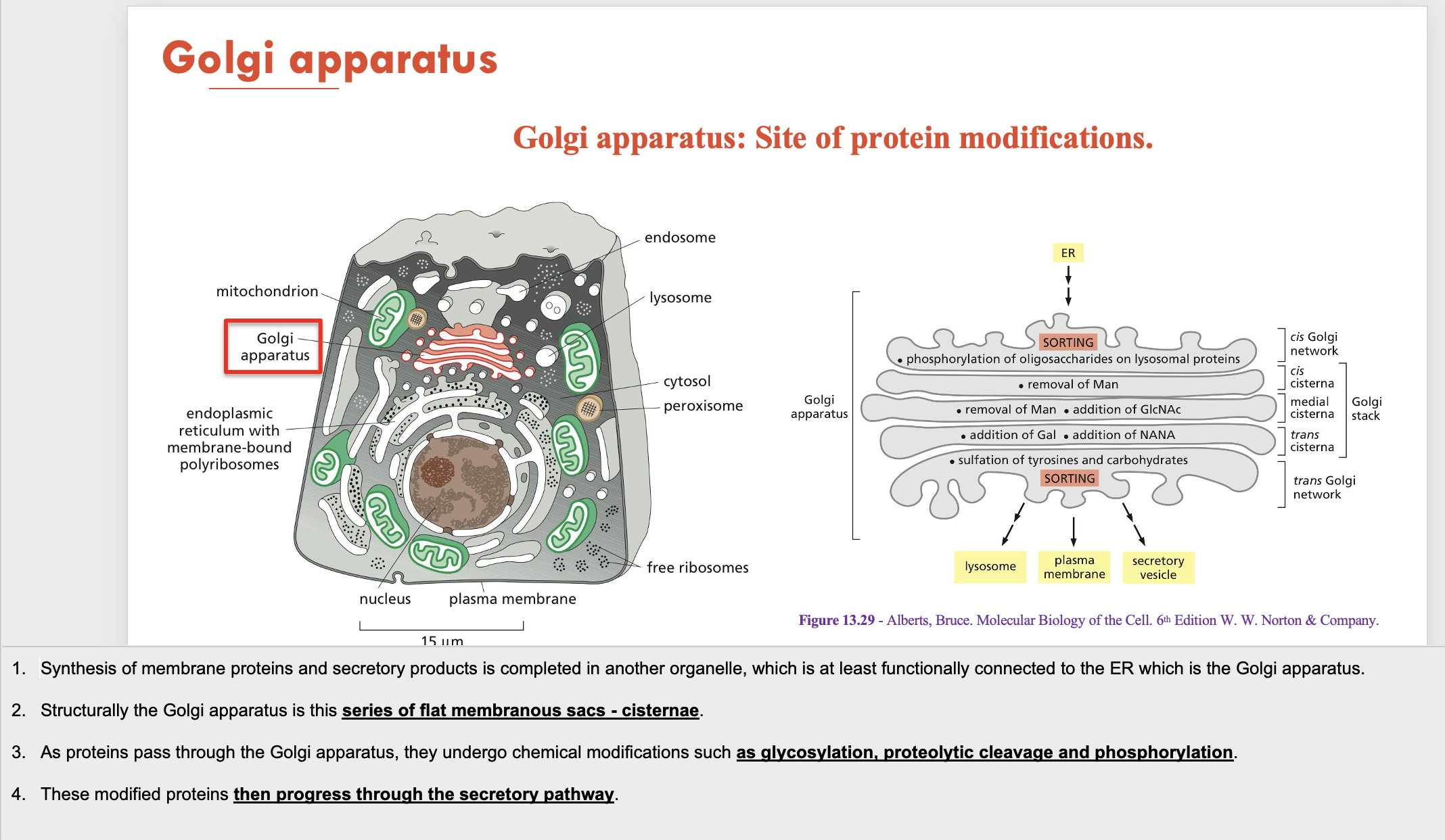
Golgi apparatus is structurally (A - composision) and is the site for (B - function). Modified proteins from the Golgi then move towards the (C). The Golgi is also responsible for the generation of (D - part of cell)
A - a series of flat membranous sacs (cisternae)
B - protein modification (glycosylation, proteolytic cleavage and phosphorylation)
C - secretory pathway
D - lysosomes
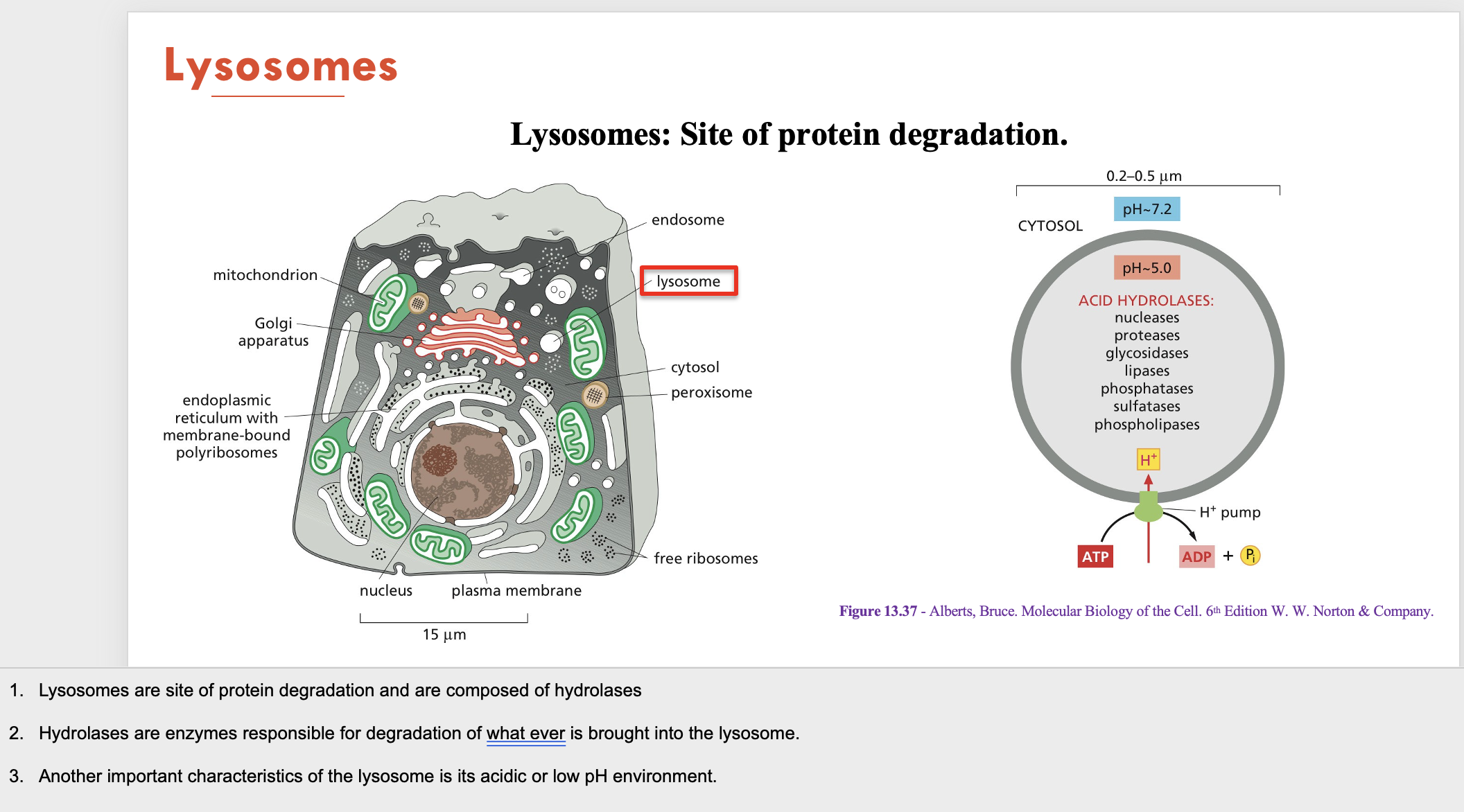
Lysosomes are composed of (A) and are the site of (B - function). Its important characteristic is having a (C)
A - hydrolases (degradation enzymes)
B - protein degradation
C - acidic environment (low pH)
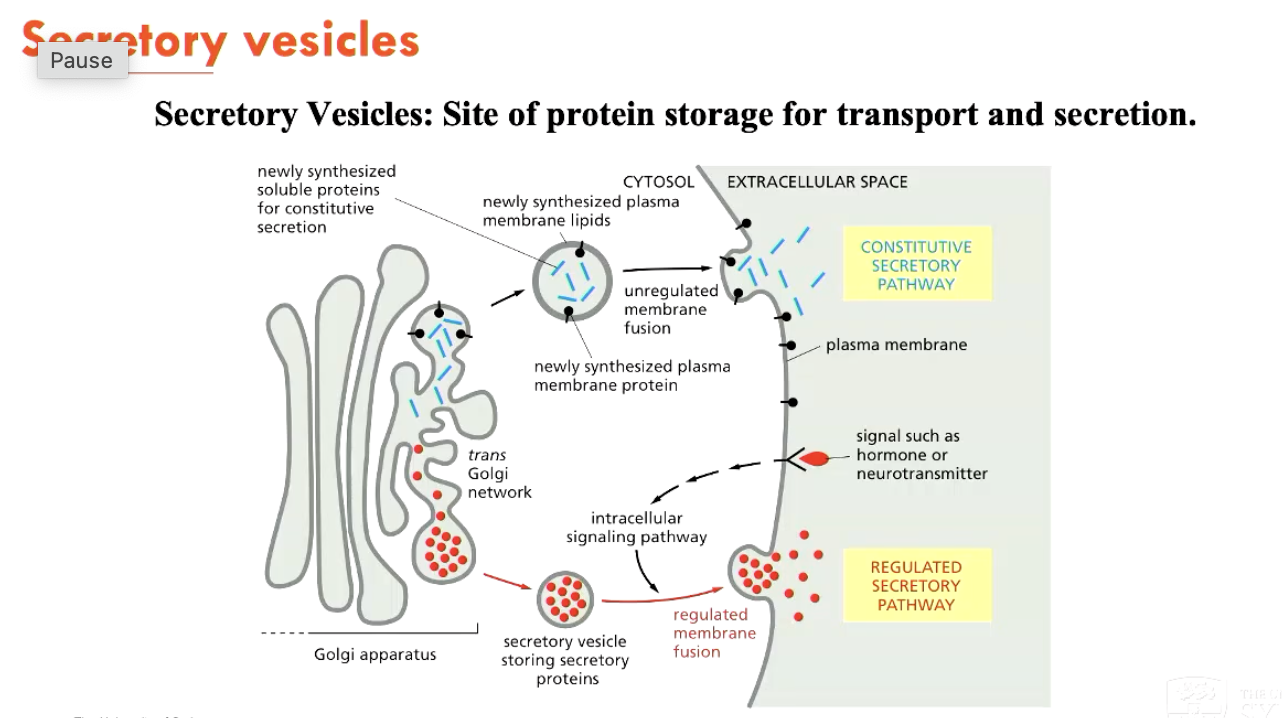
Secretory vesicles are responsible for…
Protein storage for transport and secretion
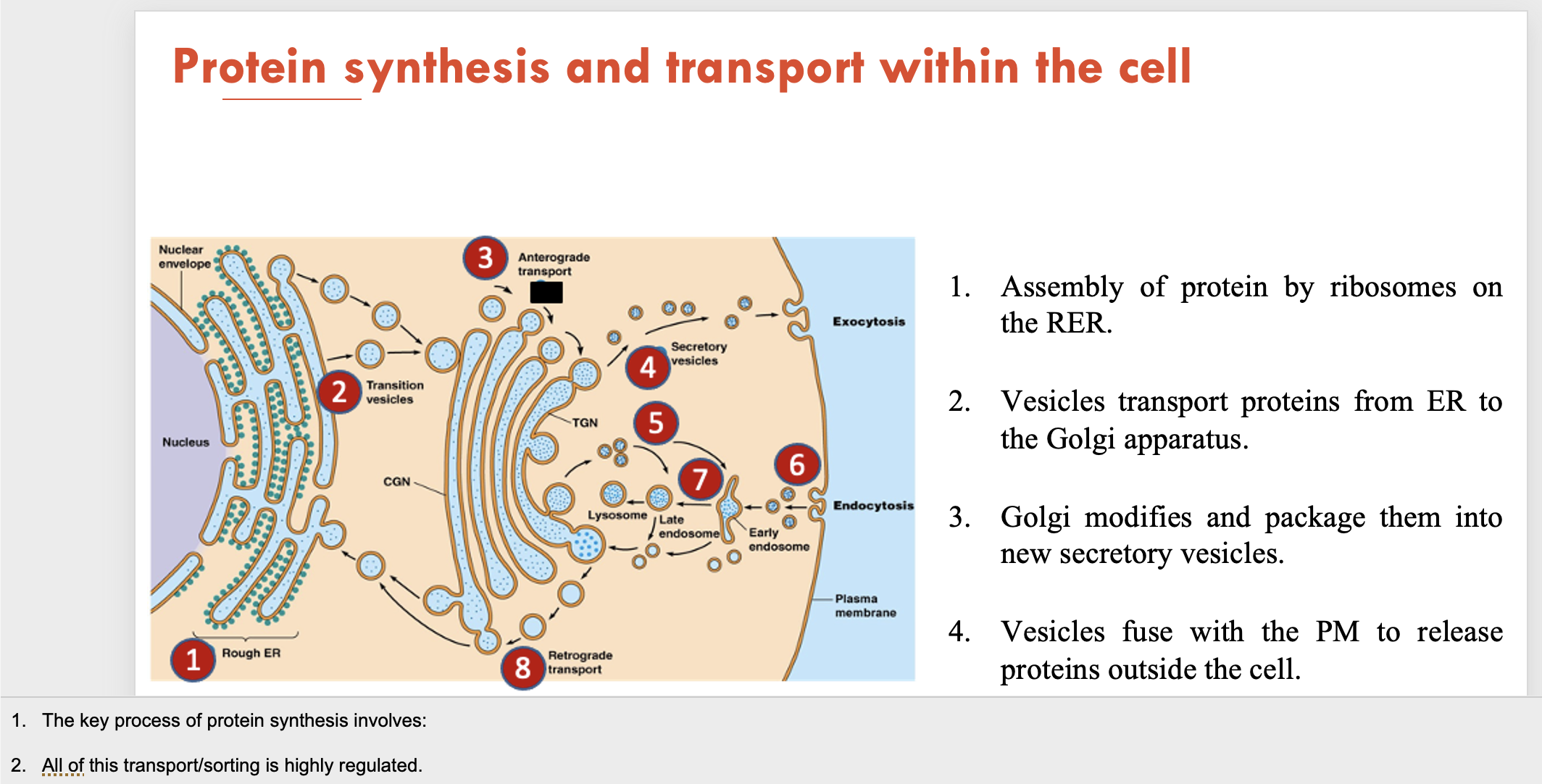
Describe the traditional pathway for protein synthesis and transport within a cell (4)
Assembly of protein by ribosomes on the RER
Vesicles transport proteins from ER to the Golgi apparatus
Golgi modifies and package them into new secretory vesicles
Vesicles fuse with the plasma membrane to release proteins outside the cell
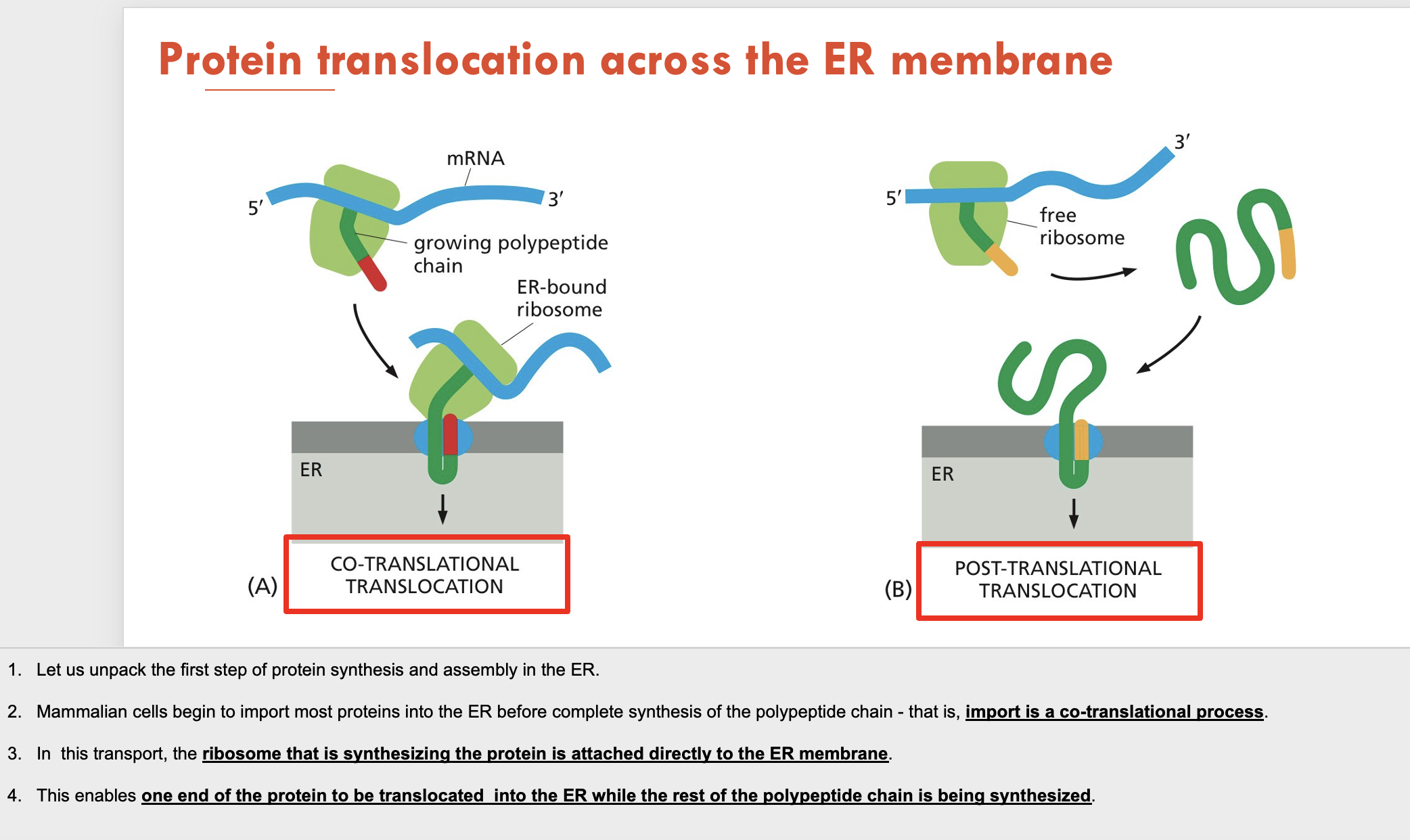
What 2 types of protein translocation can occur across the ER membrane?
Co-translational translocation: protein is synthesised while bound to the ER
Post-translational translocation: protein is synthesised in a free ribosome then moved to the ER
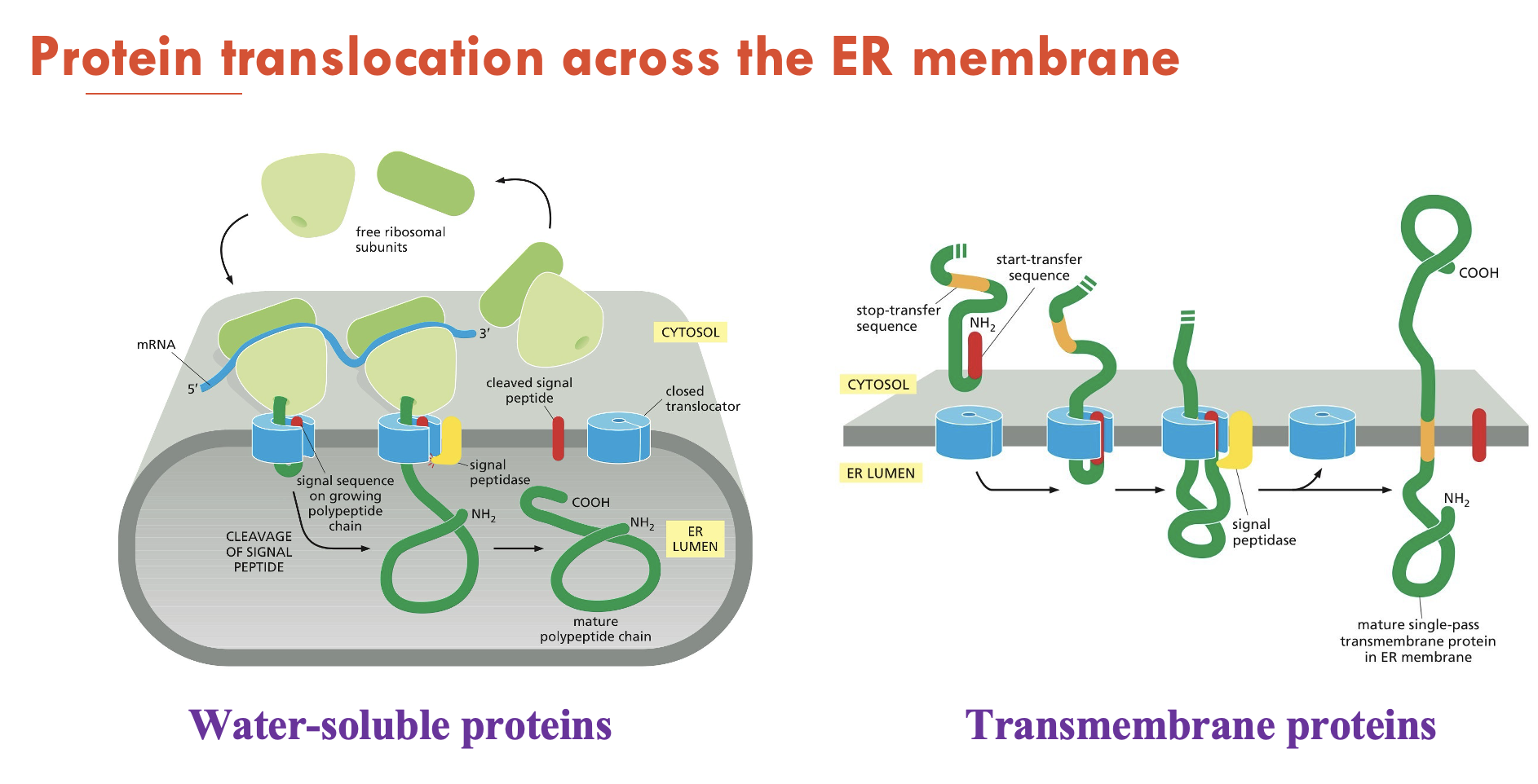
What are the 2 types of proteins that can be translocated into the ER?
Water-soluble proteins: fully translocated across the ER membrane and released into the ER lumen
Transmembrane proteins: only partly translocated across the ER membrane and become embedded in it
What 3 components help regulate protein translocation into the ER?
ER-signal sequence on the protein
Signal-recognition particle (SRP), which cycles between the ER membrane and the cytosol, and binds to the signal sequence
binding of SPR to signal peptide causes a pause in translation
SRP receptor on the ER membrane
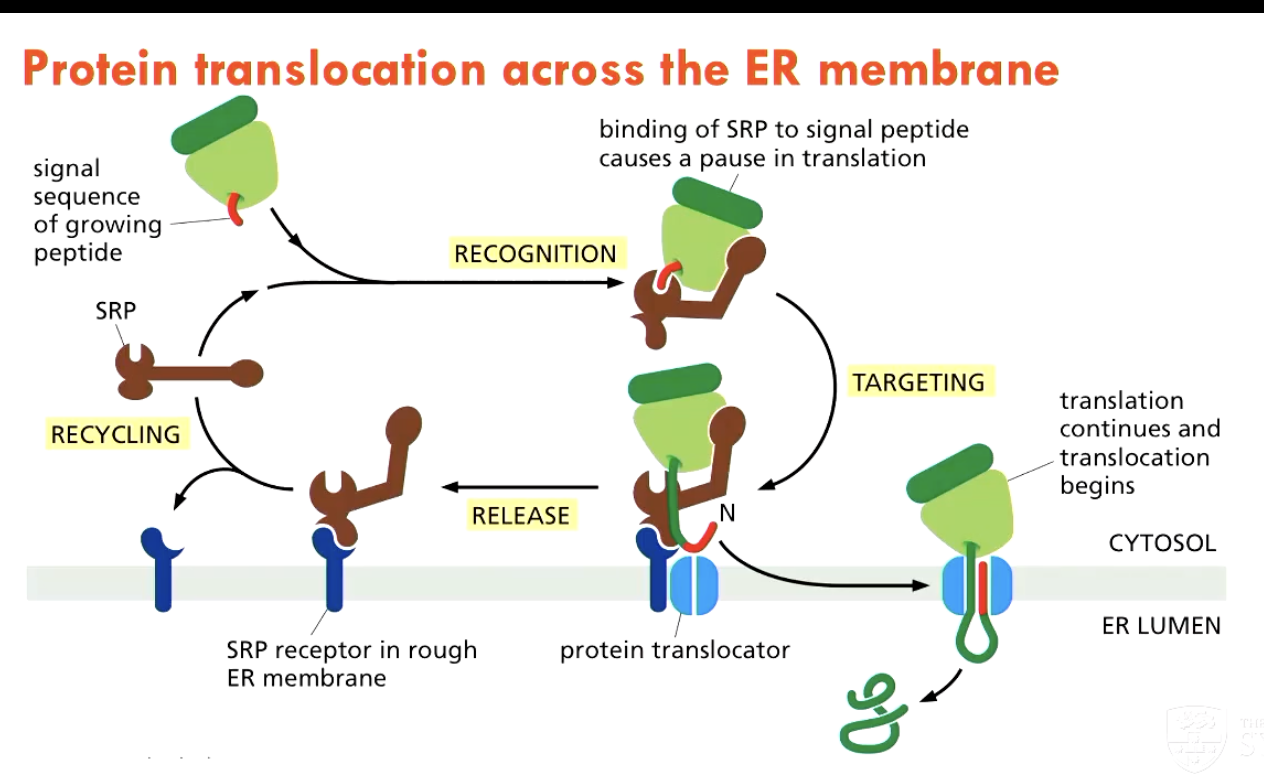
What happens to proteins in the ER lumen?
In the ER lumen, proteins fold, disulfite bonds are formed, and N-linked oligosaccharides are added
incorrectly folded proteins will be translocated back into the cytosol for degradation in proteasomes
correctly modified proteins progress into secretory pathway
How do proteins progress from the ER to the secretory pathway (3)?
Protein has an exit signal and a destination signal
Protein is linked to a cargo receptor
This signals external adaptor proteins and coat proteins (that provide the destination signal, giving the protein a target)
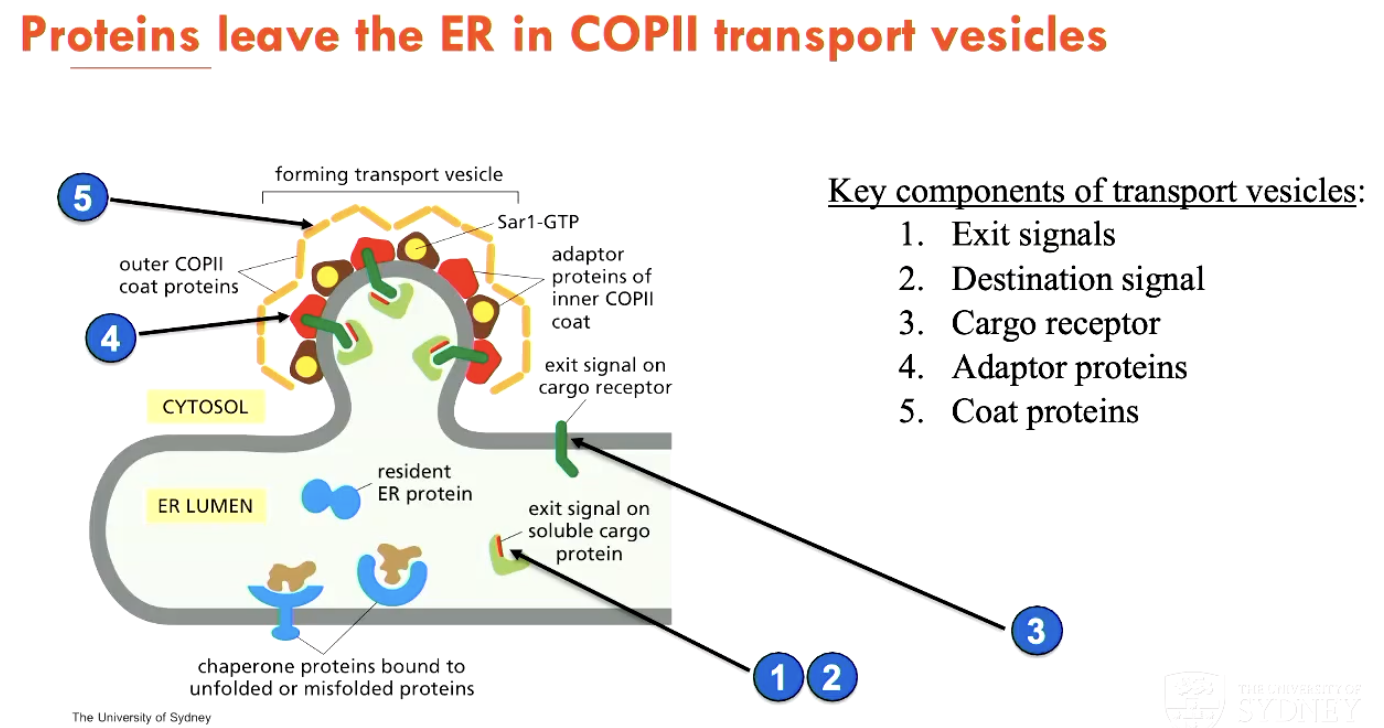
How are soluble ER-resident proteins retrieved if leaked through?
Every ER-resident protein has a KDEL attached to it, which signals for ER retention
but sometimes these proteins enter transport vehicles and are delivered to Golgi apparatus
they can be returned to the ER through a retrieval pathway involving a coat protein called COPI
What are the 3 destinations of protein sorting at the Trans Golgi Network?
Signal-mediated diversion to lysosomes (via endosomes)
Signal-mediated diversion to secretory vesicles (for regulated secretion - later release)
Constitutive secretory pathway (immediate release)
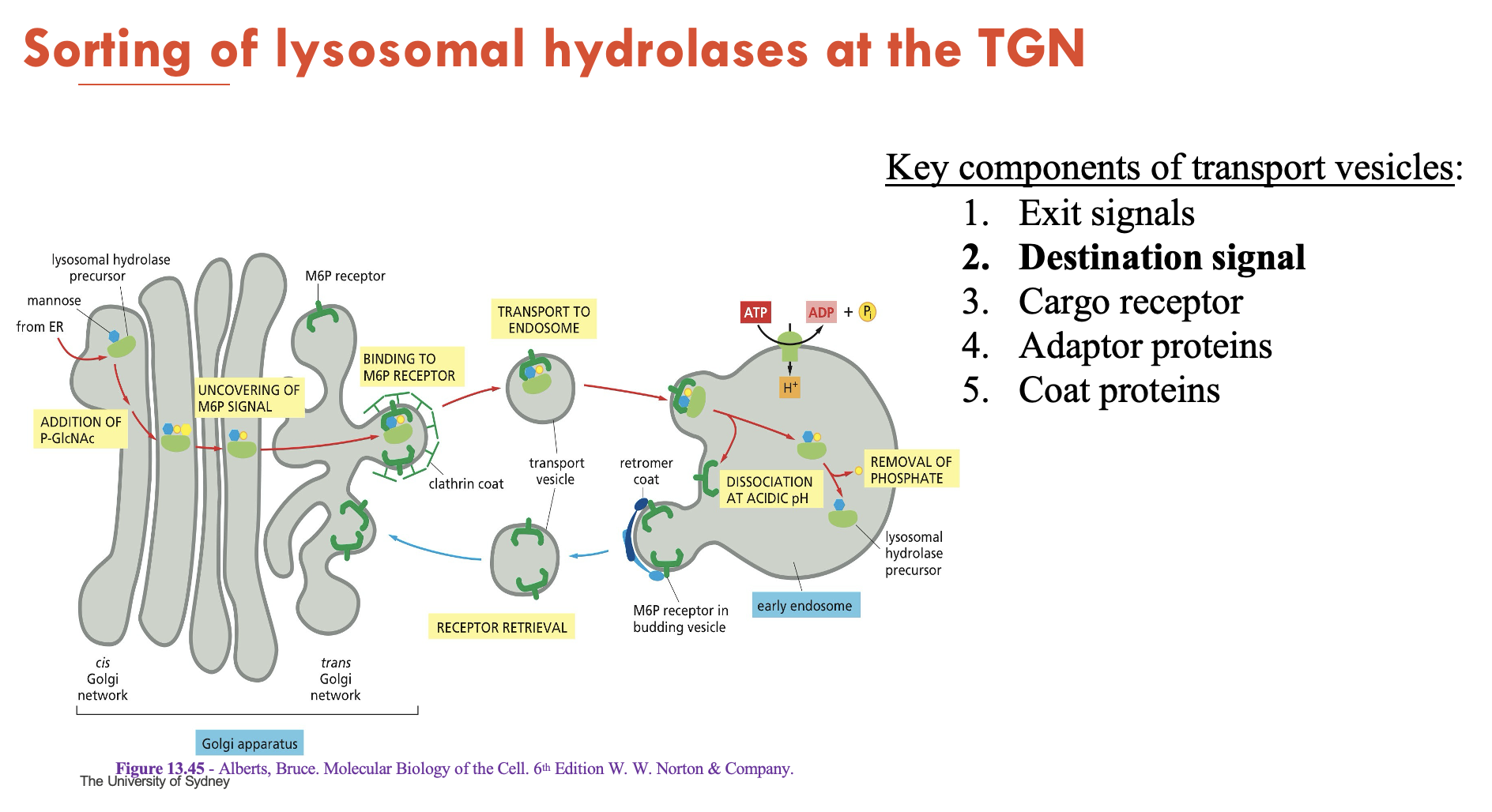
How are lysosomal hydrolases sorted at the TGN?
Tagging (cis-Golgi):
Mannose-6-phosphate (M6P) is added to hydrolases to provide a destination signal
Sorting (TGN):
M6P receptors bind M6P-tagged hydrolases on luminal side → recruit clathrin/adaptors to form clathrin coat (around vesicle) on cytosolic side
Budding:
Vesicles deliver hydrolases to endosomes
Release:
Low pH separates M6P-receptor complex:
Hydrolases travel to lysosome
Receptors are sent to TGN for recycling
Mnemonic: M6P Marks → Clathrin Captures → Endosome Escorts → Lysosome Landing
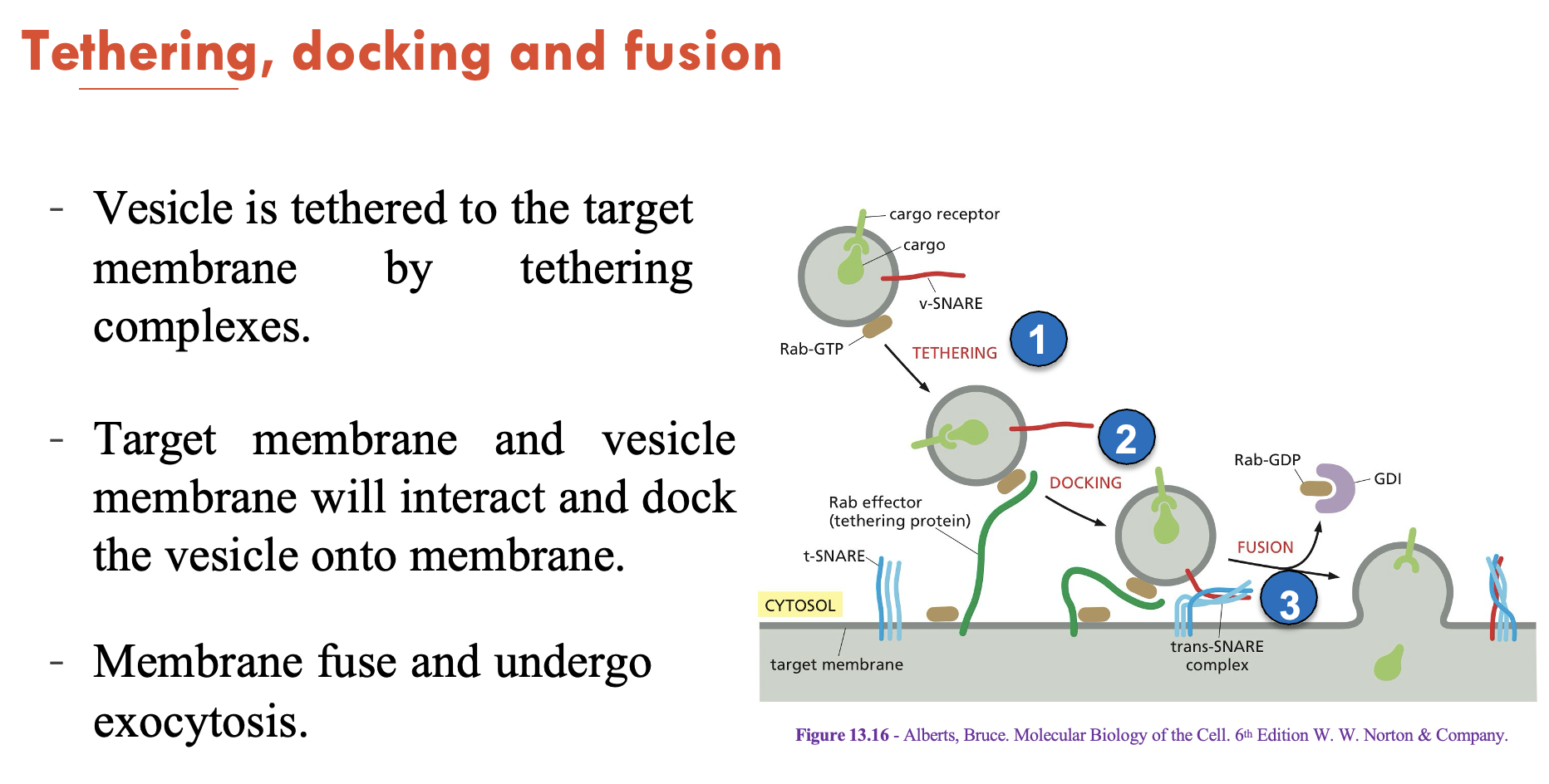
Describe the process of tethering, docking, and fusion in exocytosis
Vesicle is tethered to the target membrane by tethering complexes
Target membrane and vesicle membrane will interact and dock the vesicle onto membrane
Membrane fuses and undergoes exocytosis
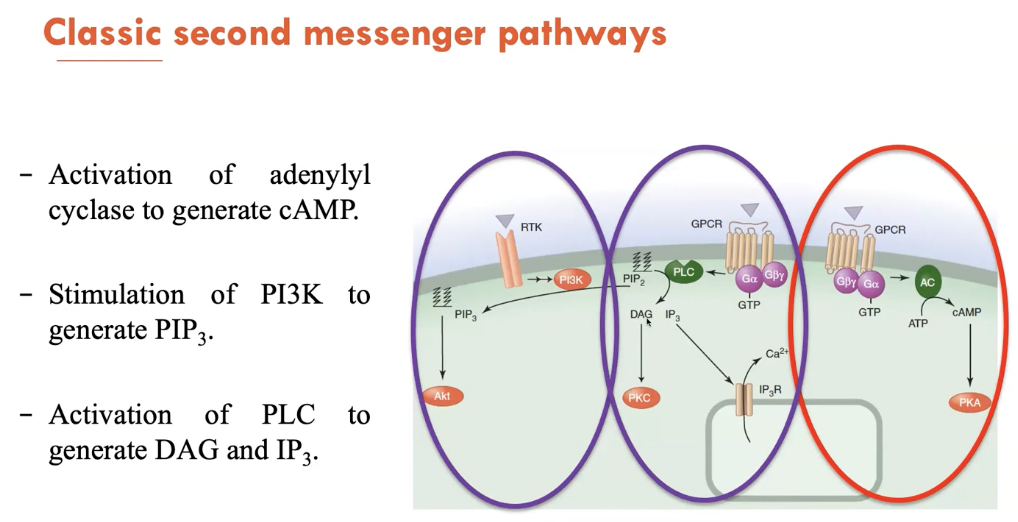
What are the 3 classic second messenger pathways?
Second messengers are small molecules and ions that are present at low levels in resting cells. 3 of its main pathways are:
Activation of adenylyl cyclase to generate cAMP
Stimulation of PI3K to generate PIP3
Activation of PLC to generate DAG and IP3
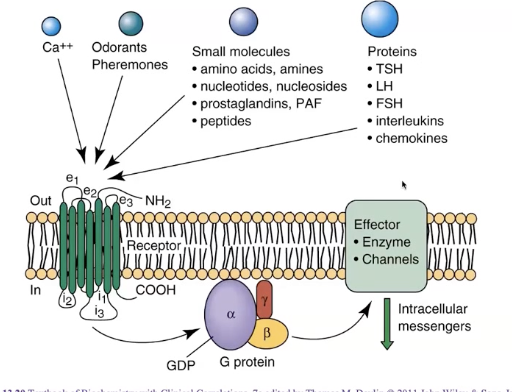
What are the features of GPCRs (3)?
largest family of cell-surface receptors in many organisms
largest and among the most efficacious class of therapeutic targets
mutations in GPCRs are linked to many diseases (e.g. Usher syndrome - variable onset deafness and blindness)
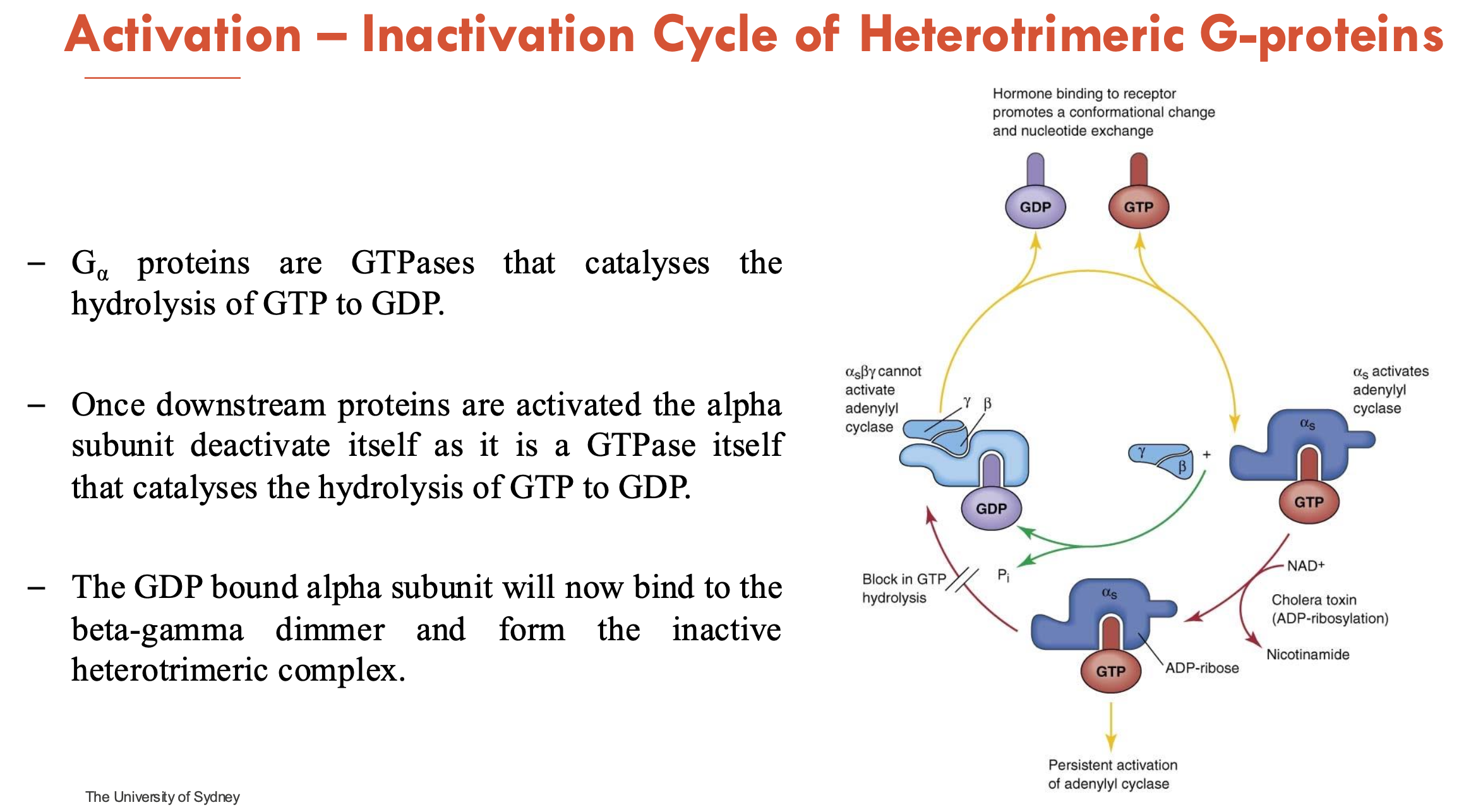
Explain the activation-inactivation cycle of heterotrimetric G-proteins
Inactive state (GDP-bound): α-subunit binds GDP; note that G-protein is a heterotrimer (αβγ)
Activation (GTP-bound)
Ligand binds GPCR → GPCR acts as a GEF (guanine nucleotide exchange factor)
GDP → GTP Exchange on α-subunit → conformational change
α-GTP dissociates from βγ → both interact with effectors (mostly enzymes or ion channels)
Inactivation (GTP hydrolysis)
α-subunit are GTPases and hydrolyses GTP → GDP (intrinsic GTPase activity)
once downstream proteins are activated, the α-subunit deactivates itself as it is a GTPase
α-GDP rebinds βγ → reforms inactive heterotrimeric complex
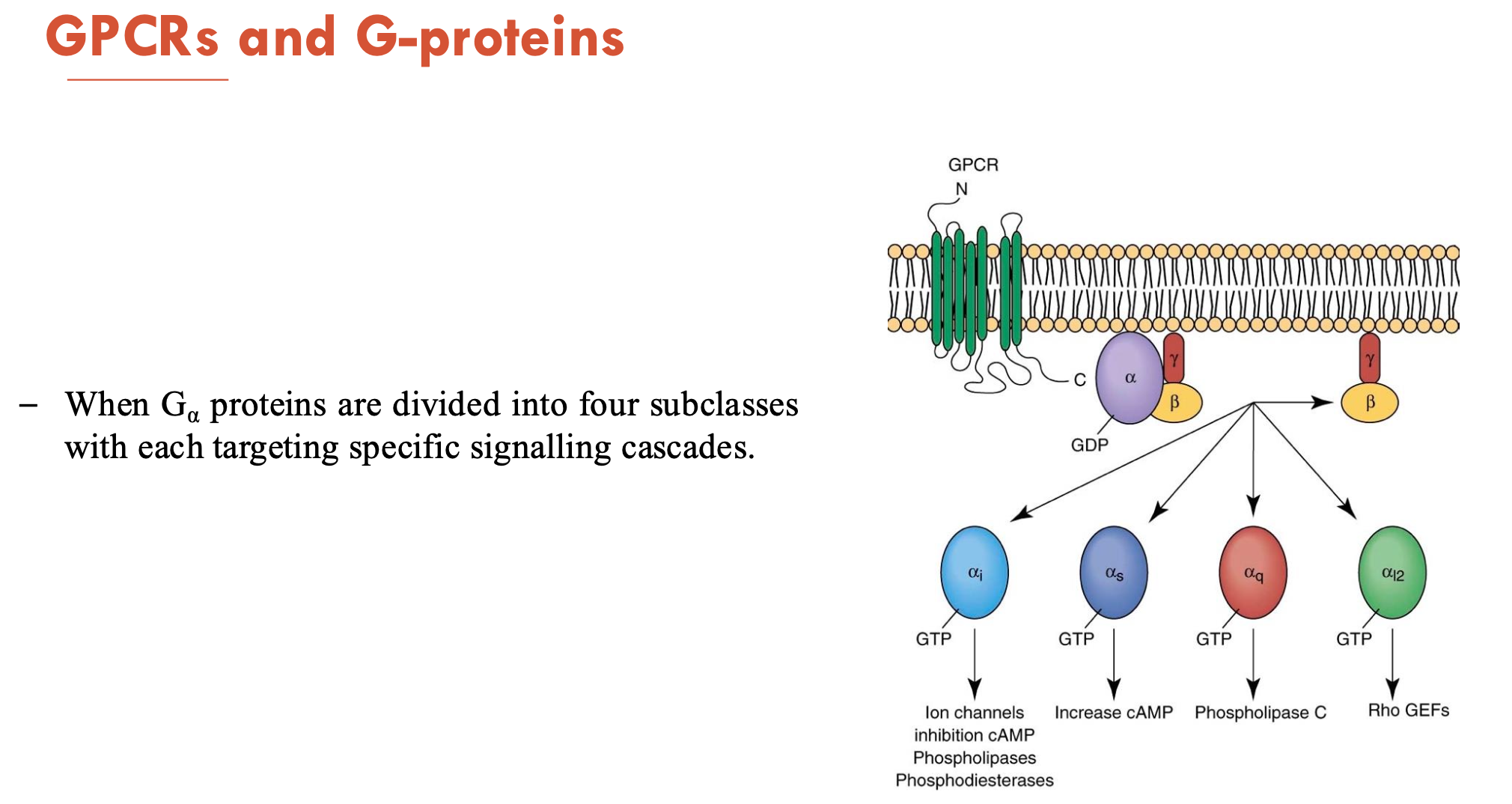
What are the 4 subclasses of G-proteins and what do they target?
αi → targets ion channels, adenylyl cyclase (→ inhibits cAMP), phospholipases, phosphodiesterases
αs → targets adenylyl cyclase (→ increase cAMP → activates PKA)
αq → phospholipase C (→ cleaves PIP2 → forms IP3)
αI2 → Rho GEFs (→ activate RhoA)
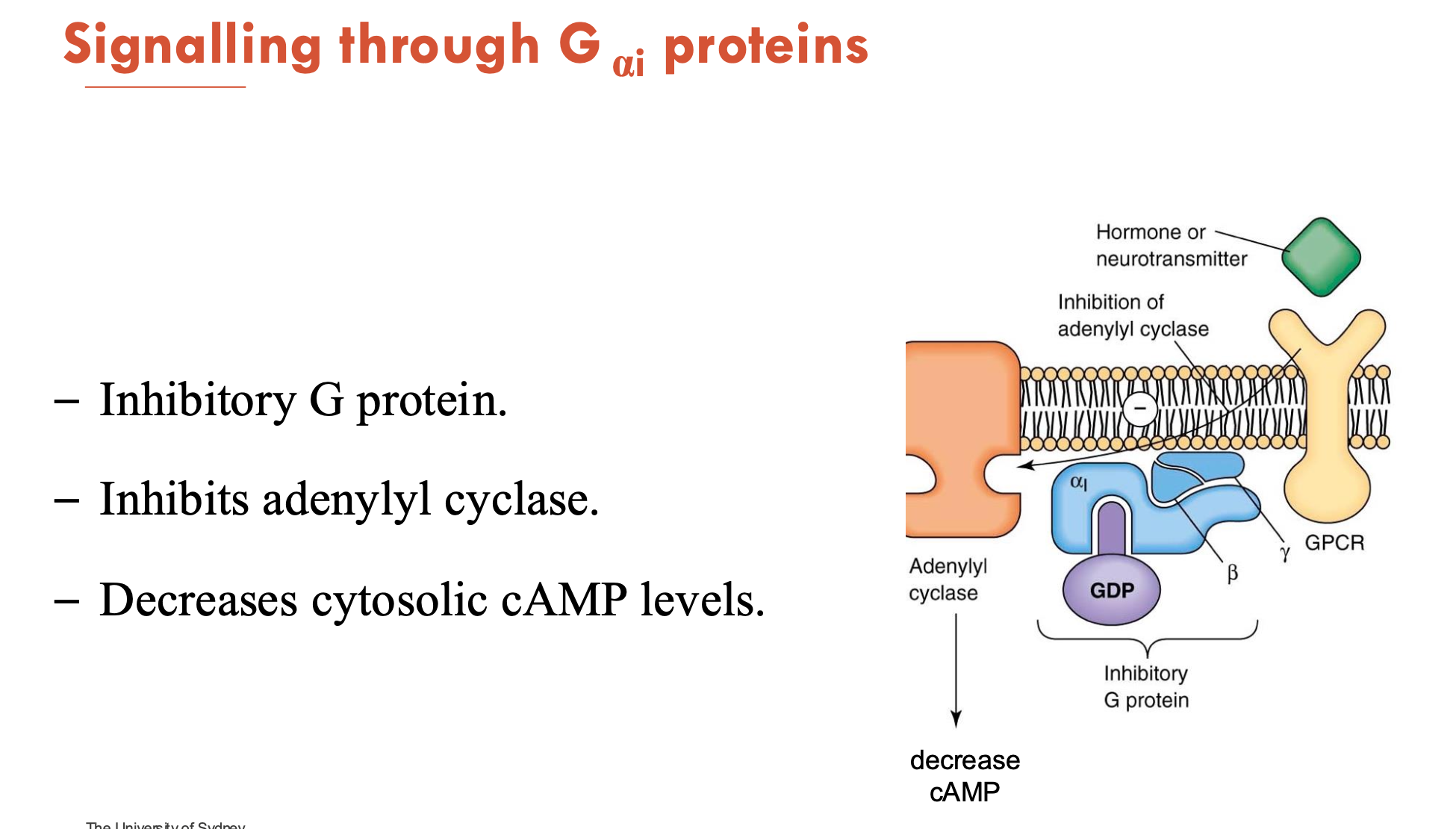
What does the signalling of Gαi proteins do?
Gαi is an inhibitory G protein
inhibits adenylyl cyclase → decreases cytosolic cAMP levels
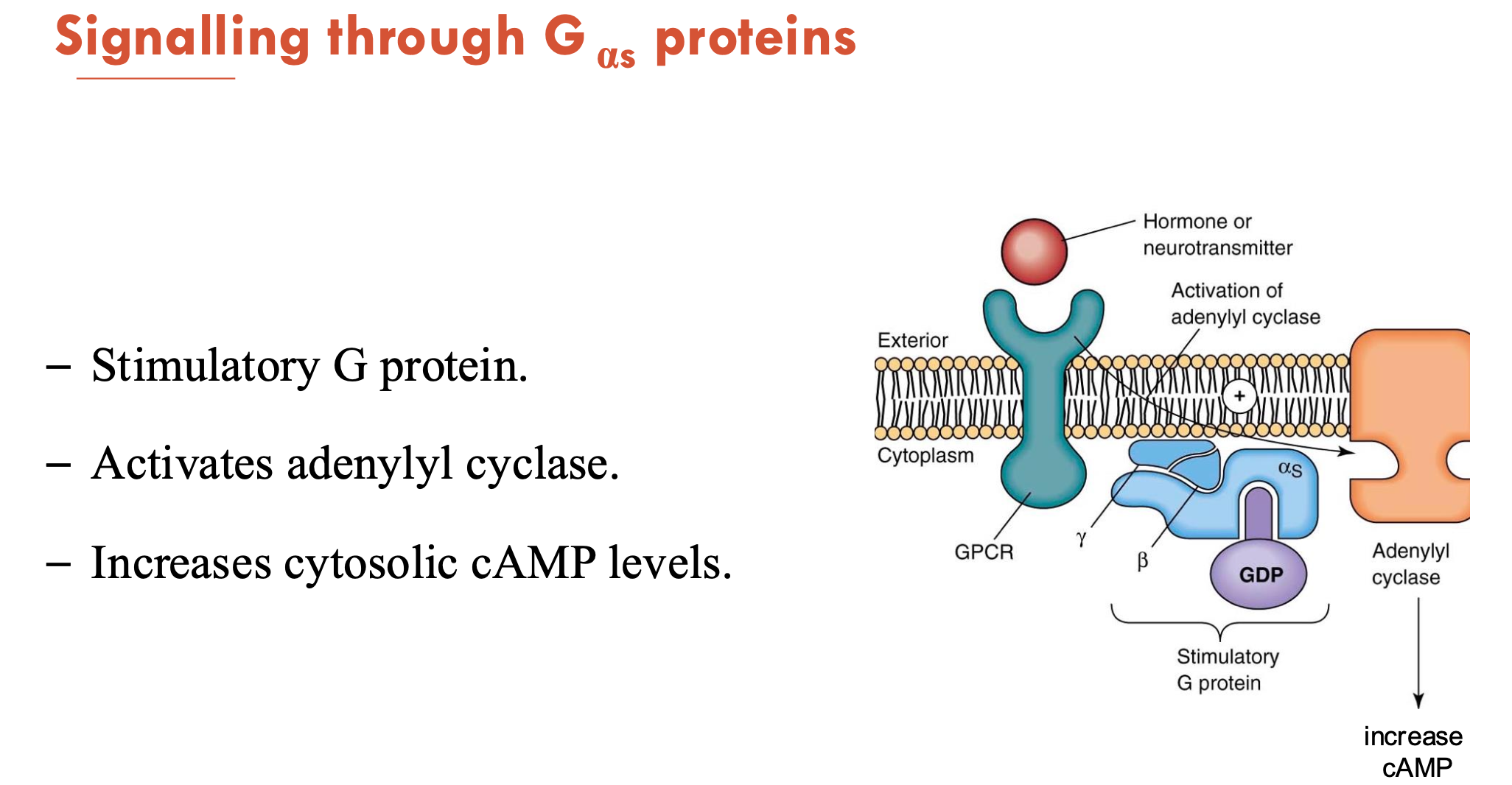
What does the signalling of Gαs proteins do?
Gαs is a stimulatory G protein
activates adenylyl cyclase → increases cytosolic cAMP levels
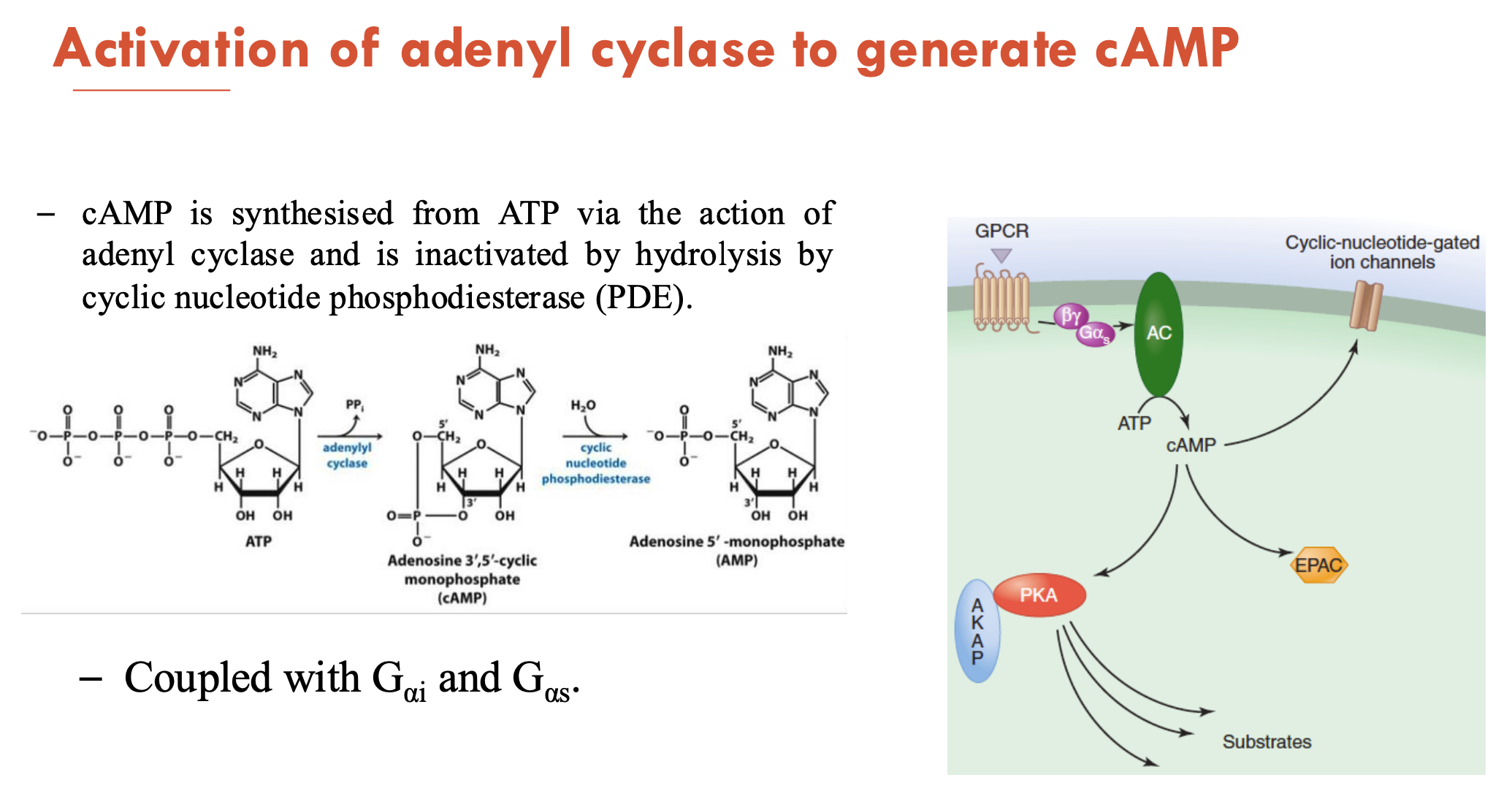
How is adenyl cyclase activated to generate cAMP?
cAMP is synthesised from ATP (via action of adenyl cyclase) and is inactivated by hydrolysis (by cyclic nucleotide phosphodiesterase (PDE))
coupled with Gαi and Gαs
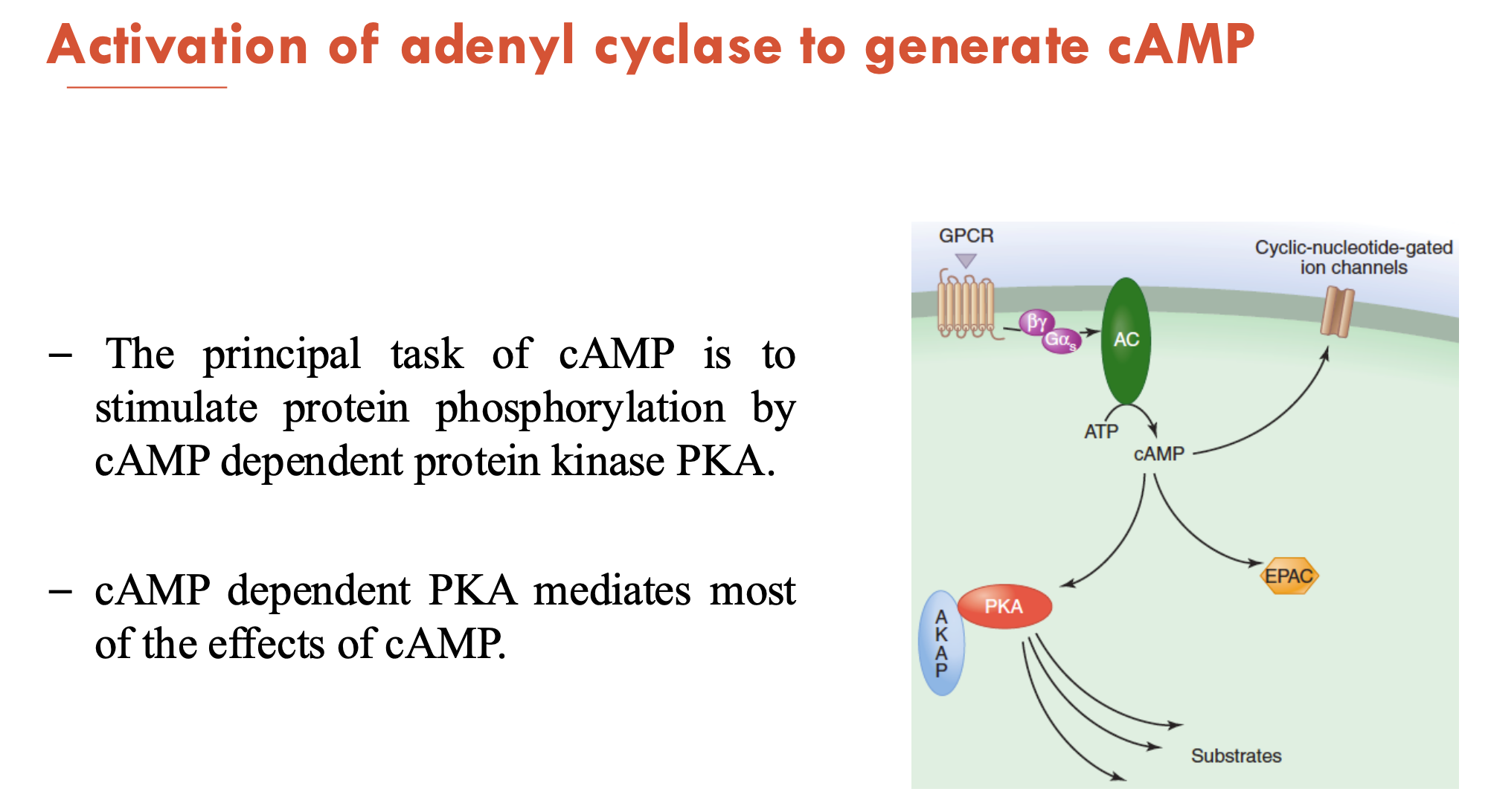
The principal task of cAMP is to (A - function) by (B - enzyme).
Increased cAMP will (C - consequence)
A - stimulate protein phosphorylation
B - cAMP-dependent protein kinase PKA (which mediates most effects of cAMP)
C - rapidly alter the transcription and expression of genes coding for important metabolic enzymes and ion channels
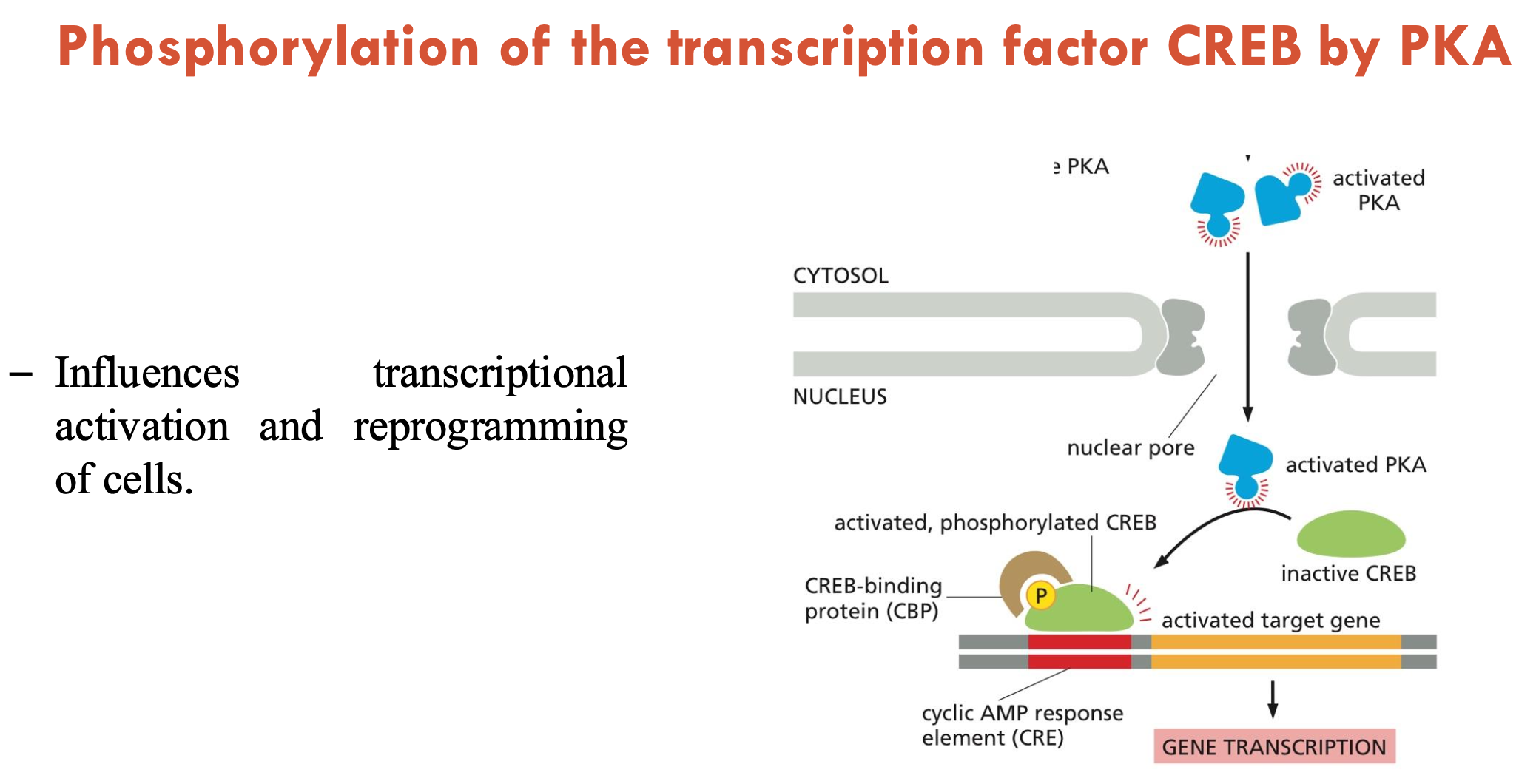
PKA goes from the (A - part of cell) into the (B - part of cell), where it binds to cAMP. PKA then phosphorylates (C), which (D - consequence).
A - cytosol
B - nucleus
C - transcription factor: CREB
D - influences transcriptional activation and cell reprogramming
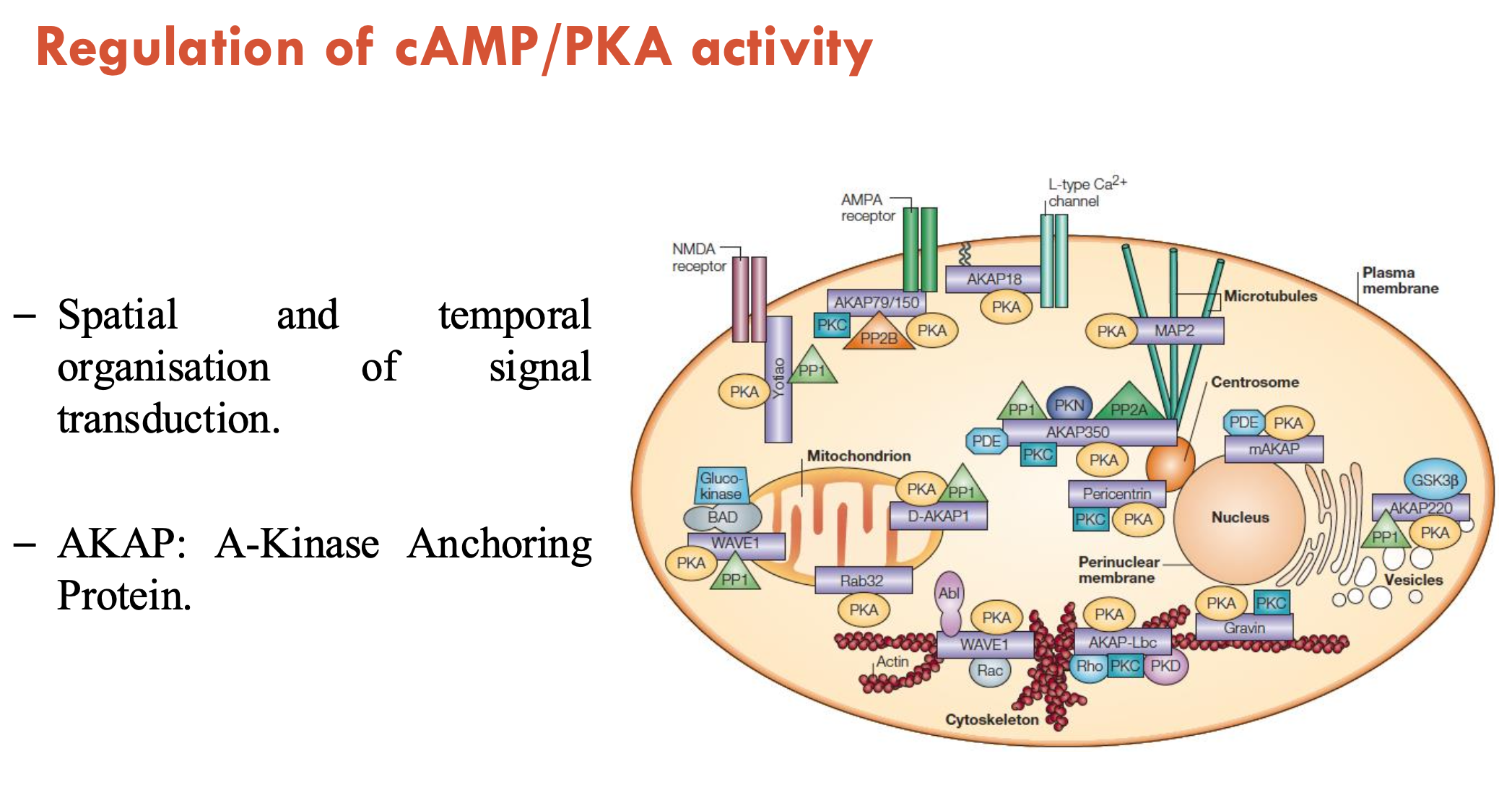
What regulates cAMP/PKA activity?
Anchoring: AKAP (A-kinase anchoring protein) achieves spatial and temporal organisation of signal transduction by localising PKA
Phosphodiesterase (PDEs): AKAPs bring PKA and PDEs together → PDEs are termination signals
there are 100 distinct PDE enzymes
PDE inhibitors are often used as therapeutic targets
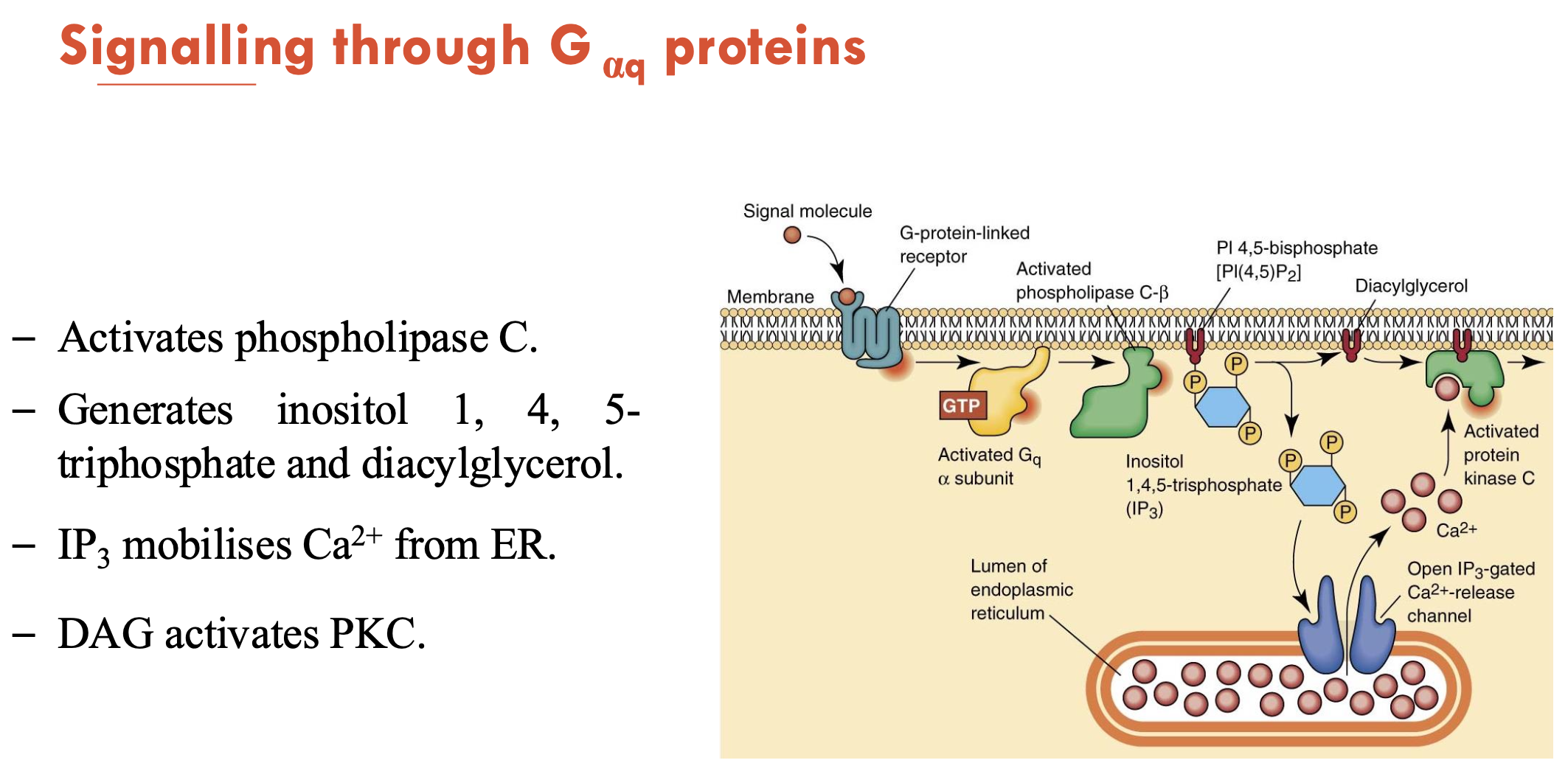
What does the signalling of Gαq proteins do?
Activates phopholipase C (PLC) → generates inositol 1, 4, 5-triphosphate (IP3) and diacylglycerol (DAG)
IP3 mobilises Ca2+ from ER
DAG/Ca2+ activates PKC → PKC phosphorylates target proteins
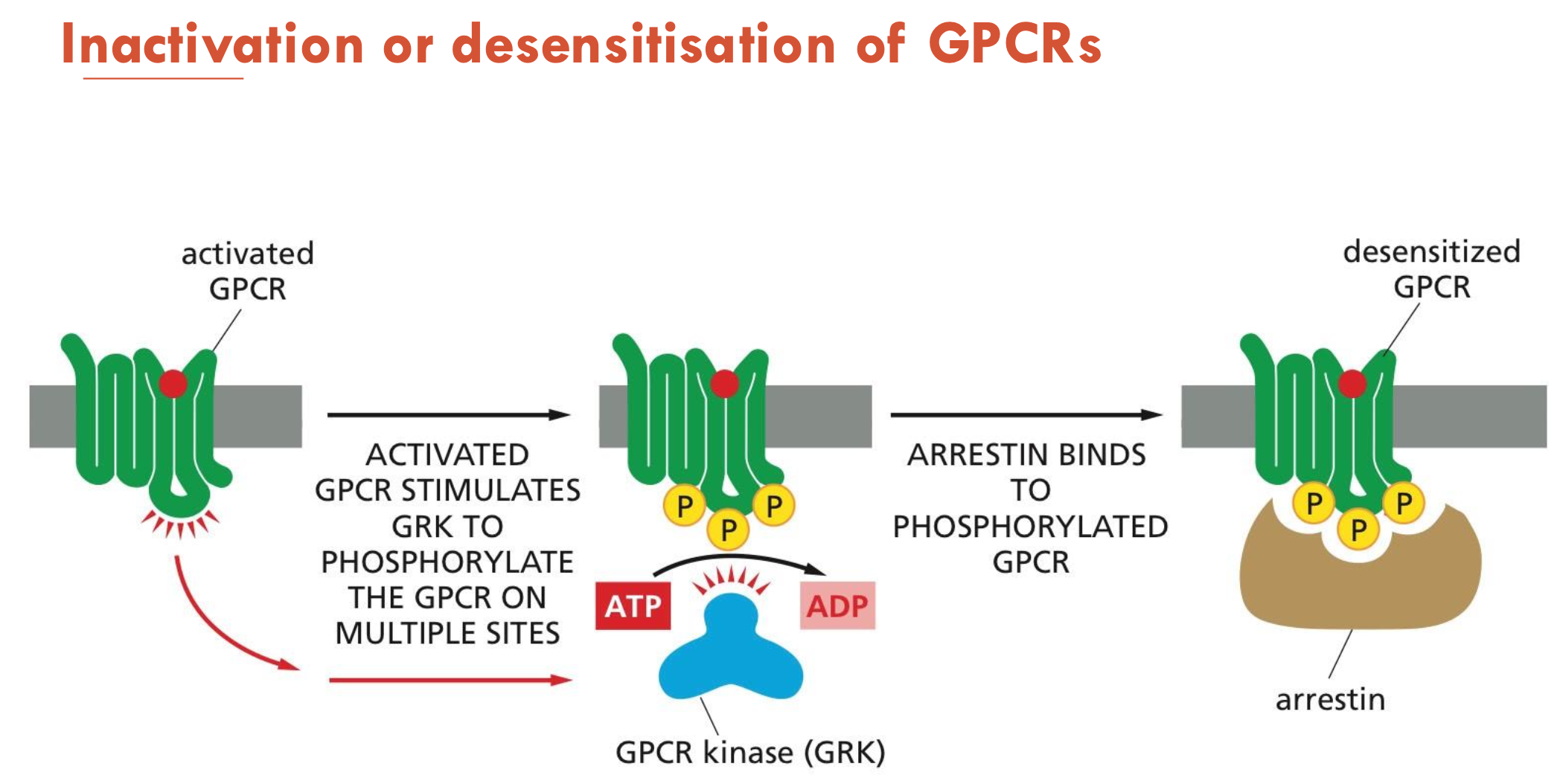
How are GPCRs inactivated or desensitised?
Activated GPCR stimulates GPK to phosphorylate the GPCR on multiple sites
Arrestin binds to phosphorylated GPCR → desensitised GPCR
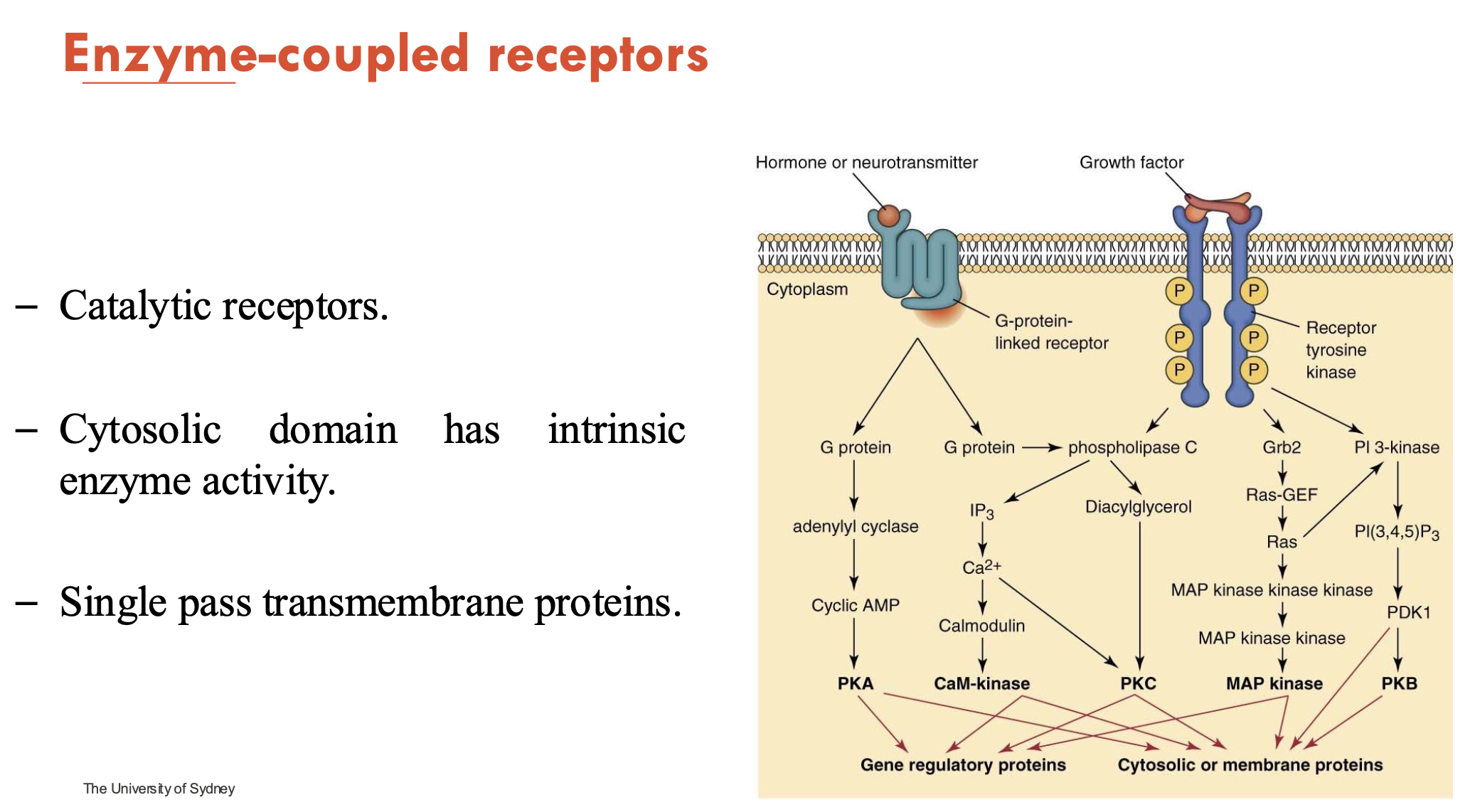
What are features of enzyme-coupled receptors?
catalytic
cytosolic domain has intrinsic enzyme activity
single-pass transmembrane proteins
most common example: receptor tyrosine kinase
How is organelle signalling compartmentalised (4)?
Mitochondria is the central hub for signalling
Nucleus is for long-term signalling
Transcription factors
Golgi is for trafficking and sorting
source for Ca2+
Endoplasmic reticulum is for calcium signalling
How is protein-protein signalling compartmentalised?
Co-localisation triggers response
rely on proteins binding and phosphorylating to each other for a response
Examples: insulin and integrins
How is second messenger signalling compartmentalised?
Through global and local/regional cAMP signalling:
Global
When cAMP is being generated in the cytosol, it has two protein targets that lead to further signalling:
PKA
EPAC2
Local/Regional
Through PDE, cAMP can be degraded (switches the signal off)
cAMPs can also be localised to specific cell regions using AKAPs (A Kinase Anchoring Proteins):
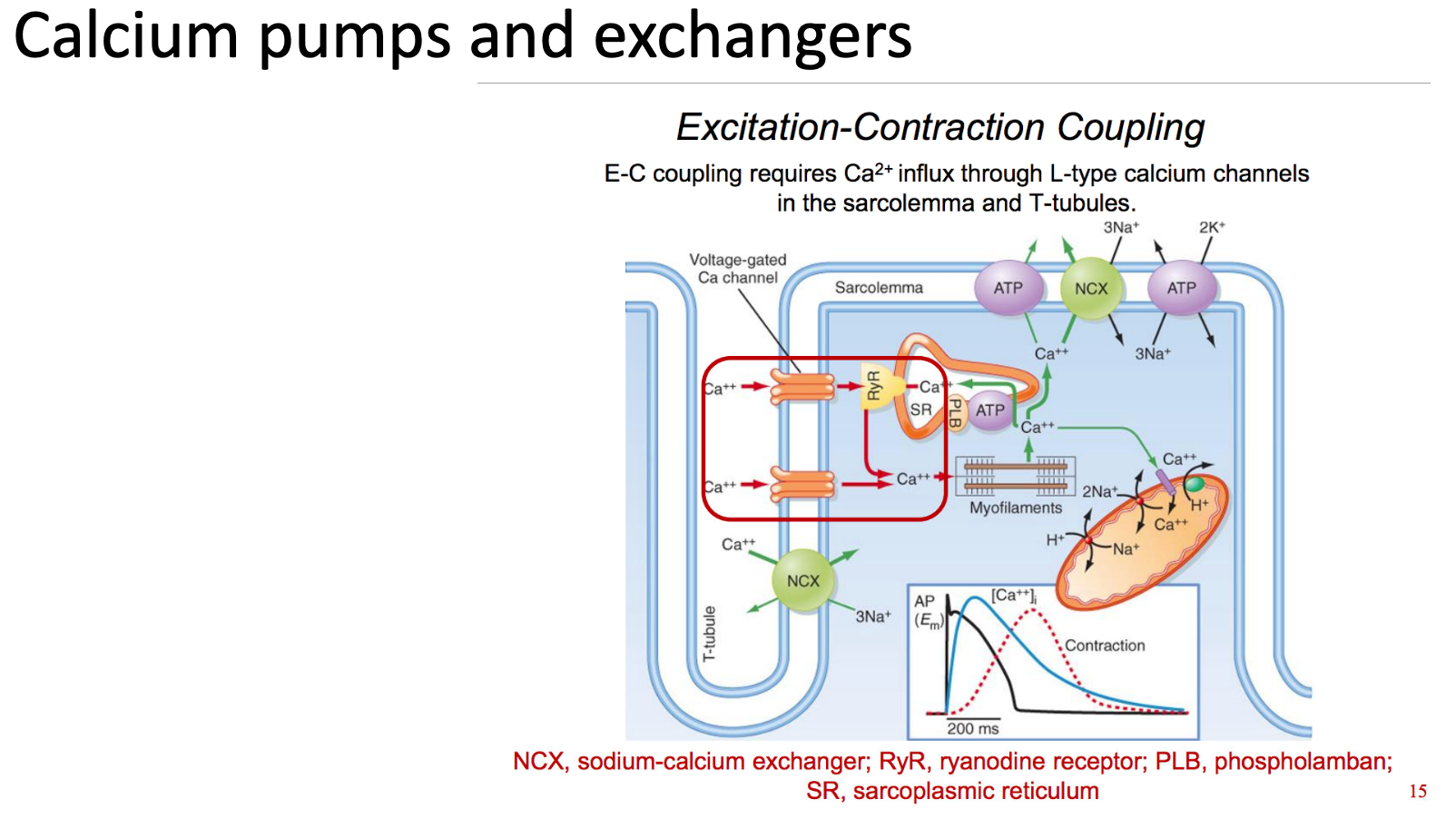
How is calcium response different to cAMP response?
Calcium is an ion, so it cannot be made up or degraded
whereas cAMP is made by ATP and can be broken down by PDE
Therefore, localised calcium responses rely on calcium being moved in and out of the cytosol.
calcium can enter the cell through voltage-gated channels or activation of ryanodine receptors
calcium response can be modulated by pumping calcium out of the cytosol, either
back into the SR/ER
into the mitochondria (temporary storage)
out of the cell
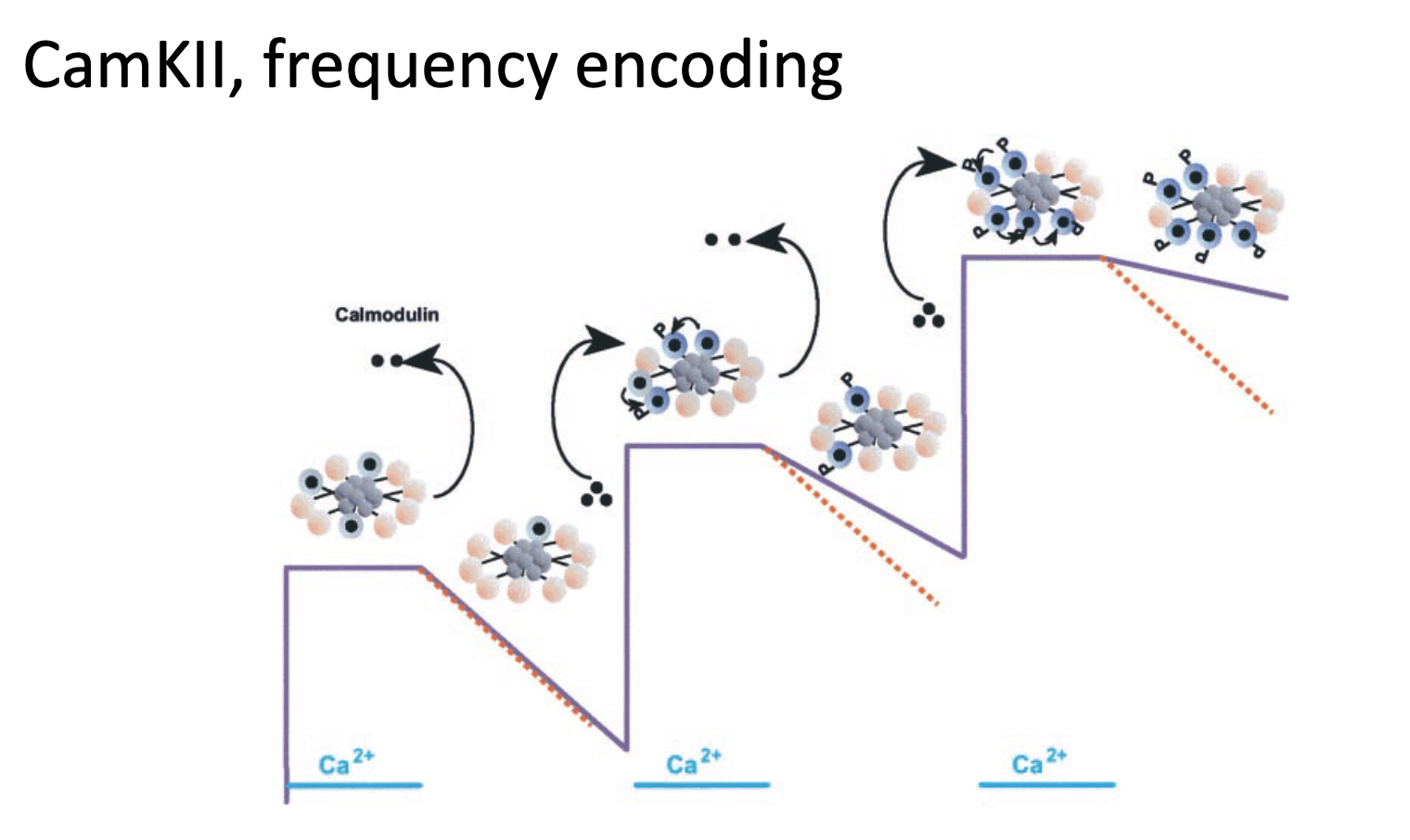
Explain what causes calcium oscillations
Calcium levels go up and down during the activation and inactivation of calcium-permeable channels
Frequency Encoding
low-frequency oscillations can be read out by TFs
high-frequency oscillations can be read out by calcium-dependent enzymes
Relaxation Delay
when calcium exits the cell, calcium-dependent response takes time → relaxation phase delays
Staircase Effect
new Ca²⁺ wave arrives before full relaxation → cumulative spike amplitude
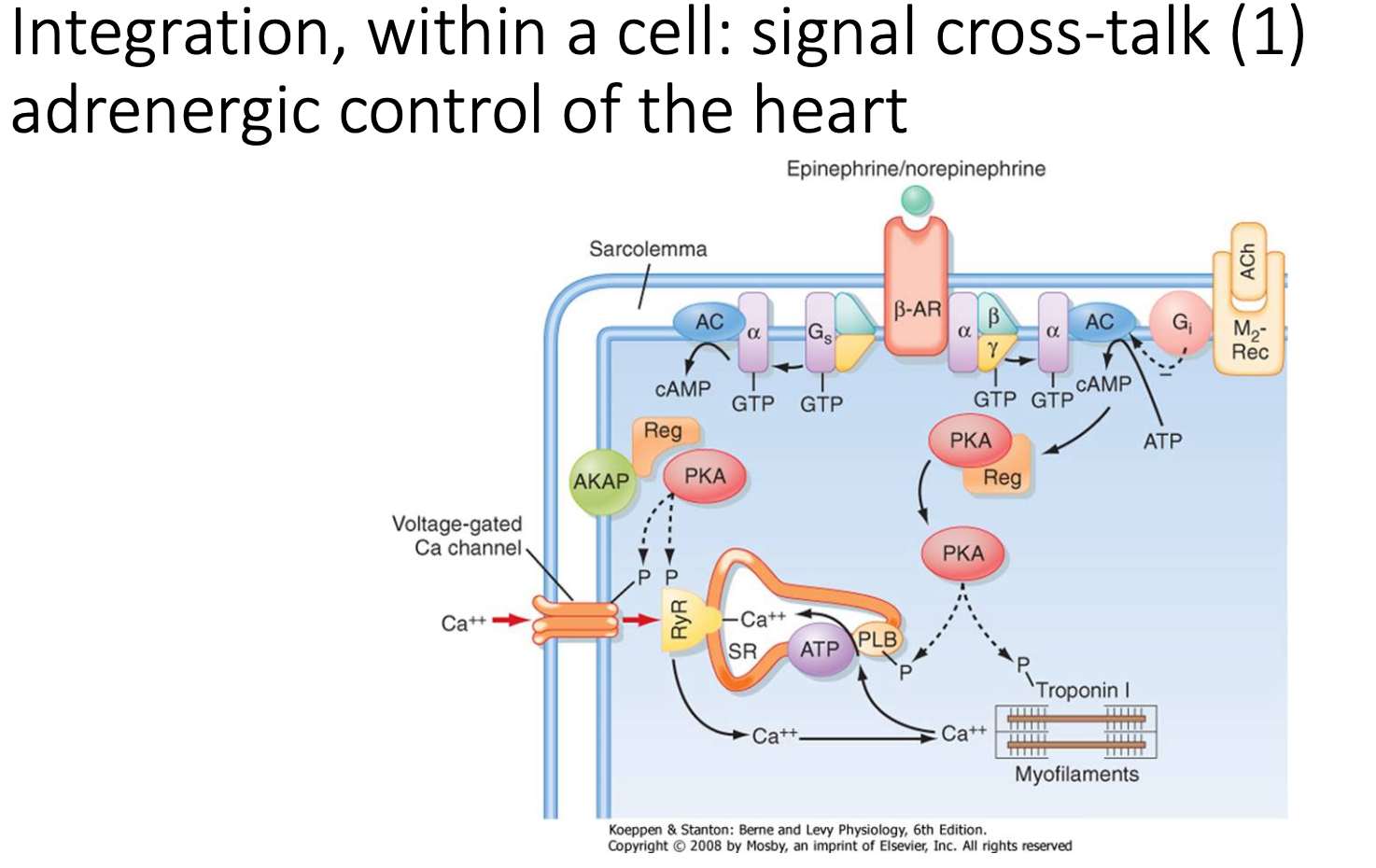
What are the signalling integration pathways of adrenergic control of the heart?
Heart contractions have APs → allows Ca2+ to enter cell from SR (drives contractions)
In sympathetic stimulation, heart depolarises quicker:
stimulation of adrenergic receptors → generates cAMP (acts on PKA and EPAC2) → intersects Ca2+ pathways (by upregulating Ca2+ channel and uptake of Ca2+ into SR) → increases rate and force of contractions
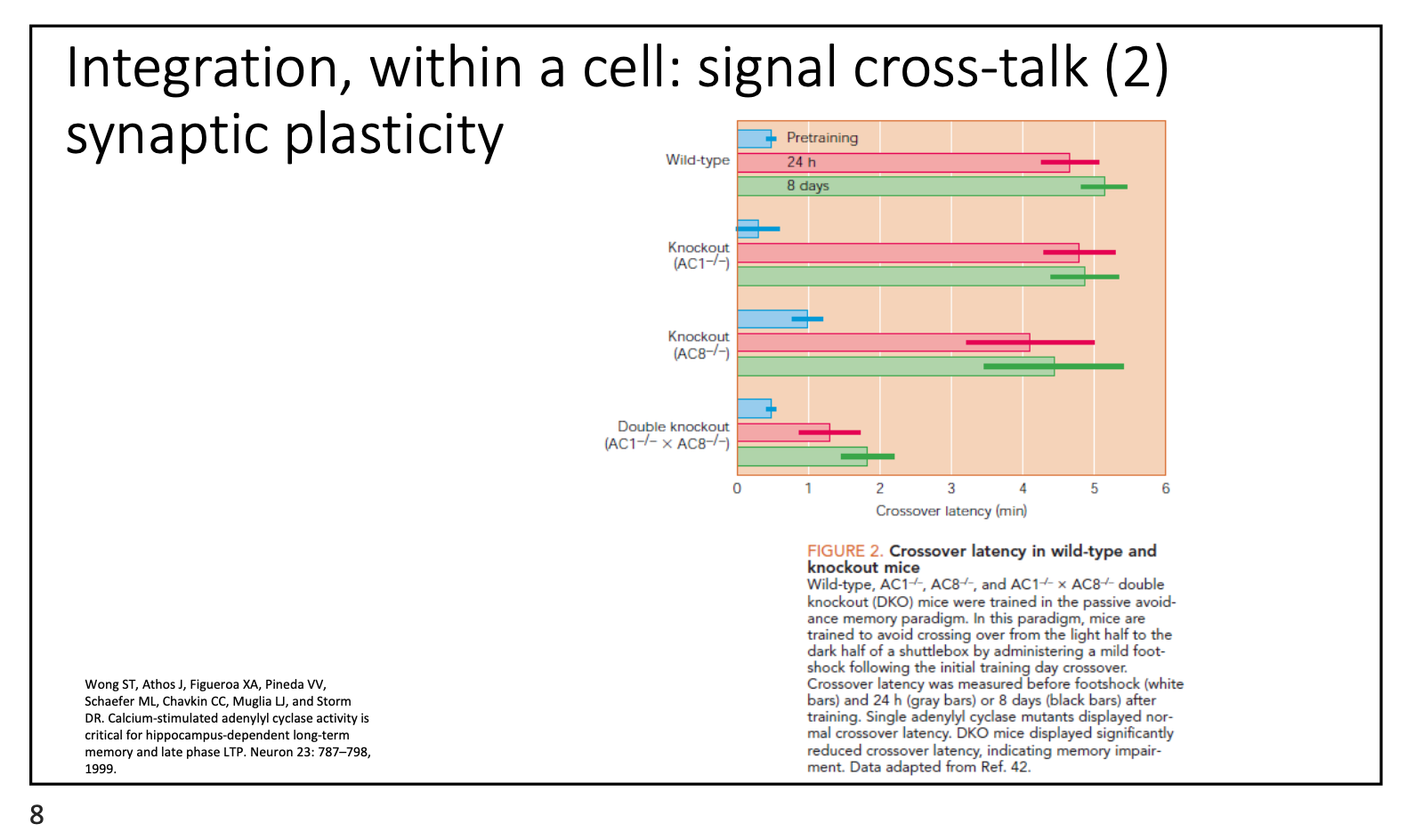
What are the signalling integration pathways of synaptic plasticity?
Adenylate cyclase helps convert ATP to cAMP
AC1 and 8 are calcium-dependent
Ca2+ enters synapses → activates AC1/8 → triggers cAMP → strengthens synapses
Knockout of AC1 & 8 → inhibits learning ability (synaptic plasticity)
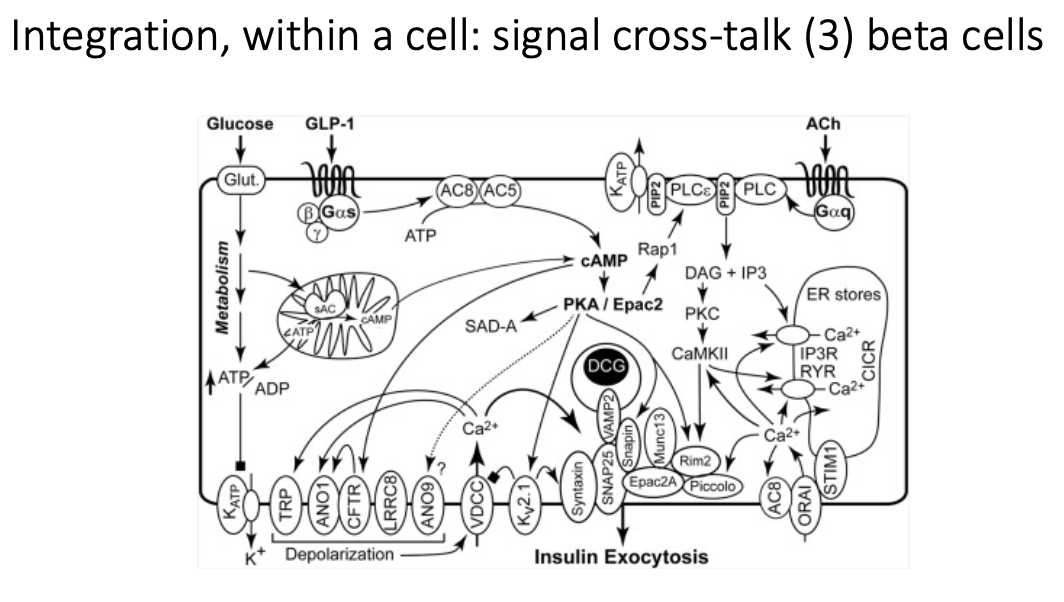
What are the signalling integration pathways of beta cells (3)?
Glucose
ATP production → Ca2+ influx → insulin secretion
GLP-1
oral glucose → Gs-coupled receptor → increases cAMP → sensitises beta cells to Ca2+ to support glucose-stiumated insulin release (incretin effect)
GIP
similar pathway to GLP-1 (Gs-coupled receptor), but acts on different receptor
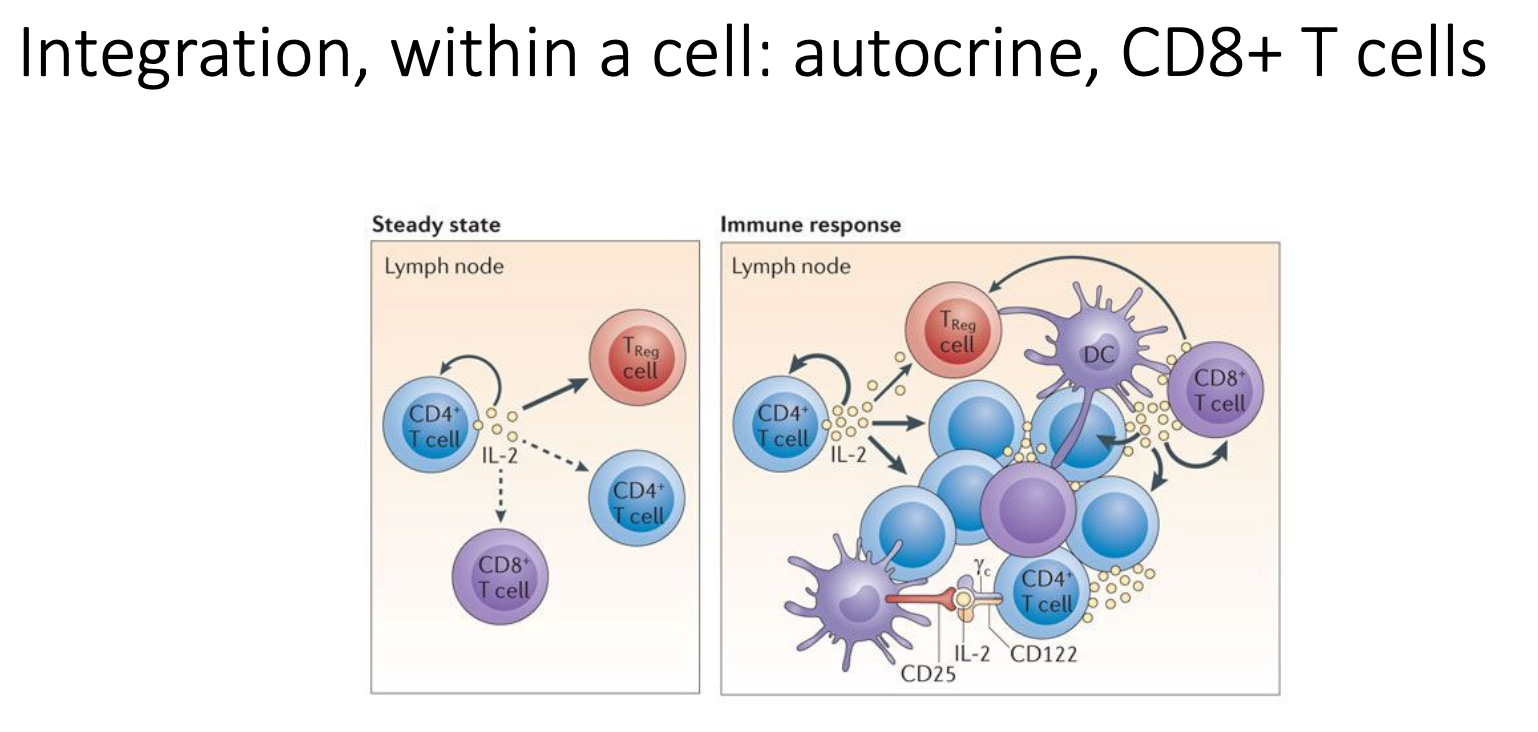
What are the signalling integration pathways of autocrine, CD8+ T cells?
Very local response - acts on target cells
ILs secreted by T cells will impact on both other inflammatory cells and themselves
leads to changes in memory T cells
What are the signalling integration pathways of gap junctions (4)?
Chemical Signalling (Calcium Waves)
Stimulation of one cell → Ca²⁺ spreads to connected cells via gap junctions
If blocked: No Ca²⁺ spread → loss of coordination
Electrical Signalling
Pancreatic beta cells synchronise insulin release via electrical coupling
Neuronal Coordination
Fast hearing signals
ATP-Mediated Signalling
Macrophages communication
What are the 3 different classes of bioactive lipids?
Eicosanoids: a broad class of lipid signalling molecules derived from arachidonic acid; lipid molecules with bits attached (such as AA or hydroxyl groups)
Endocannabinoids: a subset of eicosanoids; found in the body and mediate normal physiological functions (mainly synaptic signalling)
Phospholipids/sphingolipids: structural component of cell membrane
Note: for this lecture (lec 13), no need to know steroids
Most bioactive lipids act on…
G-protein coupled receptors
(e.g. Endocannabinoids on CB1/2 receptors; prostaglandins on 9 Ecoisanoid receptors)
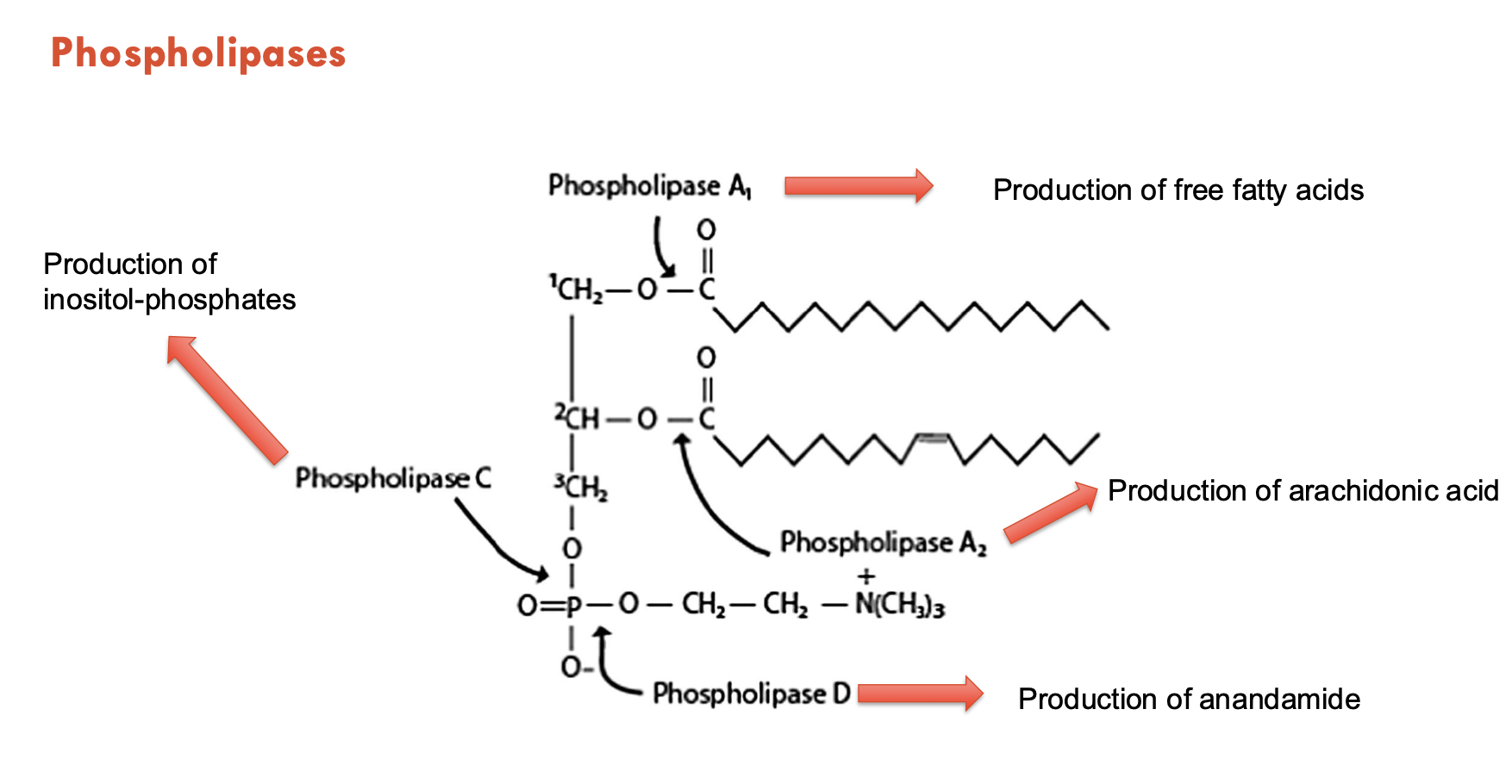
How are bioactive lipids produced?
Through phospholipases.
A prototypical membrane lipid is composed of a glycerol backbone, lipid tails (in the membrane), and a lipid head group (outside the cell).
Phospholipase A1 produces → free fatty acids
Phospholipase A2 produces → arachidonic acid
Phospholipase C produces → inositol-phosphates
Phospholipase D produces → anandamide
Arachidonic acid is (A - feature of importance).
It can be obtained either from (B) or (C).
It is also classified as (D).
A - a precursor for many bioactive lipids
B - diet (meat or eggs)
C - synthesised from linoleic acid
D - an omega-6 fatty acid (pro-inflammatory)
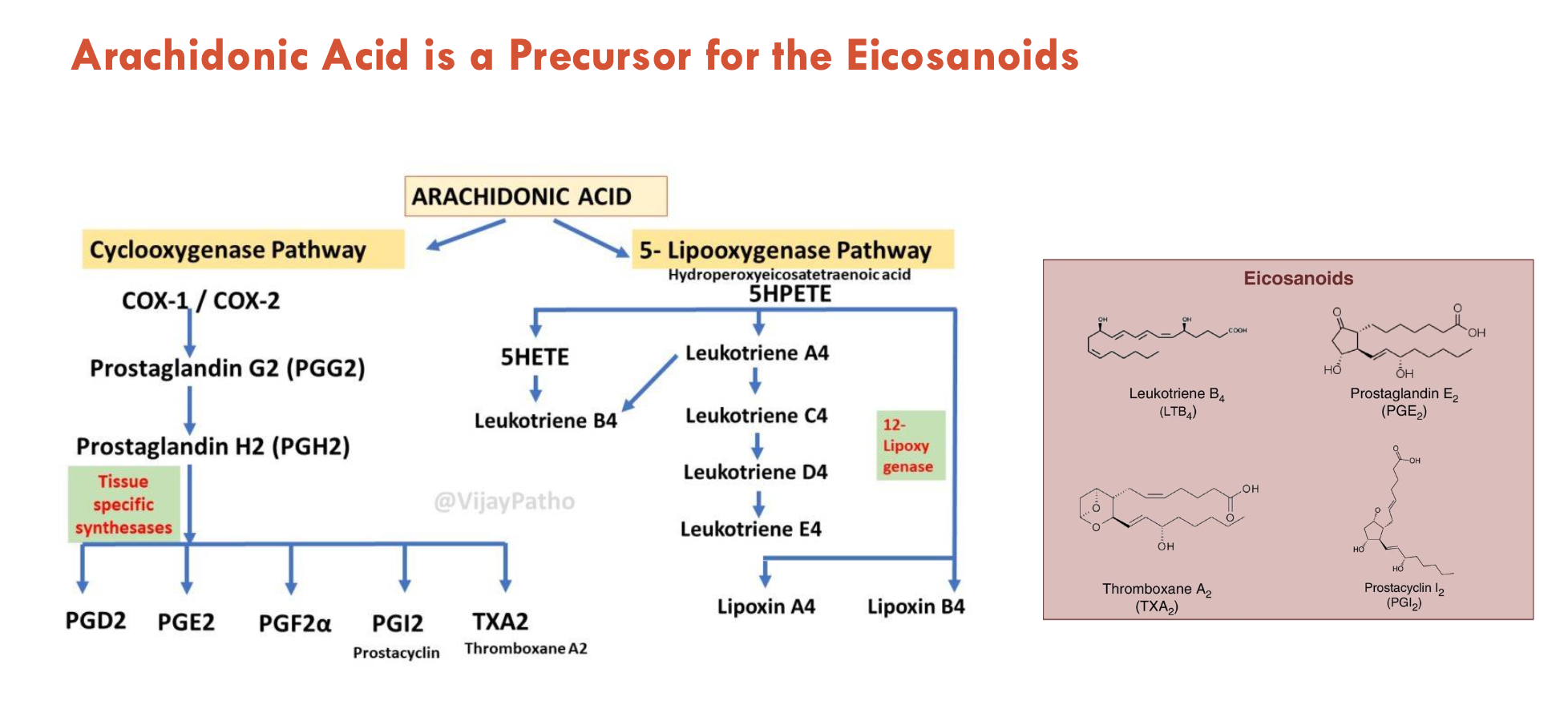
What 2 pathways allow eicosanoids to be derived from arachidonic acid?
Cyclooxygenase pathway (e.g. for prostaglandin-E2 (PGE2), thromboxane A2 (TXA2))
5-lipooxygenase pathway (e.g. for leukotriene B4)
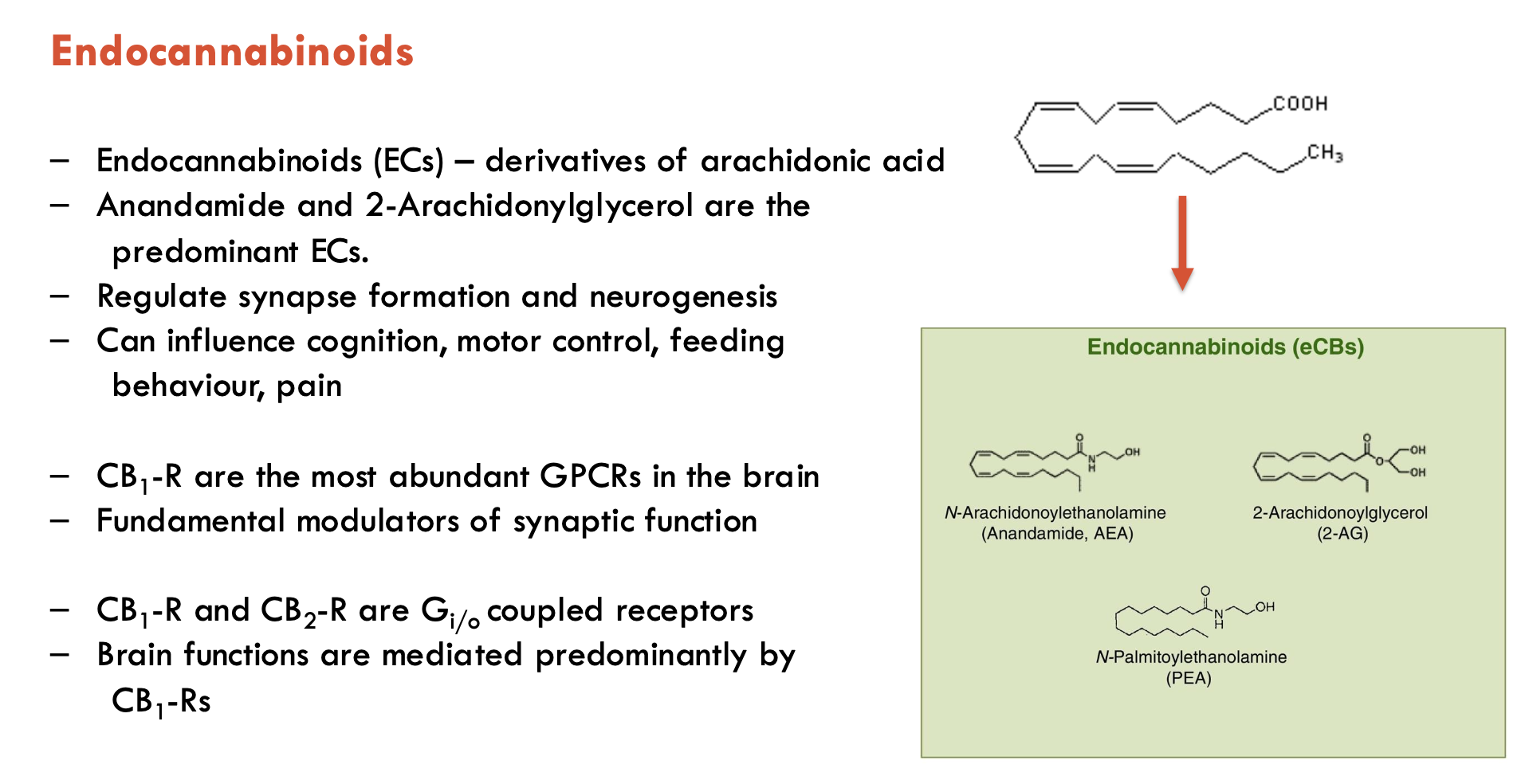
What are some features of endocannabinoids (4)?
Endocannabinoids (ECs) are derivatives of arachidonic acid
most common: anandamine and 2-Arachidonylglycerol (2-AG)
regulate synapse formation and neurogenesis
can influence cognition, motor control, feeding behaviour, pain
act through CB1 receptors (CB1-R)
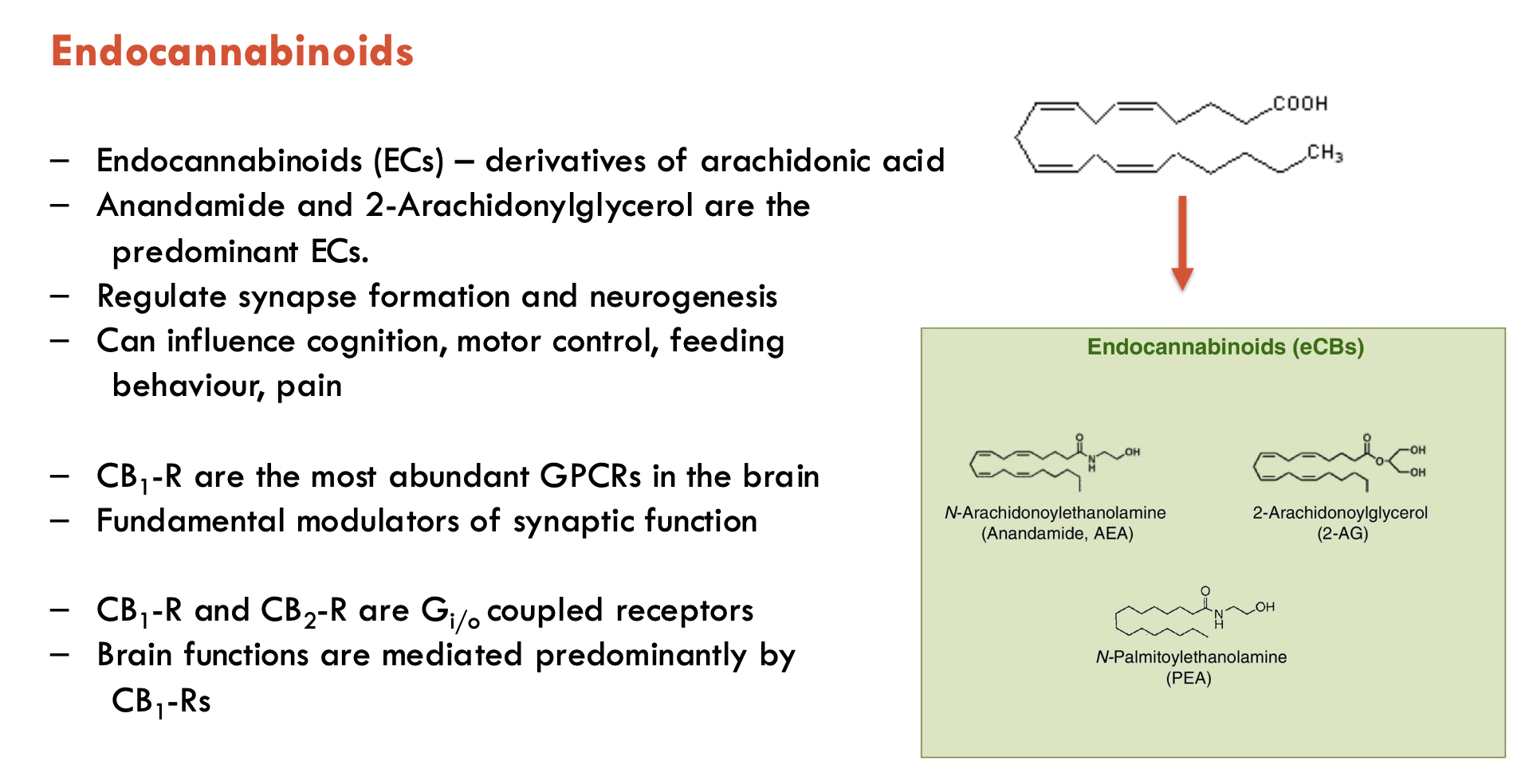
What are some features of CB1 receptors (4)?
CB1 receptors (CB1-R) are targets for endocannabinoids
most abundant GPCRs in the brain
fundamental modulators of synaptic function
brain functions are mediated predominantly by CB1-Rs
CB1-R and CB2-R are Gi/o-coupled receptors
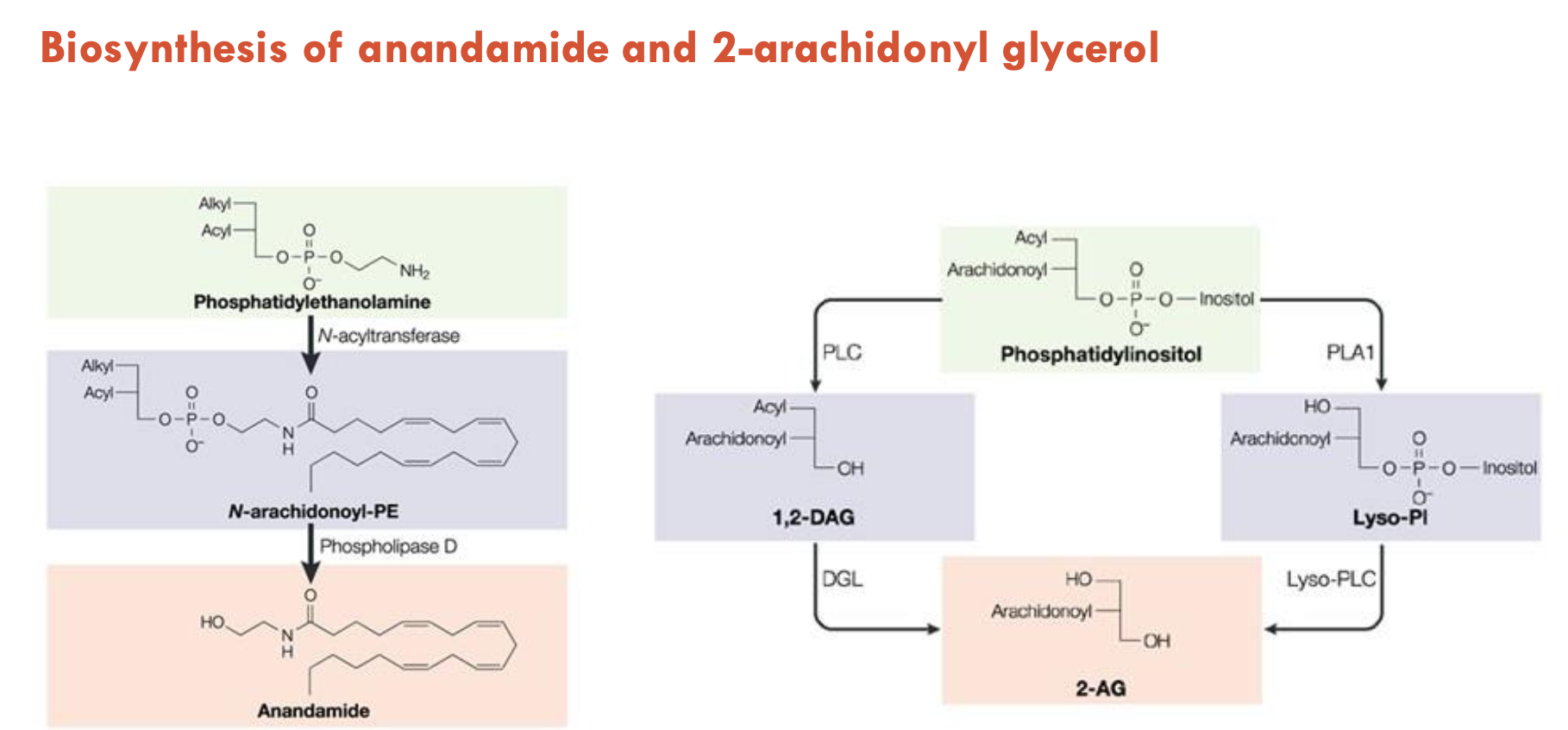
How are anandamide and 2-AG biosynthesised?
Anandamide
Base molecule: Phosphatidylethanolamine (PE) = 2 lipids + amine group on a glycerol backbone
Step 1: N-acyltransferase adds arachidonic acid to the amine group → forms N-arachidonyl-PE (NAPE), where 3 lipids are now attached
Step 2: Phospholipase D cleaves NAPE → releases anandamide
2-Arachidonyl Glycerol (2-AG)
Base molecule: Phosphatidylinositol (PI) = 2 lipids + inositol on a glycerol backbone
There are two pathways to create 2-AG:
PLC Pathway:
Phospholipase C (PLC) cleaves off inositol → 1,2-DAG
DGL cleaves off acyl and replaces it with a hydroxyl group → 2-AG
PLA1 Pathway:
Phospholipase A1 (PLA1) can remove the acyl tail —> Lyso-Pl
Lyso-PLC removes inositol-phosphate group → 2-AG
What is a special feature of anandamide (endocannabinoid) transport?
Dual solubility: eCBs are soluble in both aqueous and membrane environments
EC transporters may be involved in both release and re-uptake of ECs
Cholesterol (in the membrane) regulates EC transport, likely by:
direct interaction b/w EC and cholesterol OR
cholesterol regulating transporter activity
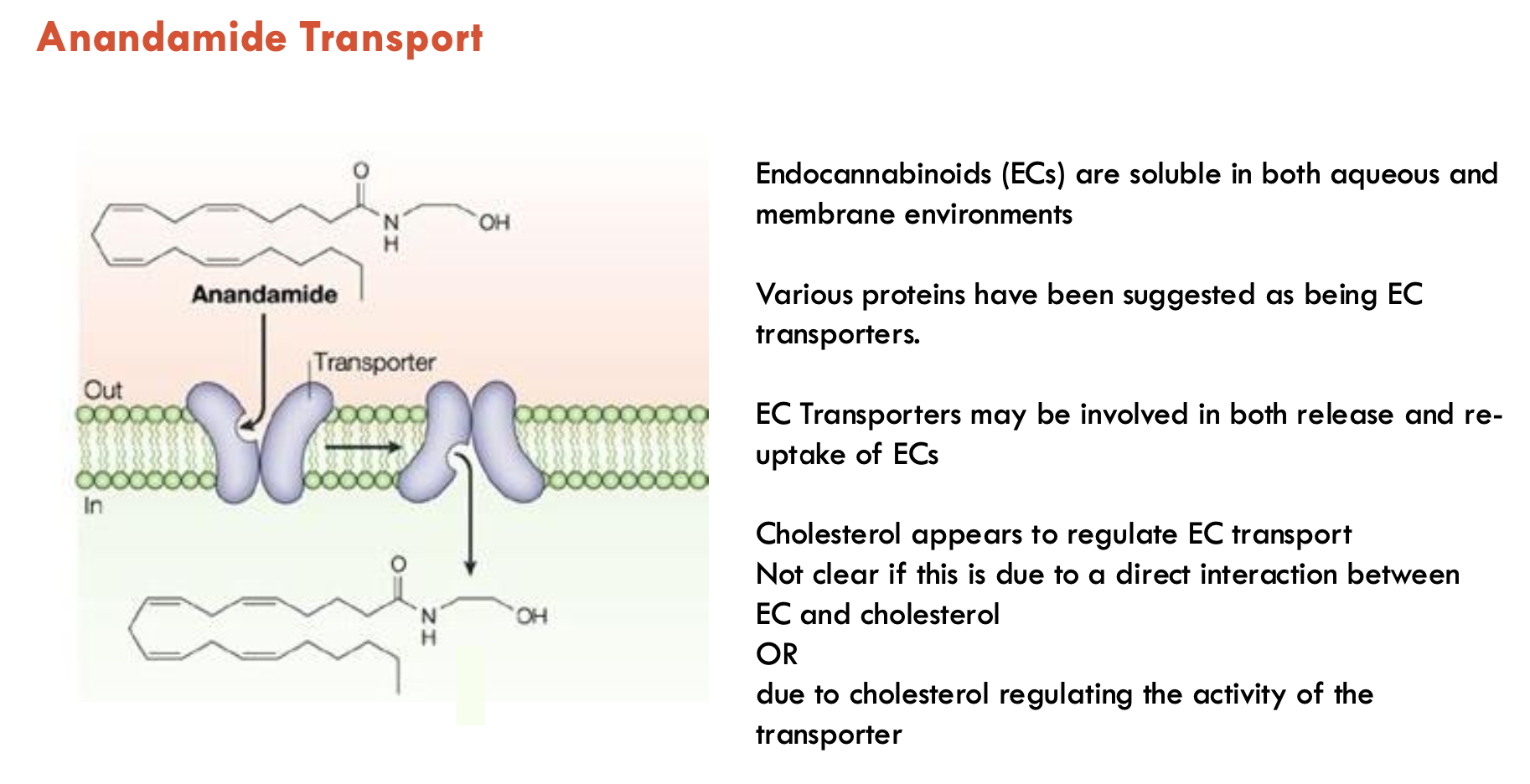
What enzymes degrade endocannabinoids (2)?
Fatty Acid Amide Hydrolase (FAAH): breaks down anandamide
Monoacylglycerol lipase (MAGL): breaks down 2-AG
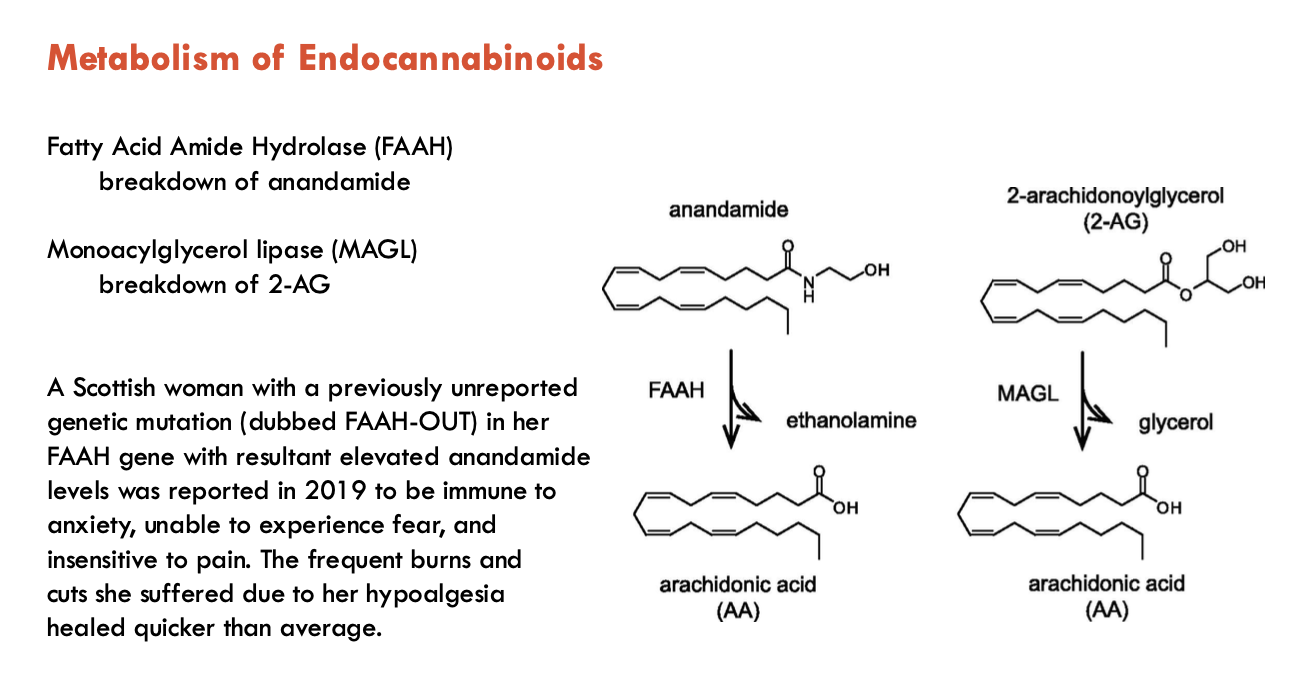
What is the location of enzymes involved in biosynthesis and breakdown?
Biosynthesis enzymes
PLC, DGL (for 2-AG biosynthesis) are found postsynaptically
N-acyltransferase, N-acylphosphatidyl-ethanolamine PLD (for anandamide biosynthesis) are found postsynaptically
Transporters (EMT) are found both pre- and post-synaptically
Breakdown enzymes
FA amide hydrolase - postsynaptic
Monoacylglycerol lipase - presynaptic
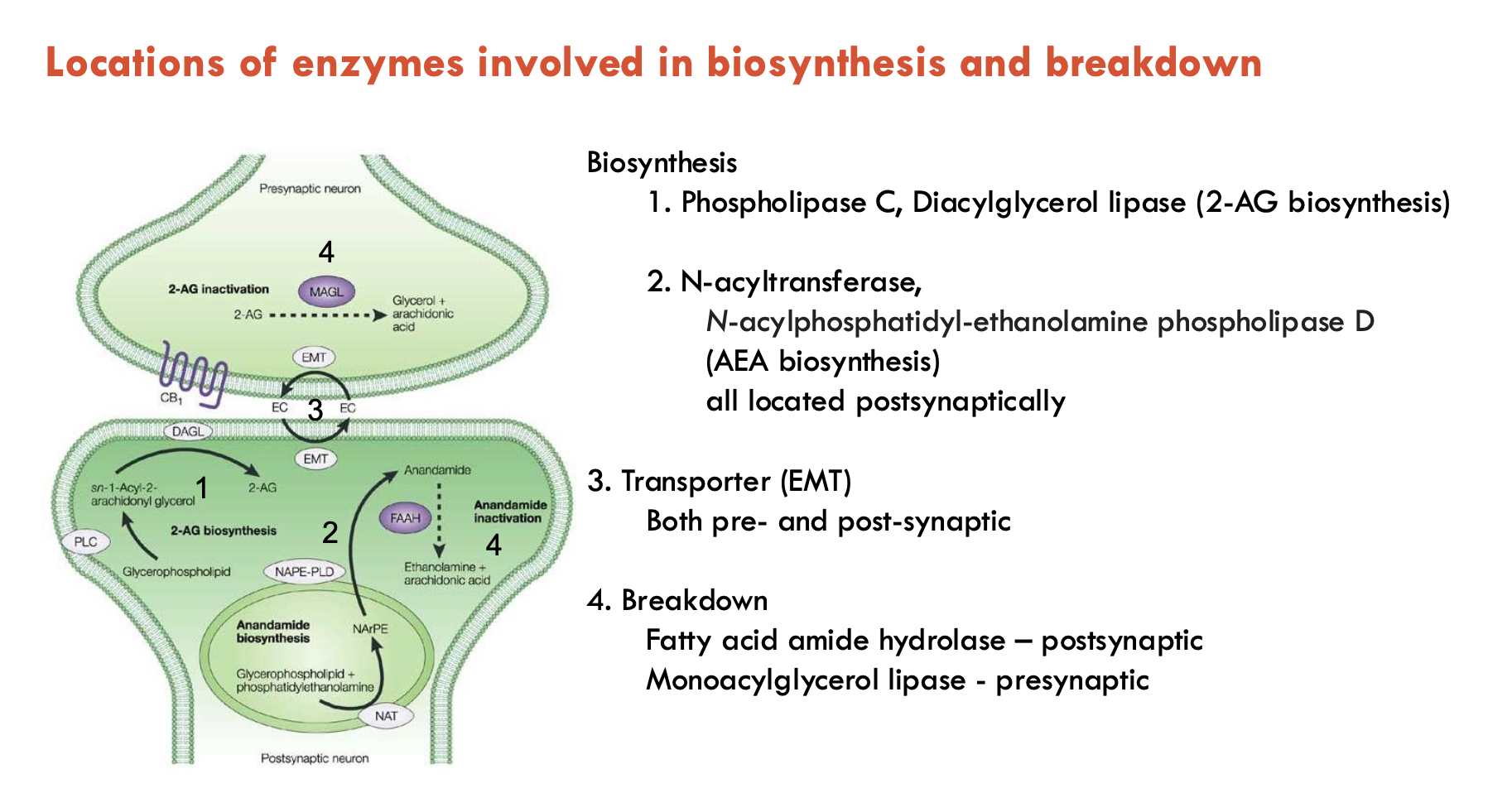
What happens during cannabinoid receptor activation?
Activation of CB1/2 stimulates the following, which lead to changes in gene expression:
Gi/o heteromeric proteins → inhibits adenylate cyclase → reduce PKA activity
Mitogen → activates protein kinase
Stimulation of Gi/o by CB1 receptors inhibits voltage-dependent Ca2+ channels and stimulates K+ channels → inhibits NT release
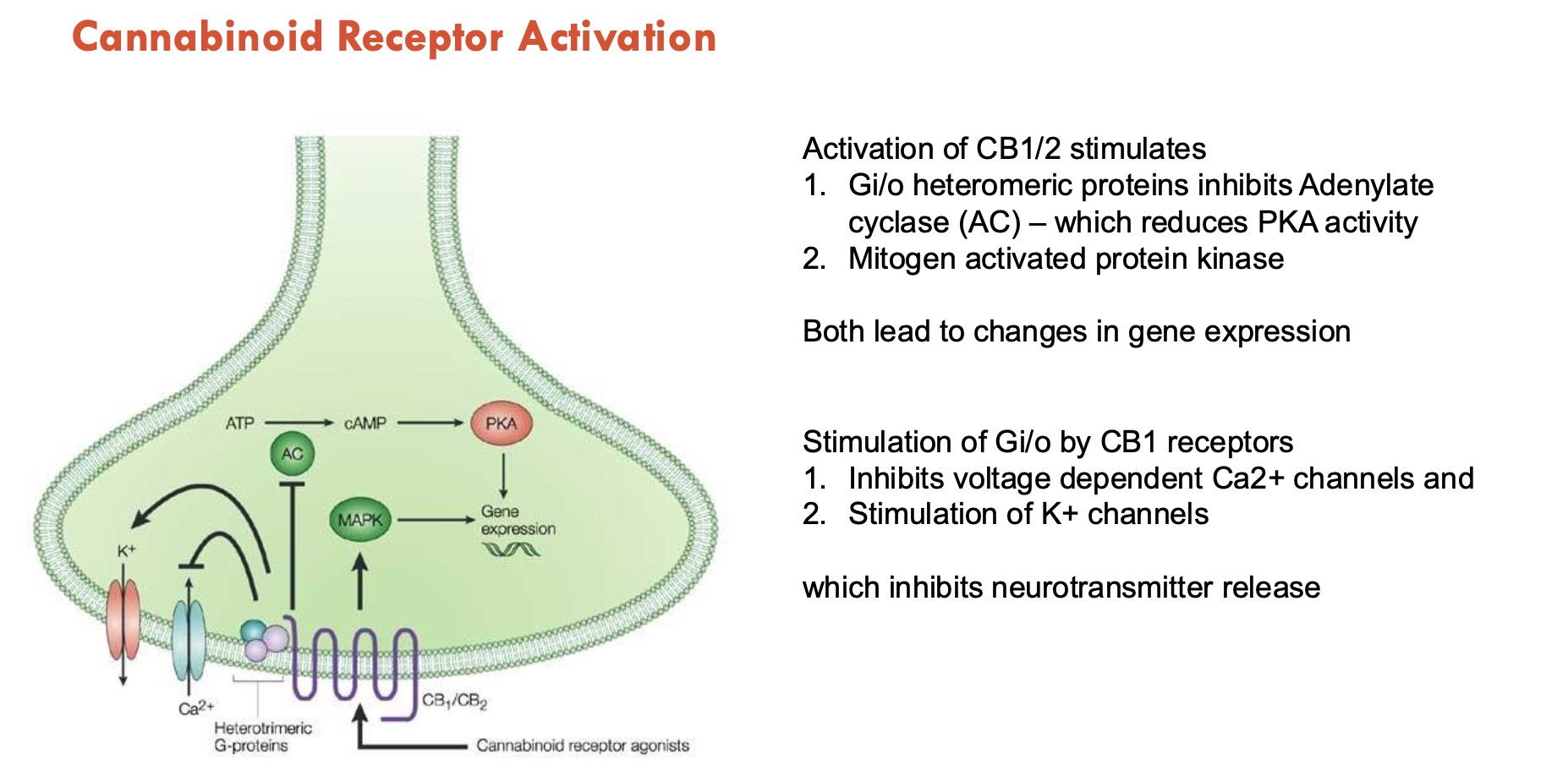
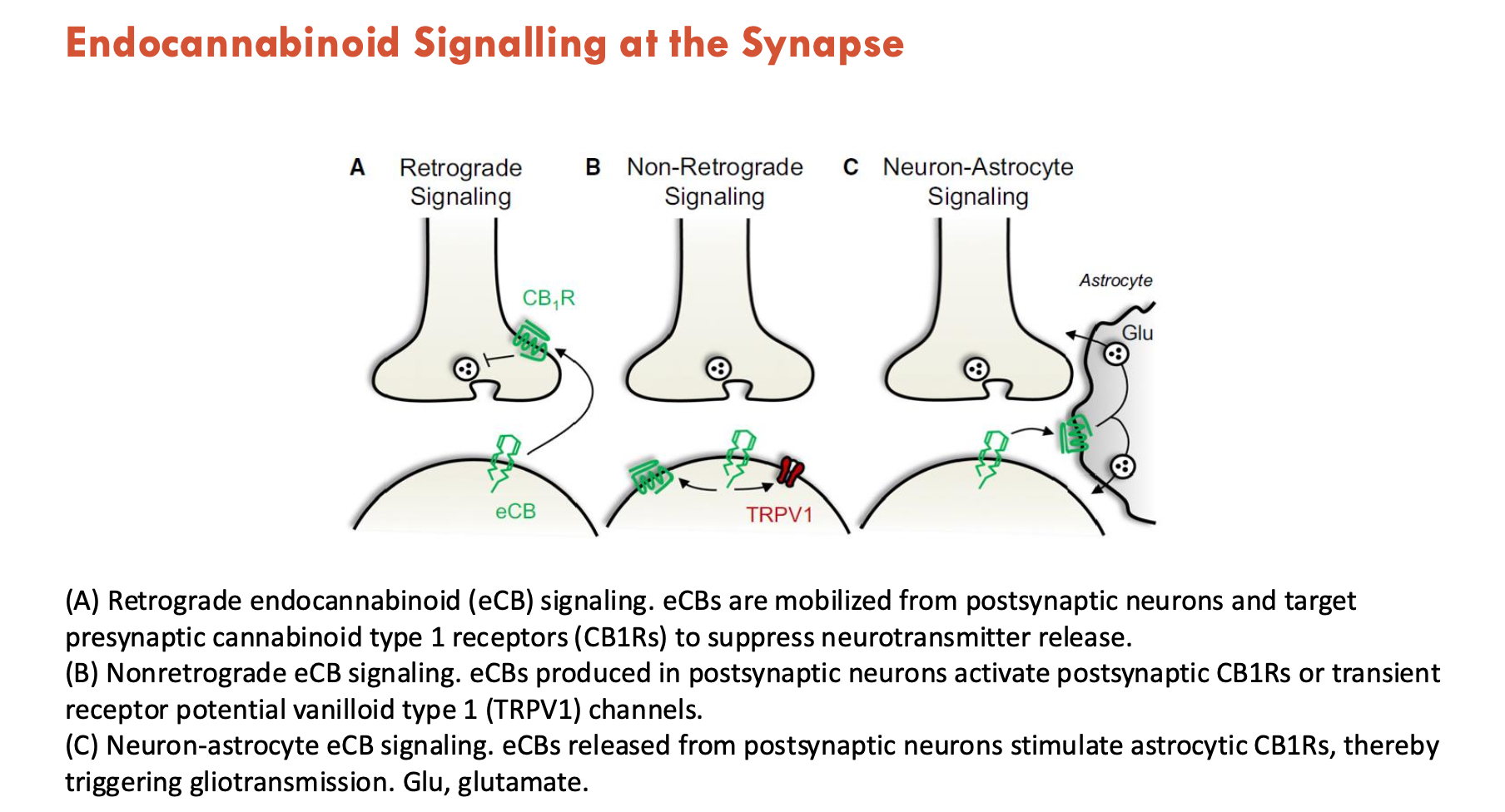
How do endocannabinoids signal at the synapse (3)?
Retrograde endocannabinoid (eCB) signalling: eCBs are mobilised from postsynaptic neurons and target presynaptic cannabinoid type 1 receptors (CB1Rs) to suppress NT release
Nonretrograde eCB signalling: eCBs produced in postsynaptic neurons activate postsynaptic CB1Rs or transient receptor potential vanilloid type 1 (TRPV1) channels
Neuron-astrocyte eCB signalling: eCBs released from postsynaptic neurons stimulate astrocytic CB1Rs → triggers gliotransmission (via GABA, glutamate)
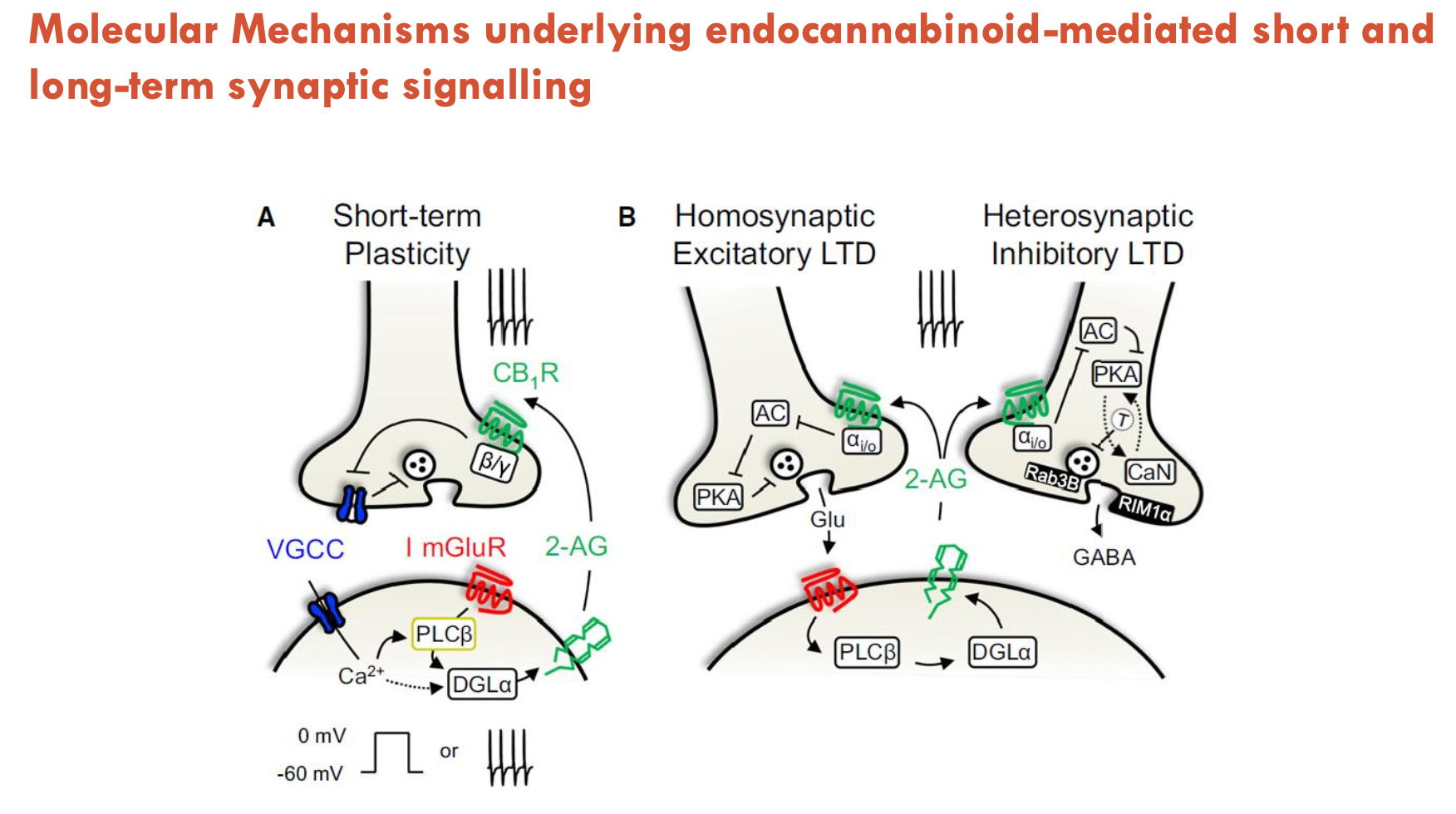
What molecular mechanisms underlie endocannabinoid-mediated short and long-term synaptic signalling?
1. Short-Term Plasticity (Retrograde Signalling):
Glutamate release → activates postsynaptic mGluRs → produces 2-AG
2-AG travels retrogradely (via anandamide transporter) → binds presynaptic CB1 receptors → inhibits NT release (e.g., glutamate/GABA).
2. Long-Term Depression (LTD):
Homosynaptic LTD (excitatory):
Repeated long-term glutamate release → prolonged CB1R activation → reduce glutamate release long-term (weakens glutamatergic synapses, self-inhibition)
Heterosynaptic LTD (inhibitory):
2-AG also binds CB1Rs on GABAergic terminals → suppresses GABA release (disinhibits neighbouring synapses)
Look into role of Tyrosine Phosphorylation in PDGF and Insulin Receptors
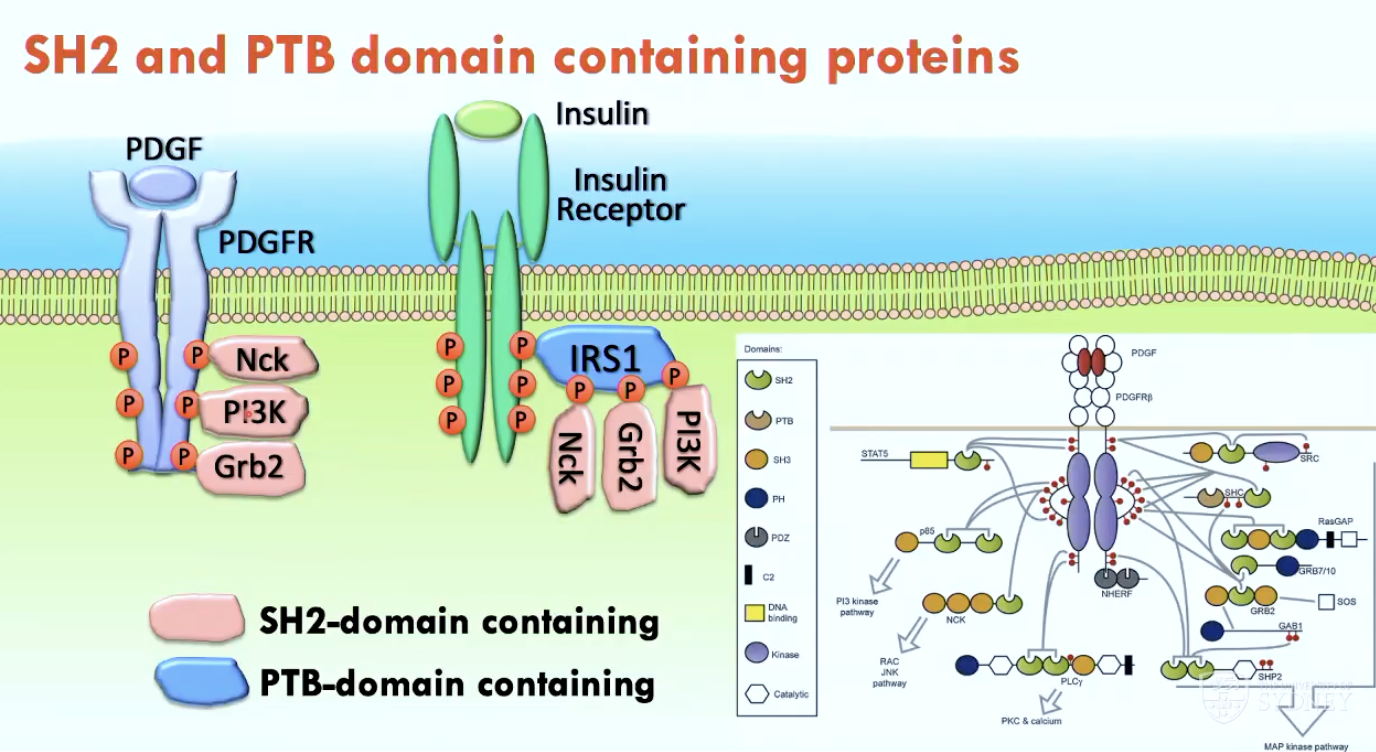
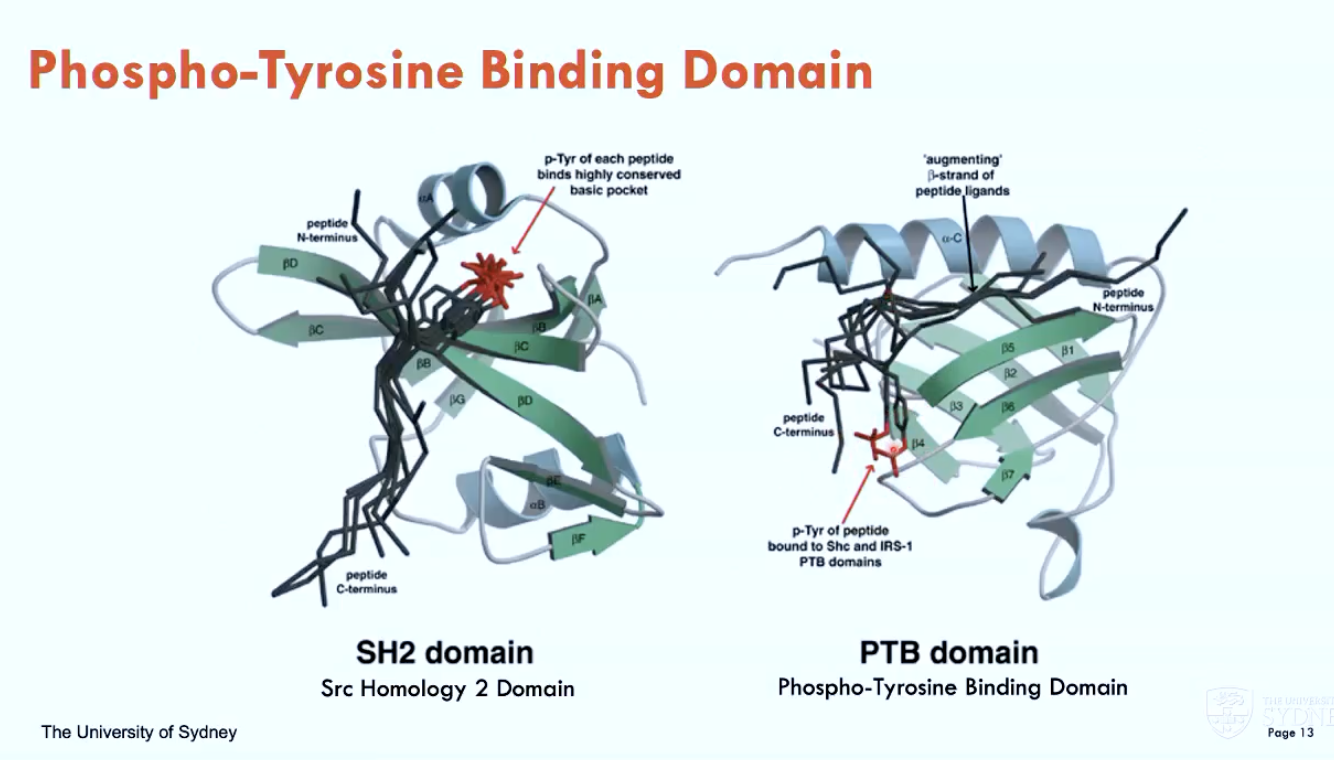
What are two phospho-tyrosine binding domains?
SH2 domain (Src Homology 2)
PTB domain (phospho-tyrosine binding)
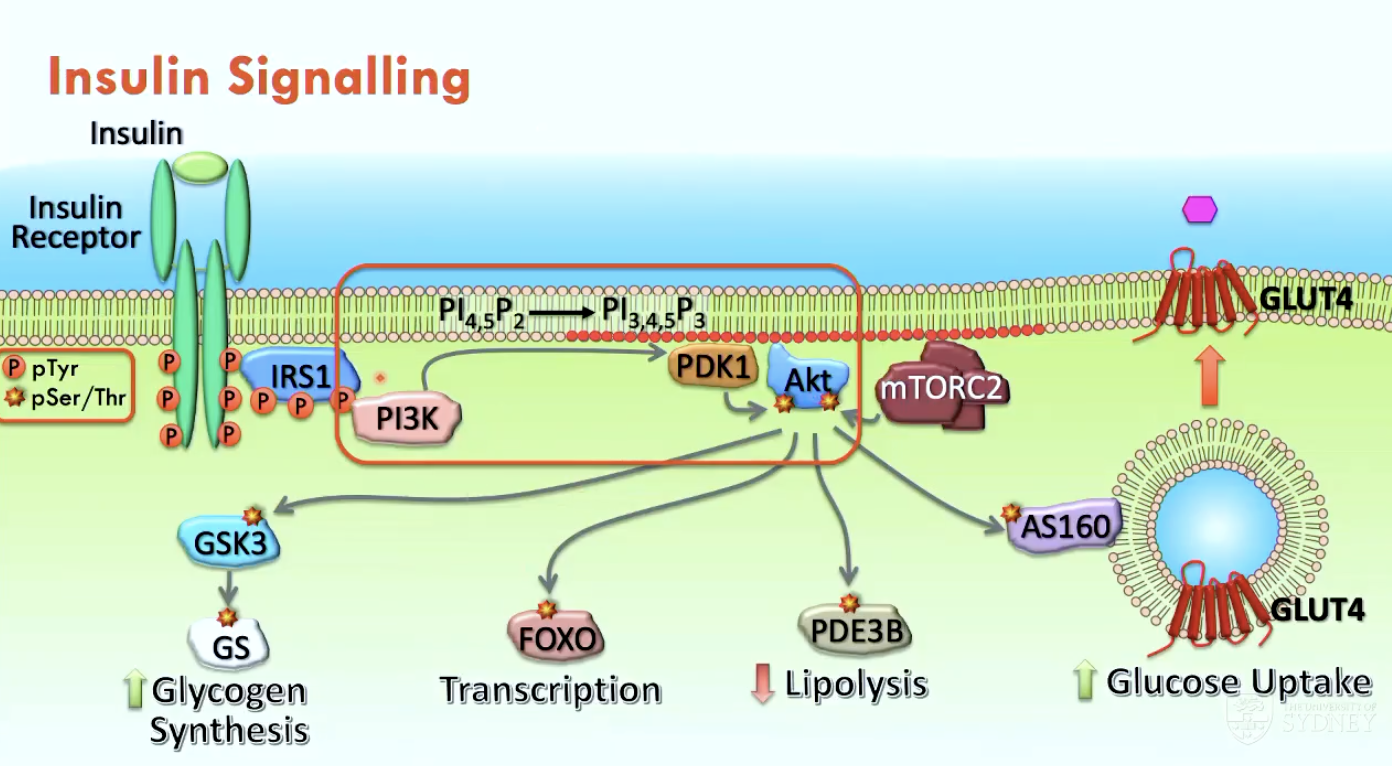
What occurs in insulin signalling (4)?
Insulin binds to the insulin receptor → activates tyrosine kinase activity
PI3K Activation: SH2 domain of PI3K binds to a phosphorylated tyrosine residue on IRS1, which is recruited to the membrane.
PIP3 Production: PI3K converts PIP2 into PIP3, which serves as a docking site for proteins with a pleckstrin homology (PH) domain.
Akt Activation: Akt binds to PIP3 via its PH domain → becomes activated by serine/threonine kinases that come in close proximity
Activated Akt kinase (Ser/Thr) drives metabolic effects:
phosphorylates AS160 → opens GLUT4 channels to uptake glucose
phosphorylates PDE3B → reduces lipolysis
phosphorylates FOXO → translocates FOXO into cytoplasm → stops glucose-producing genes
phosphorylates GSK3 → increases glycogen synthesis
Describe Green Fluorescent Protein (GFP)
GFP tags proteins or domains, allowing for live-cell fluorescent microscopy
GFP-tagged PH-domain helps visualise insulin signalling.