tutorial 7: isolation, cloning and transformation of plasmid DNA-Day 2 gel electrophoresis
1/9
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
10 Terms
how big are the pKAN and pUC18 plasmids? what does this mean for tehir locations on the gel?
The pKan plasmid has a molecular weight of 4,194 bp, whereas the pUC18 plasmid is only 2,686 bp.
Therefore, the pUC18 plasmid will appear lower on the gel image relative to pKan
after electrophoresis, there may be multiple bands of varying intensities and migration distances rather than one band. why might this be?
These may correspond to different plasmid topologies, or conformations in which a plasmid may exist.
Plasmid conformations include: supercoiled, circular, and linear forms
(plasmid with cuts in both strands) among othersThe size differences will also influence their migration rates.
The covalently closed circular topology is the most compact and will migrate furthest, followed by the linear and open circular forms
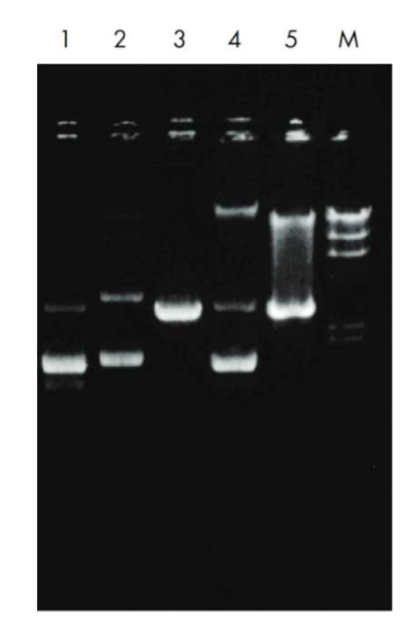
describe what plasmid structure is in each lane
Lane 1: Supercoiled (lower band) and open circular form (ds DNA, nicked in one of the strands to released supercoils; upper band) of the high-copy plasmid pUC18 with an additional band of denatured supercoiled DNA migrating just below the supercoiled form.
Lane 2: Multimeric forms of supercoiled plasmid DNA which may
be observed with some host strains, and should not be mistaken for
genomic DNA. Multimeric* plasmid DNA can easily be distinguished from genomic DNA by a simple restriction digestion: linearization of a plasmid sample displaying multimeric bands will yield a single defined band with the size of the linearized plasmid monomer (see lane 3).Lane 3: Linearized form of the plasmid shown in lane 2 after restriction digestion with EcoRI.
Lane 4: Sample contaminated with bacterial chromosomal DNA, which may be observed if the lysate is treated too vigorously Genomic DNA contamination can be identified by digestion of the sample with EcoRI. A smear is observed, in contrast to the linear band seen after digestion of multimeric plasmid forms.
Lane 5: Eco RI digestion of a sample contaminated with bacterial genomic DNA which gives a smear above the plasmid DNA.
what are multimers? how do they migrate on a gel?
fused products of several plasmids recombined together. These plasmids are
several times as large as the individual plasmid and therefore run very slowly in agarose (regardless of their conformation)
describe a Nicked, Relaxed Circular Plasmid
DNA found in the supercoiled form is not easily accessed by replication machinery.
During replication, cellular topoisomerases nick one strand of the DNA helix and relax the superhelical tension, thus allowing polymerases to gain access to the DNA.
This large floppy circle is the slowest migrating form in an agarose gel
describe a linear plasmid
Linearized DNA occurs when the DNA helix is cut in both strands at the same place.
Linear DNA generally migrates between the nicked circle and the supercoiled forms. However, it may also migrate the same distance as nicked circle — it migrates as predicted by the length of the DNA (as compared to the MW markers).
You can identify the linear DNA form on an agarose gel by comparing uncut plasmid DNA with a sample of the plasmid that has been linearized using a restriction enzyme.
If you get linear DNA when you are hoping for supercoiled (e.g. after a plasmid prep) it is due to nuclease contamination or harsh treatment during purification.
describe a supercoiled plasmid
Supercoiled DNA is the native confirmation found in vivo and occurs when extra twists are introduced into the double helix strand.
Supercoiled DNA migrates faster than predicted in an agarose gel due to its conformation
describe circular, single stranded plasmid
During alkaline lysis plasmid preps, plasmids are denatured because the hydrogen bonds are disrupted by the alkaline conditions.
But the covalently-closed circular strands remain intact and topologically constrained and when the pH is returned to neutral the hydrogen bonds reform and the supercoiled DNA is re-formed.
However, if the alkaline lysis step is overly harsh (e.g. it is incubated for too long) the DNA can become permanently denatured and give you single
stranded closed circles that migrate ahead of all of the other forms of the plasmid in a gel
Why is the RedSafe Nucleic Acid Stain added?
RedSafe™ Nucleic Acid Staining Solution is a new and safe nucleic acid staining solution. It is an alternative to the traditional ethidium bromide (EtBr) stain for detecting nucleic acid in agarose gels. It emits green fluorescence when bound to DNA or RNA.
This new stain has two fluorescence excitation maxima when bound to nucleic acid, one centered at 309 nm and another at 419 nm. In addition, it has one visible excitation at 514 nm.
The fluorescence emission of RedSafe™ bound to DNA is centered at 537 nm
What is the role of the running dye?
Tracking dye ( DNA Loading or Running Dye, a charged, low molecular weight compound) is usually loaded into each well at the start of the run to monitor the progress of molecule movement on the gel.
gel loading dye is a colored dye that is mixed with a DNA sample prior to running electrophoresis.
Loading dyes serve a few purposes:
They color your sample, making it easier to tell if you were successful in loading your gel.
They are dense and keep the DNA in your sample from diffusing away in the buffer.
They migrate through the gel alongside the DNA, helping you track the rate of migration of your sample through the gel.