bio u3o2 (biochem pathways)
1/97
Earn XP
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
98 Terms
anobolic reaction
reactions which atoms and small molecules are joined together to make a larger molecule
e.g. glucose to glycogen
endergonic
catabolic reaction
reactions in which larger molecules are broken down into smaller molecules
e.g. digestion of food
spontaneous and exergonic
endergonic reactions
reactions that require the input of energy to proceed
e.g. photosynthesis
exergonic reaction
reaction that do not require an initial input of energy to proceed
the importance of enzymes in relation to biochemical pathways
The biochemical pathway is a series of chemical reactions, each controlled and regulated by an enzyme (specific)
Enzymes act as catalysts to lower the activation energy of chemical reactions
the enzyme brings the initial substrate molecule(s) to a final product.
Without enzymes, biochemical reactions would occur too slowly to maintain life
enzyme
enzymes are proteins acting as catalysts that speeds up the rate of a chemical reaction by lowering its activation energy, and is not consumed in the reaction
how does the specificity of enzymes’ shape link to its function?
enzyme specificity is why enzymes can control each step in a biochemical pathway, as there are a lot of enzymes, with each acting on one or a small number of substrates
the active site on an enzyme is where the substrate(s) would attach and is specific for a particular substrate
product molecules are released as their changed shape is no longer specific to the active site and no longer binds to it
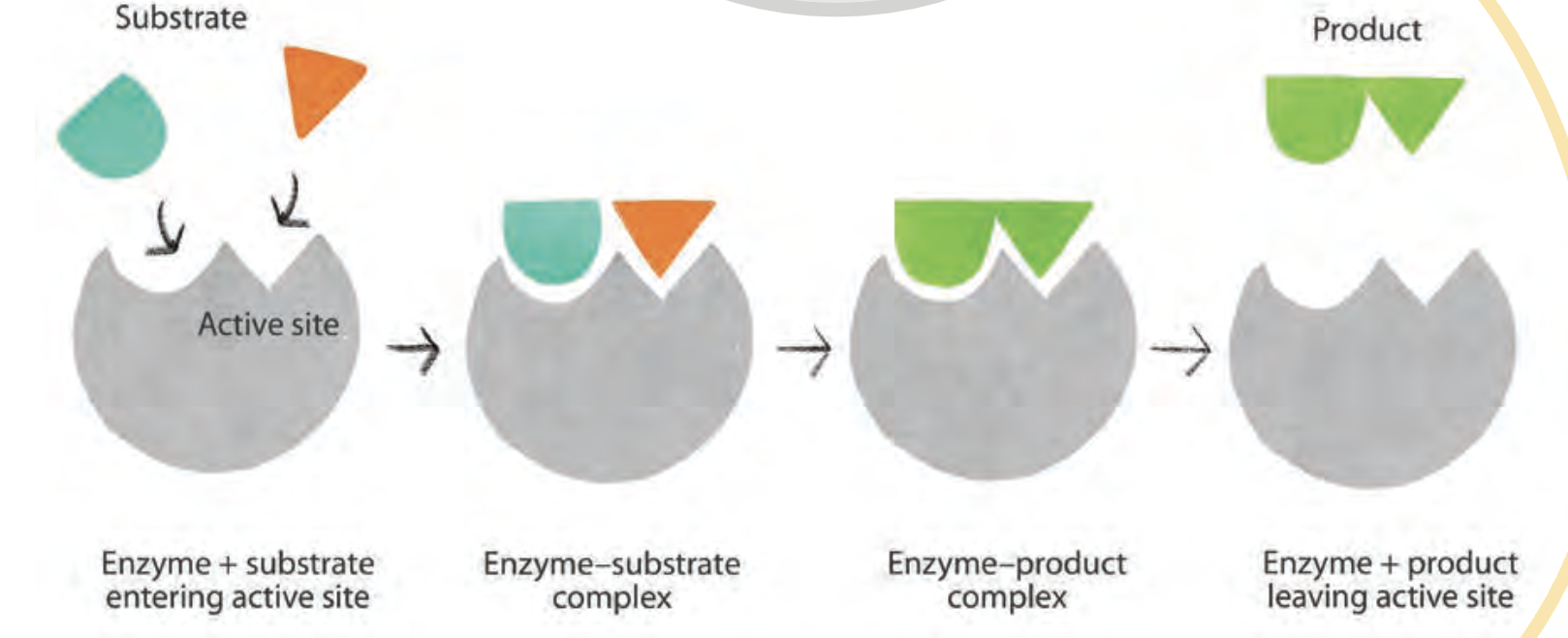
Identity the components of the enzyme-substrate complex
substrate
enzyme
product
active site
allosteric site
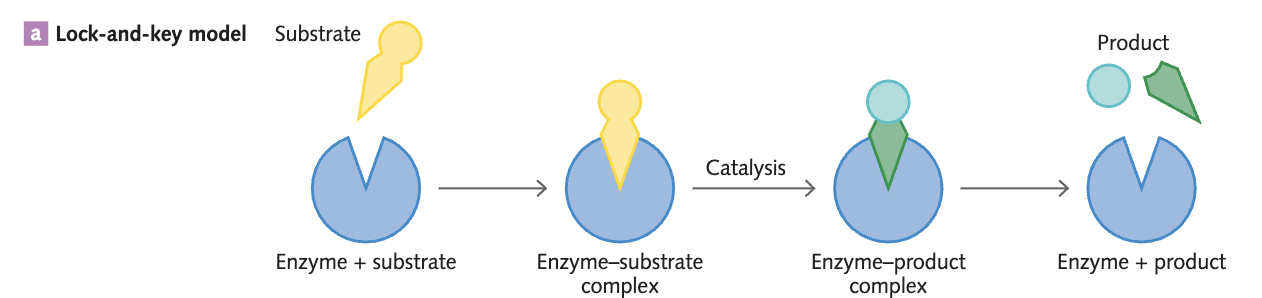
The lock-and-key model in terms of enzyme action
The lock-and-key model shows the binding into the active site of an enzyme, where the substrate’s shape is complementary to the shape of the specific active site on the enzyme.
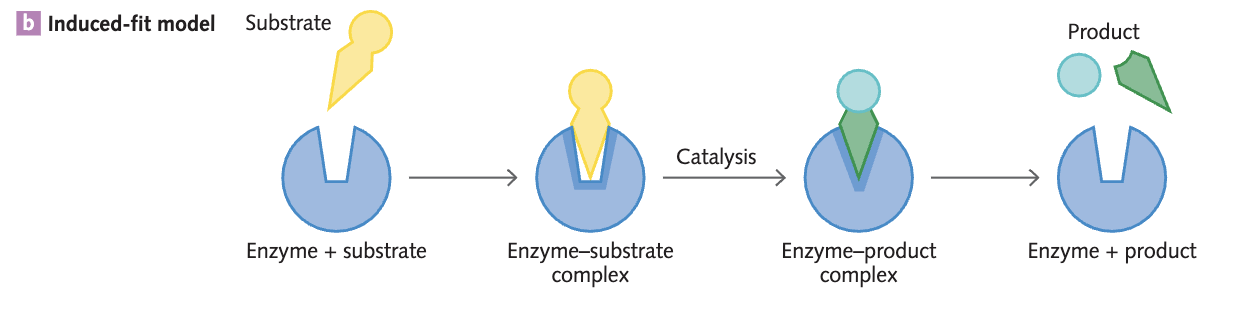
The induced-fit model in terms of enzyme action
The induced-fit model shows how the bonds that form between an enzyme and its substrate can modify the active site’s shape so that the substrate can fit into the active site
activation energy
energy required to initiate a reaction
coenzymes
non-protein
organic
assists enzyme activity by carrying groups of atoms to or from the reaction
required in cellular respiration and photosynthesis
e.g. ATP, NADH, NADPH, FADH2
cofactors
inorganic
assists enzyme activity by helping the enzyme to fold properly or to facilitate the reaction
e.g. Mg2+, Zn2+
coenzymes involved in photosynthesis
ADP + Pi → ATP
NADP+ → NADPH (light dependant)
The primary coenzyme involved in photosynthesis is NADP⁺.
During the light-dependent reactions, NADP⁺ acts as an electron carrier, accepting electrons and hydrogen ions produced when water molecules are split by sunlight. This conversion forms NADPH.
NADPH then carries the high-energy electrons to the Calvin cycle (light-independent reactions), where it provides the reducing power needed to convert carbon dioxide (CO₂) into glucose and other carbohydrates.
coenzymes involved in cellular respiration
The coenzymes involved in cellular respiration are: NAD+ and FAD. They form NADH and FADH₂ respectively and carry electrons to the electron transport chain from glycolysis and the Krebs cycle where the molecules are generated.
how are coenzymes involved photosynthesis and cellular respiration
Photosynthesis:
ATP and NADPH are produced during the light-dependent reactions and used up in the Calvin cycle
NADP+ and ADP (+Pi) are loaded in the light-dependent reactions to produce NADPH and ATP. The NADPH is then unloaded and used in the light-independent reaction (calvin cycle)
ADP + Pᵢ and NADP⁺ are de-energised energy carriers that will be required to absorb energy released from the splitting of water
Cellular respiration:
The NADH produced in glycolysis is used in the ETC. The unloaded form, NAD+, accepts 1 H+ and 2e-.
FAD is loaded in glycolysis and krebs cycle, where it accepts 2 H+ and 2e- to form FADH2, which is unloaded in the ETC
in glycolysis, 4 ATP produced 2 ATP consumed
how do coenzymes faciliate reactions
Coenzymes are loaded and unloaded to move energy, protons, and electrons between reactions in the cell through the acceptance/donation
e.g. NADP+ (unloaded) accepts H+ to produce NADPH (loaded) in photosynthesis
NADPH is required to provide energy and hydrogen ions for the formation of glucose
NAD+ accepts 1 H+ ion and 2 e- and is involved in all stages of cellular respiration
FAD → FADH2, FAD accepts 2 H+ ions and 2 e- and is only used in the ETC
The hydrogen ions (H+) from NADH and FADH2 are moved to the intermembrane space of the mitochondria.
how the cycling of energy in terms of electrons helps to create loaded
coenzymes
The unloaded form of a coenzyme accepts an electron, a proton, or a chemical group.
The loaded coenzyme stores chemical energy in the bonds between the coenzyme and the chemical group it accepts.
Coenzyme’s relationship with electrons in the electron transport chain (ETC)
NADH and FADH2 carry electrons as well as H+ ions, releasing both together
the H⁺ ions are moved to the intermembrane space of the mitochondria
Electrons are moved along the chain of protein complexes
Hydrogen ions, oxygen gas (O2) and electrons combine to form water (oxygen is the final e- acceptor): 4 H⁺ + 4 e⁻ + O₂ → 2 H₂O
Competitive inhibitor and how it works
Competitive inhibitors compete directly with the substrate for space in the active site and prevent the substrate from binding, either lowering the rate or stopping the reactions.
The shape of the inhibitor molecule is complementary to the shape of the active site of the enzyme
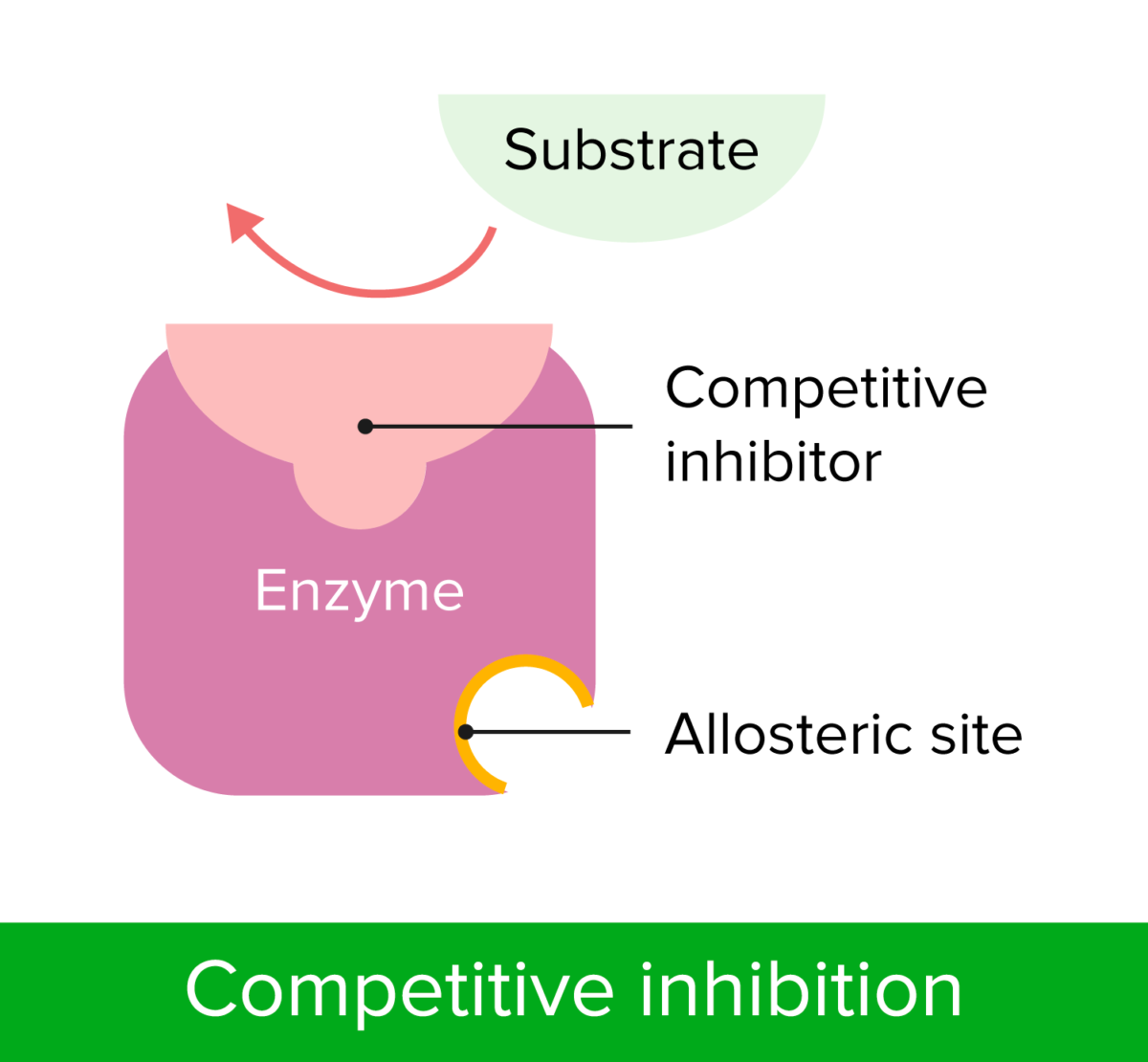
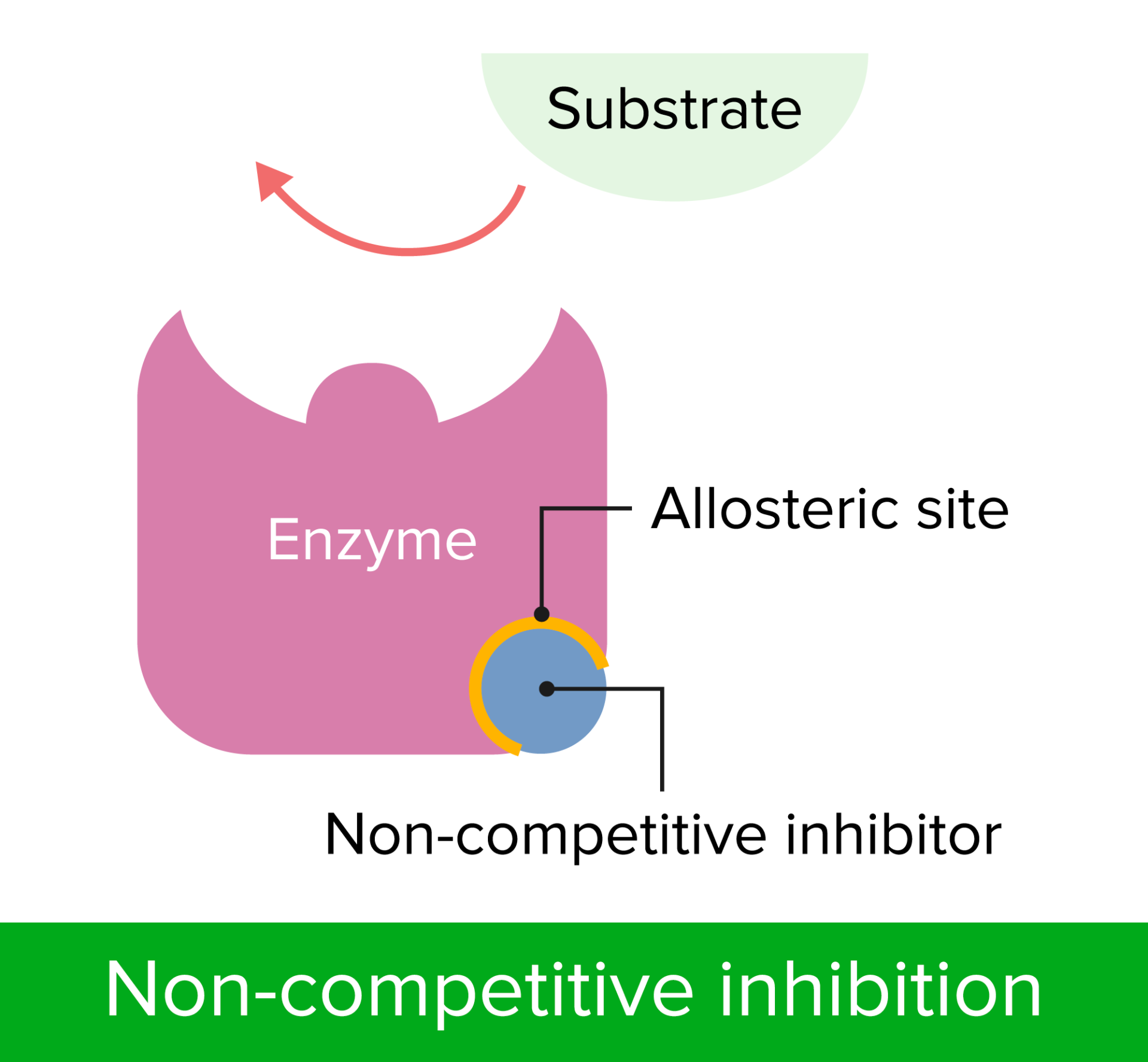
Non-competitive inhibitor and how it works
Non-competitive inhibitors are molecules that bind to a part of the enzyme that is not at the active site, called the allosteric site.
This alters the structure of the enzyme in such a way that the active site of the enzyme changes shape and no longer has the complementary shape for the substrate to bind.
Feedback inhibition and how it works
type of non-competitive inhibition
When the output of a process is in excess, it will act as an inhibitor by binding to the allosteric site of the enzyme to alter its shape and slow the rate of reaction
conserves energy and resources
Factors affecting enzyme activity
temperature
pH
substrate concentration
Inhibitors (competitive, non-compe
catalyst importance in biochemical pathways
Enzymes act as catalysts to lower the activation energy of chemical reactions. As a result of lowering the activation energy, enzymes make chemical reactions more likely to occur (called making them ‘energetically favourable’), which makes the reactions appear to run more quickly. Without enzymes, chemical reactions would proceed too slowly.
reversible inhibition
Reversible inhibitors can temporarily prevent enzyme activity when binding to the enzyme to prevent the formation of the product. When product in the cell decreases as it is used up or removed from the cell, the inhibitory effect no longer existsand the biochemical reaction can begin again.
where can inhibitors bind to enzymes
active site - competitive inhibitors
Competitive inhibitors are molecules that bind to the active site of an enzyme, directly competing with the substrate and preventing it from attaching.
allosteric site - non-competitive inhibitors
Non-competitive inhibitors bind to a site other than the enzyme's active site, called the allosteric site, causing a change in the enzyme's shape (no longer shape specific to enzyme) and reducing its activity
purpose of photosynthesis
Photosynthesis is the process by which green plants, algae, and some bacteria convert light energy into chemical energy (light energy → chemical energy).
It mainly occurs in the chloroplasts of plant cells. During photosynthesis, plants take in carbon dioxide from the air and water from the soil. Using sunlight, they transform these into glucose and oxygen. This process is essential for producing food and oxygen, supporting life on Earth.
word and chemical equation of photosynthesis
6CO₂ + 6H₂O → C₆H₁₂O₆ + 6O₂
carbon dioxide + water → glucose + oxygen
chlorophyll location and function
chlorophyll is the green pigment in the organelle chloroplast
chlorophyll absorbs light in the thylakoid membranes and allows it to convert into energy used to split a water molecule
irreversible inhibition
When a compound binds to the amino acids that make up the enzyme, changing the structure
reversible inhibition
When the enzyme is not permanently inhibited or damaged.
There are two types: competitive and non-competitive
compounds are bonded non-covalently
denature vs interrupted enzyme
denatured enzyme has completely altered the structure of the protein
enzyme activity can be interrupted by inhibition
An enzyme’s active site changes during denaturation; it is not the substrate’s shape that changes.
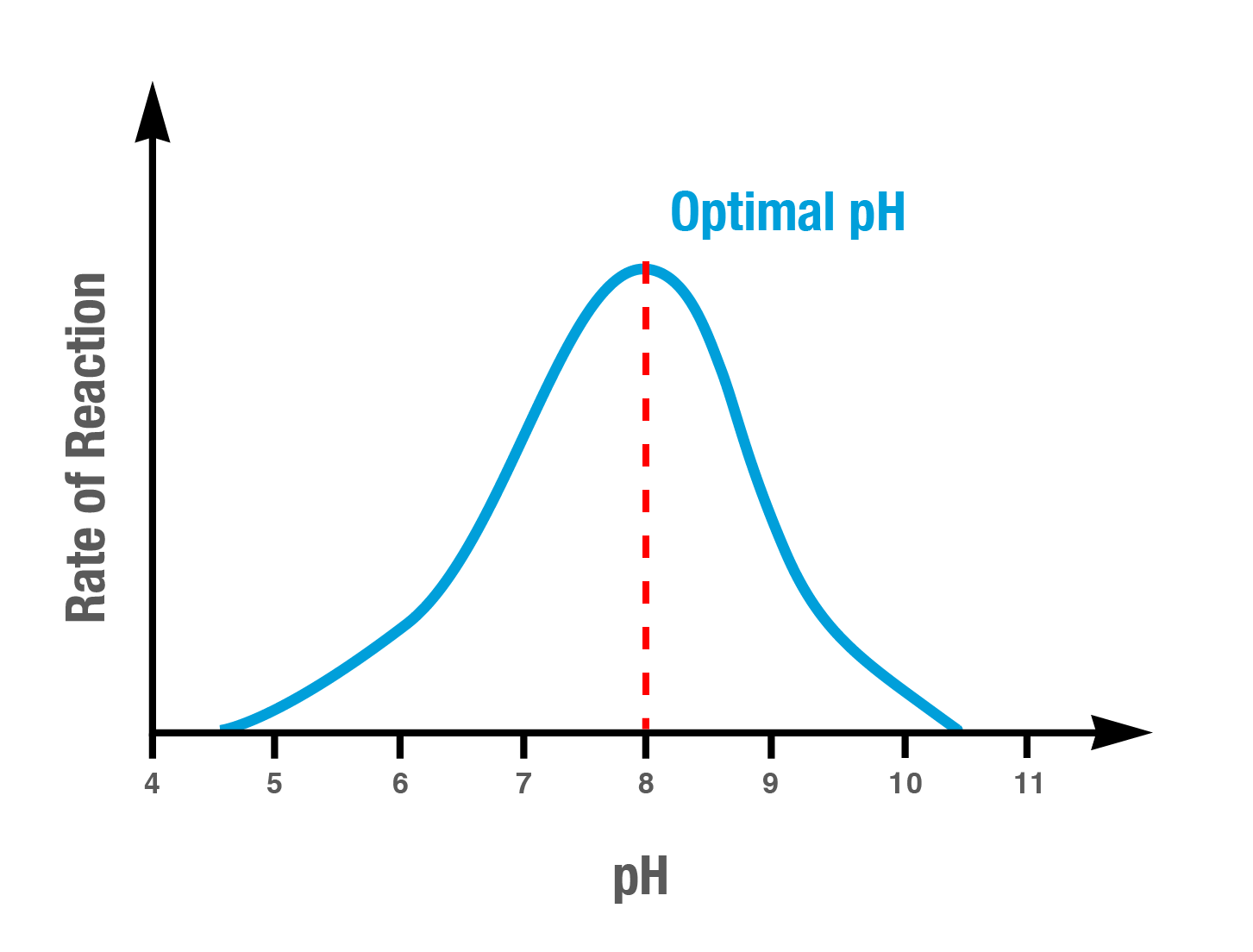
how pH affects enzyme activity
Each enzyme has an optimal pH range in which it functions most effectively, outside the opimal range leads to reduced activity or denaturation of the enzyme.
too high or low pH levels can cause enzymes to denature, losing their 3D structure and the shape of the active site. This loss of structure can prevent the substrate from binding properly.
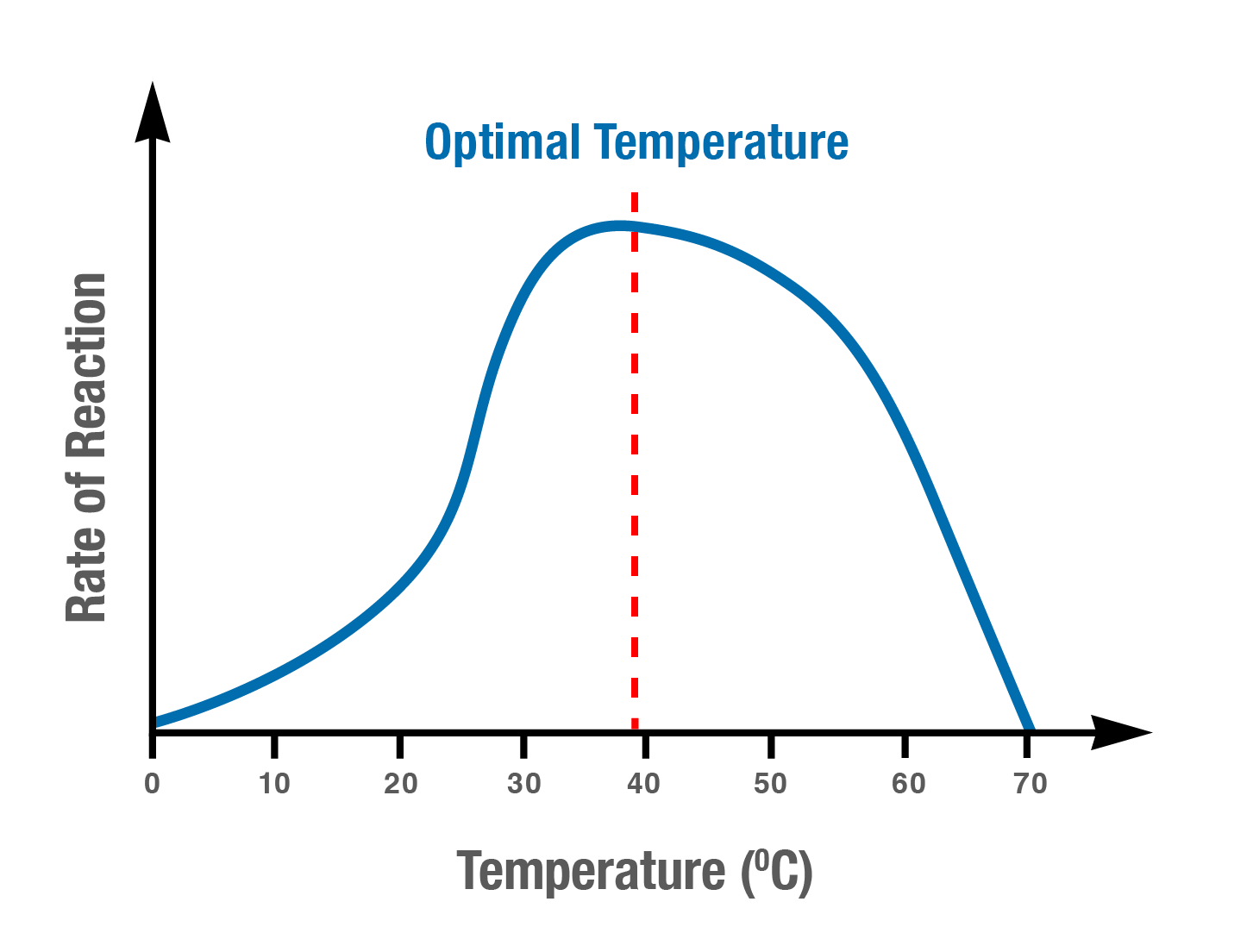
how temperature affects enzyme activity
Collision theory
temperature can affect the rate of enzyme-catalysed reactions as the substrate molecule moves faster in high temperatures and slower in low temperatures
at low temperatures, molecular movement slows, and fewer collisions occur between the substrate and enzyme molecules. The reaction rate decreases
at high temperatures, the molecules begin to move faster as they gain energy. More collisions occur between the substrate and enzyme molecules. The reaction rate increases.
Optimal temperature
Different enzymes have different optimum temperatures
enzyme’s activity rates are highest at its optimum temperature
Denaturation
If the temperature gets too high, the bonds that determine the three-dimensional shape of the enzyme proteins will break. As a result, the protein loses its functional shape. It becomes permanently denatured and the substrate can no longer fit into the active site. The enzyme’s activity stops.
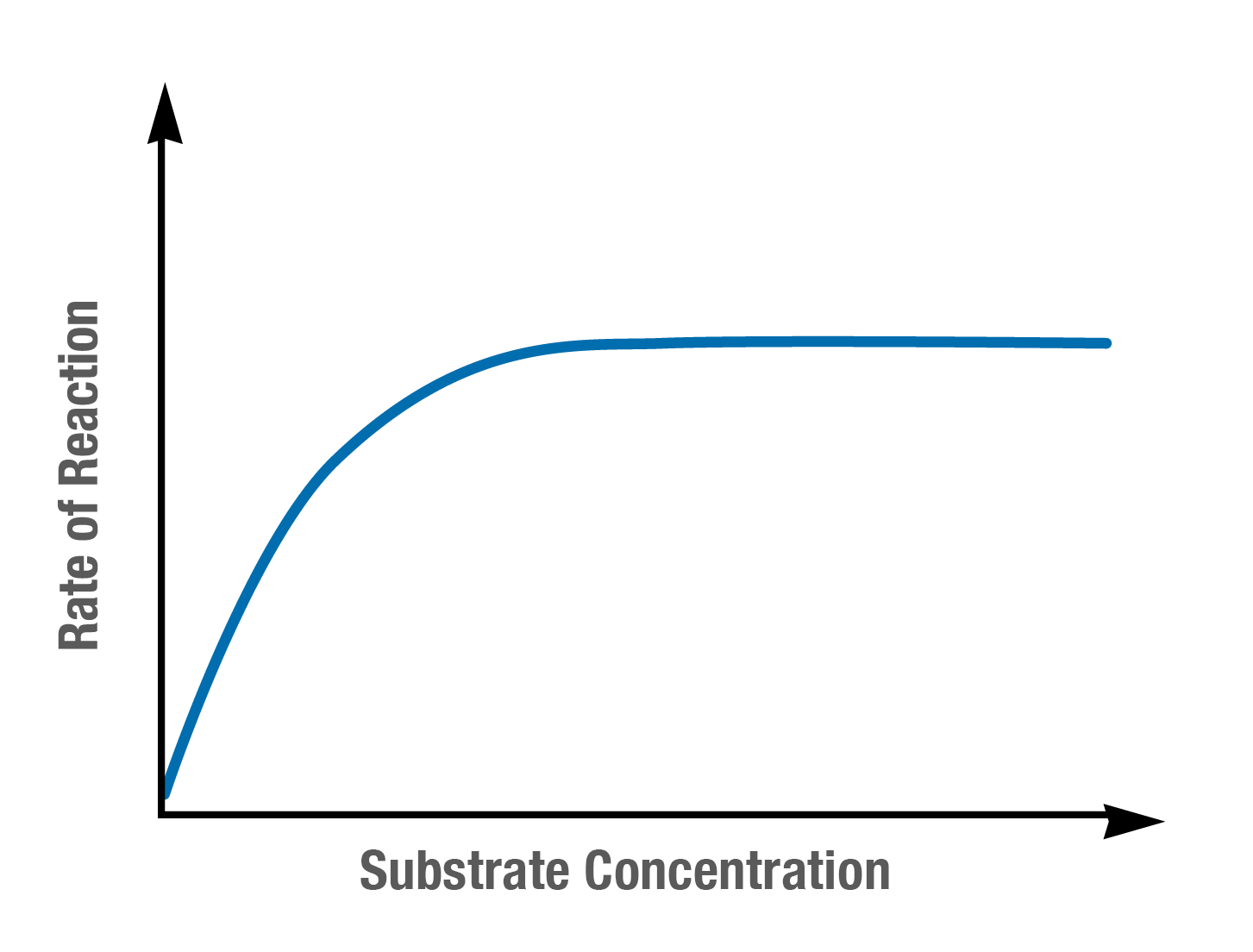
how substrate concentration affects enzyme activity
As substrate concentration increases, the rate of an enzyme-catalysed reaction initially rises as more substrate molecules are available to bind to enzyme active sites. This continues until the enzyme’s saturation point is reached.
how enzyme concentration affects enzyme activity
Increasing enzyme concentration can increase the rate of reaction, as there are more enzyme molecules available to catalyse the reaction, leading to more frequent enzyme-substrate interactions.

location of light dependent reaction
thylakoid membrane of the chloroplast grana.
input and output of light dependent reaction
Input:
H2O
ADP + Pi
NADP+ (unloaded coenzyme)
light (only occurs in the presence of light)
Output:
O2
ATP
NADPH (loaded coenzyme)
location of the light-independent reaction/Calvin cycle
the stroma of the chloroplast.
input and output of the light-independent cycle/Calvin cycle
Input:
CO₂
NADPH (loaded coenzyme)
ATP
extra: rubisco is the enzyme catalysing carbon fixation
Output:
C6H12O6
O2
NADP+ (unloaded coenzyme)
ADP + Pi
Carbon fixation process in C3 plants
C3 uses the most common carbon fixation method, where the Calvin Cycle's first step is producing a 3-carbon compound called 3-PGA.
rubisco is the enzyme used to catalyse the carbon fixation reaction between CO₂ and RuBP
3-PGA which is converted to glucose
at higher temperatures, rubisco will bind with O₂ instead of CO₂, in a process called photorespiration. Therefore, C3 plants are found in temperate climates.
enzyme used in carbon fixation?
Rubisco
Glucose formula and origin of its components
Formula: C₆H₁₂O₆
Components’ origin: RuBP, CO2, H+ ions from NADPH
differences in leaf structure of C3, C4 and CAM plants
C3:
Calvin Cycle's first step is producing a 3-carbon compound called 3-PGA, taking place in the palisade and spongy mesophyll cells.
In warm weather conditions, Rubisco (which normally captures CO2) reacts with O2 instead, reducing CO2 capture efficiency in hot weather. Stomata would need to be open for longer periods to obtain sufficient CO₂, which increases water loss via transpiration
C4:
distinct leaf anatomy (special partitioning in mesophyll cells and bundle sheath cells)
carbon fixation occurs in 2 steps:
CO2 is catalysed by the enzyme PEP carboxylase to form the 4C molecule malate.
malate moves out the mesophyll cell to the bundle sheath cells, which surround the vascular bundle in C4 plants
Bundle sheath cells form an airtight container, allowing CO₂ to be concentrated inside. This keeps CO₂ concentrations higher
CAM:
stomata only opens at night to minimise water loss
CAM plants do not transport malate away from the mesophyll cells but store it during the night and then liberate carbon dioxide from these molecules and use it in the Calvin cycle during the day. The malate is stored in vacuoles overnight, allowing the plant to conserve CO₂ for use during the day.
This use of malate requires four more ATP molecules than the C3 pathway, so these plants tend to grow more slowly than other plants.
C4 plant carbon fixation
CO2 is fixed in the mesophyll cell in an additional biochemical pathway. This pathway is catalysed by the enzyme PEP carboxylase, which has a higher affinity for CO₂ than O₂. The CO₂ is joined to form a 4C molecule: malate.
malate moves out the mesophyll cell to the specially adapted bundle sheath cell
in the bundle sheath cell is where malate breaks down to CO₂, which fixed by Rubisco and made into sugars via the Calvin Cycle
In this way the carbon dioxide gradient stays low in mesophyll cells so that it will continue to diffuse in from the outside, even when the stomata are almost closed. This partitioning also means that C4 plants move the Calvin cycle into an area with a high carbon dioxide concentration.
CAM plants carbon fixation
Day
During the day, the stomata close to further minimise water loss. CO2 is stored as malate
The stored malate is converted back into CO₂ in the mesophyll cells. This released CO₂ then enters the Calvin cycle, where it is fixed into sugars.
Night
CAM plants utilise the enzyme PEP Carboxylase to fix CO₂ into malate
Having high concentrations of CO₂ during the day prevents the competition between CO₂ and oxygen for RuBisCO activity.
Why do C4 plants store carbon dioxide in bundle sheath cells? Mention the production of a C4 compound.
CO2 is converted into a C4 compound, malate, in C4 plants. This is catalysed by the enzyme PEP carboxylase.
PEP carboxylase has a much higher affinity for CO₂ than RuBisCO and does not catalyse reactions with oxygen, which helps prevent photorespiration.
malate is moved into the bundle sheath cell, which forms an airtight container, enabling a higher CO₂ concentration
this way rubisco is less likely to engage in photorespiration
overall calvin cycle
rubisco catalyses the reaction between CO2 and RuBP to form 3-PGA, which ultimately forms glucose
NADPH and ATP, provide energy for this process to occur
ATP and NADPH are converted back to ADP +Pi and NADP+
Role of Rubisco in photorespiration
When RuBisCO binds with O₂, this process is called photorespiration.
how does photorespiration affect the rate of photosynthesis
As the temperature increases, O₂’s affinity to rubisco is higher. The rate of photorespiration increases and photosynthetic efficiency decreases, resulting in lowered glucose production. Another factor that will affect the photosynthetic cells is the production of a waste product (ammonia) from photorespiration, which is toxic and must be broken down by the cells using cell energy from cellular respiration, further reducing their glucose supply.
c4 plants adaptations and examples
C4 plants are found in temperate, tropical, warmer regions
Bundle sheath cells allow these cells to have a higher concentration of CO₂.
Increased CO₂ level means that when it enters the Calvin cycle, the chances of RuBisCO reacting with O₂ instead of CO₂ are reduced.
Efficient photosynthesis (low photorespiration) if hot, dry and low CO₂ concentrations.
examples: Maize, Sugarcane, Millet, Sorghum, Native Grasses
CAM plants photosynthetic adaptations and examples
CAM plants are found in hot, arid and drought regions
At night, CAM plants open their stomata, utilising the enzyme PEP carboxylase to fix CO₂ into malate. This helps reduce water loss, as cooler nighttime temperatures decrease evaporation.
The malate is stored in vacuoles overnight, allowing the plant to conserve CO₂ for use during the day.
the stomata is closed to prevent competition between CO₂ and O₂ so there is less photorespiration.
Imalate/organic acids are transported out of the vacuole and broken down to release CO₂which enters the Calvin Cycle
examples: Cacti, Succulents, Orchids, Pineapples
basic structure and location of Rubisco
location: calvin cycle
Constituting ~30% of the total protein in a plant leaf, it is considered one of the most abundant enzymes on Earth.
basic structure: idk it looks like a donut i need to ask what folding structure it is it might be quaternary
advantages and disadvantages of c3 plants
Advantages
Efficient photosynthesis if cool, moist, and high CO₂ concentrations.
doesn’t cost much energy
Disadvantages
high photorespiration if in warmer conditions
decreased rates of photosynthesis
advantages and disadvantages of c4 plants
Advantages
Efficient photosynthesis if hot and low CO₂ concentrations.
Low photorespiration
Ignore Rubisco O2 affinity in heat
Disadvantages
High water usage
Malate synthesis needs energy, therefore more ATP is needed to produce glucose
advantages and disadvantages of cam plants
Advantages
minimised water loss
can survive in very dry conditions
High rate of photosynthesis due to high CO₂ (though slowed)
Disadvantages
more energy cost as it operates less efficiently with the converting stored acids back to CO₂
only undergoes calvin cycle at night
factors that affect the rate of photosynthesis
light availability
water availability
temperature
carbon dioxide concentration
how does light availability affect rate of photosynthesis
Light is essential for photosynthesis to occur, as light splits water in the light-dependent stage. As the light intensity increases, chlorophyll in plant cells can absorb more light, thus increasing the rate of photosynthesis. The rate of photosynthesis will reach a maximal point at the light saturation point.
Plants also absorb different wavelengths of light to a varying degree. For plants that have chlorophyll as their light trapping pigment, red and violet wavelengths are most absorbed, whilst green light is least absorbed (reflects the colour and is not absorbed).
how does water availability affect rate of photosynthesis
Adequate water availability is essential for photosynthesis to occur, as water is split in the light-dependent stage, producing hydrogen and oxygen molecules and energy for the Calvin cycle. Lowered water supply causes stomatal closure, decreasing rate of photosynthesis by reducing CO₂ availability.
how does temperature affect rate of photosynthesis
Ideal temperatures are essential for photosynthesis to occur at optimum levels.
At low temperatures, enzymes like RuBisCo, have low rates of collisions to catalyse the reaction between RuBP and carbon dioxide. Therefore, the rate of photosynthesis is low, and less glucose is produced.
As temperatures increase, Rubisco and its substrates have higher rates of collisions to catalyse the reaction. Therefore, the rate of photosynthesis increases, and more glucose is produced.
However, if temperatures exceed the enzyme’s optimal working conditions, the rate of photosynthesis decreases rapidly as the enzyme begins to denature.
how does carbon dioxide concentration affect rate of photosynthesis
Adequate carbon dioxide availability is essential for photosynthesis, as it is a key input in the light-independent stage.
As the rate of carbon dioxide increases, the rate of photosynthesis will also increase. The rate of photosynthesis will reach a maximal point.
Absorbance spectra of chlorophylls
Chlorophylls absorb blue and red wavelengths and reflect green wavelengths, which is why plants appear green.
how temperature contributes to increased reaction rates
According to the collision theory, molecules must collide for chemical reactions to occur.
At low temperatures, the enzymes for photosynthesis and the substrate molecules move slowly, and so few collisions occur between the molecules, and the rate of photosynthesis is low.
As the temperature increases, the molecules have more kinetic energy, so they move faster and collide and interact more frequently, increasing the rate of reaction.
how limiting factors alter the rate of photosynthesis
Light
low light intensity or shorter days in winter can limit the optimum light level
CO2 concentration
as CO2 is a key input of photosynthesis, it is the main limiting factor
Enzyme concentration:
too few enzymes to catalyse reactions in the Calvin cycle for a given CO2 concentration
limited coenzymes, such as NADPH, restricting the Calvin cycle.
Purpose of cellular respiration
Cellular respiration is an exergonic reaction that cells use to make useable energy in the form of ATP.
word and chemical equation of cellular respiration
Aerobic respiration
6O₂ + C₆H₁₂O₆ → 6CO₂ + 6H₂O + 30/32 ATP
oxygen + glucose → carbon dioxide + water + ATP
Anaerobic respiration
Lactic acid in animal cells
C₆H₁₂O₆ → 2C₃H₆O₃ + 2ATP
glucose → lactic acid + ATP
ethanol in yeast
C6H12O6 → 2CH3CH2OH + 2CO2+ 2ATP
glucose → ethanol + carbon dioxide + 2ATP
location of glycolysis
cytosol
input and outputs of glycolysis
Input
glucose C₆H₁₂O₆
ADP + Pi
NAD+
Output
pyruvate
ATP
NADH
ATP output in anaerobic respiration
In anaerobic respiration, glycolysis occurs, resulting in a net yield of 2 ATP per glucose molecule.
Without oxygen, pyruvate is converted into lactic acid or ethanol. This step does not yield any energy.
krebs cycle input and output
Input:
pyruvate
NAD+
FAD
ADP + Pi
Output:
CO₂
NADH
FADH2
2 ATP
purpose of the krebs cycle/citric acid cycle
break down of pyruvate, which is toxic if built up
producing loaded electron carriers essential for ATP production in the next stage (i.e. NADH and FADH₂)
2 ATP is formed
brief overview of krebs cycle
pyruvate produced in glycolysis are transported to the mitochondrial matrix
pyruvate is converted into acetyl CoA (2 carbon), which is further broken down into CO2
ATP is formed from the energy released in the reactions
coenzymes NAD+ and FAD are loaded to become NADH and FADH2
CO2 is released as a waste product
location of krebs cycle
Mitochondrial matrix
ATP output of the electron transport chain (ETC)
26-28 ATP gained, depending on the type of cell
input and output of the electron transport chain (ETC)
Input:
O2
ADP + Pi
NADH
FADH2
Output:
H₂O
ATP
NAD+
FAD
location of the the electron transport chain (ETC)
mitochondrial cristae
purpose of the electron transport chain (ETC)
transfer electrons from NADH and FADH₂ to oxygen, forming water as a byproduct
forming ATP (26-28)
summary of the electron transport chain (ETC)
NADH and FADH2 from the Krebs cycle and glycolysis enter the ETC
hydrogen ions from NADH and FADH₂ move through the series of protein molecules embedded in the inner mitochondrial membrane
ATP synthase uses the energy of this proton gradient to synthesise ATP from ADP + Pi.
H⁺ ions, O₂, and e⁻ combine to form H₂O
O₂ is the final electron acceptor
The net ATP yield from the ETC is 26 or 28 ATP molecules.
significance of oxygen availability in terms of the electron transport chain (ETC)
The electron transport chain can only operate if there is a supply of oxygen. Oxygen binds with hydrogen ions and electrons to form water. The cell will switch to anaerobic respiration of there is inadequate oxygen, which produces less energy.
input and output of lactic acid fermentation
Input
Glucose
ADP + Pi
Output
Lactic Acid
2 ATP
input and output of alcoholic fermentation
Input
Glucose
ADP + Pi
Output
ethanol
ATP
CO2
relationship between glycolysis and anaerobic respiration
If oxygen is absent, the Krebs cycle and the electron transport chain shut down.
The cell will rely on glycolysis to maintain a supply of ATP for its energy needs.
The products of glycolysis (pyruvate and NADH) are removed by reacting NADH with pyruvate in both alcoholic and lactic acid fermentation.
Anaerobic respiration yield of ATP
2 ATP per glucose molecule
Compare the efficiency of ATP production in aerobic and anaerobic respiration
ATP production by anaerobic respiration is faster than the more complex reactions of aerobic respiration.
if the demand for ATP outweighs the supply from aerobic respiration, eukaryotic cells shift to anaerobic respiration.
Compare the yield of ATP production in aerobic and anaerobic respiration
Aerobic respiration yields 30/32 ATP molecules per glucose molecule.
Anaerobic respiration yields 2 ATP molecules per glucose molecule.
Cellular respiration limiting factors
temperature: frequency of collisions of substrate and enzyme increased as temperature increases, denaturation at high temperatures
glucose concentration: input of the reaction
oxygen concentration: determines if anaerobic or aerobic respiration will occur
enzyme availability, pH, inhibitors
Food availability’s impact on glucose available
cellular respiration requires an ongoing supply of glucose
phototrophs can produce glucose, but autotrophs require intake of glucose (food)
Glucose can be stored in specialised cells: glycogen in animals and starch in plants
If glucose levels are low, cells may rely on alternative energy sources such as fatty acids or amino acids, but these processes may yield less ATP or be less efficient.
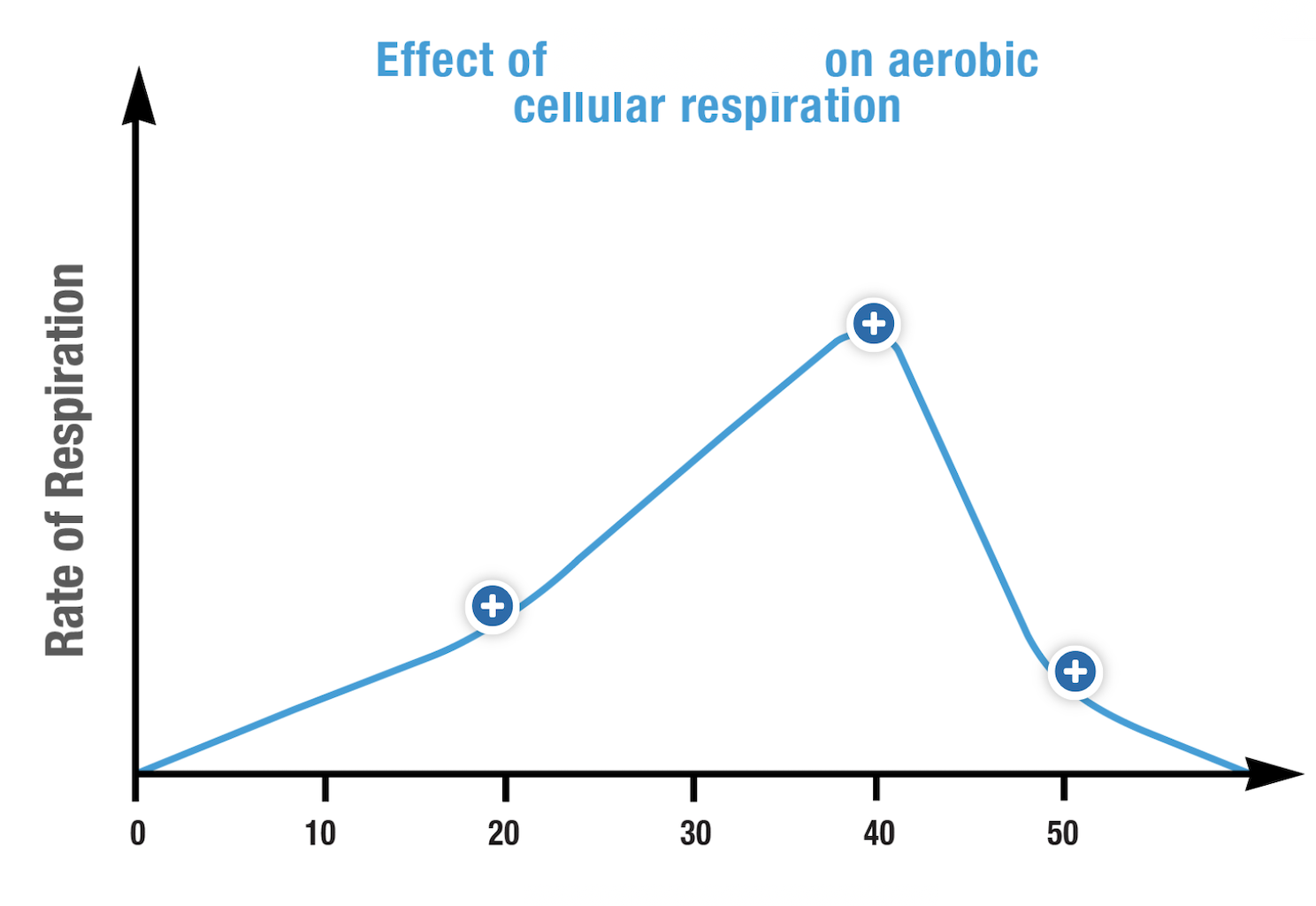
What is the dependent variable represented in this graph?
temperature
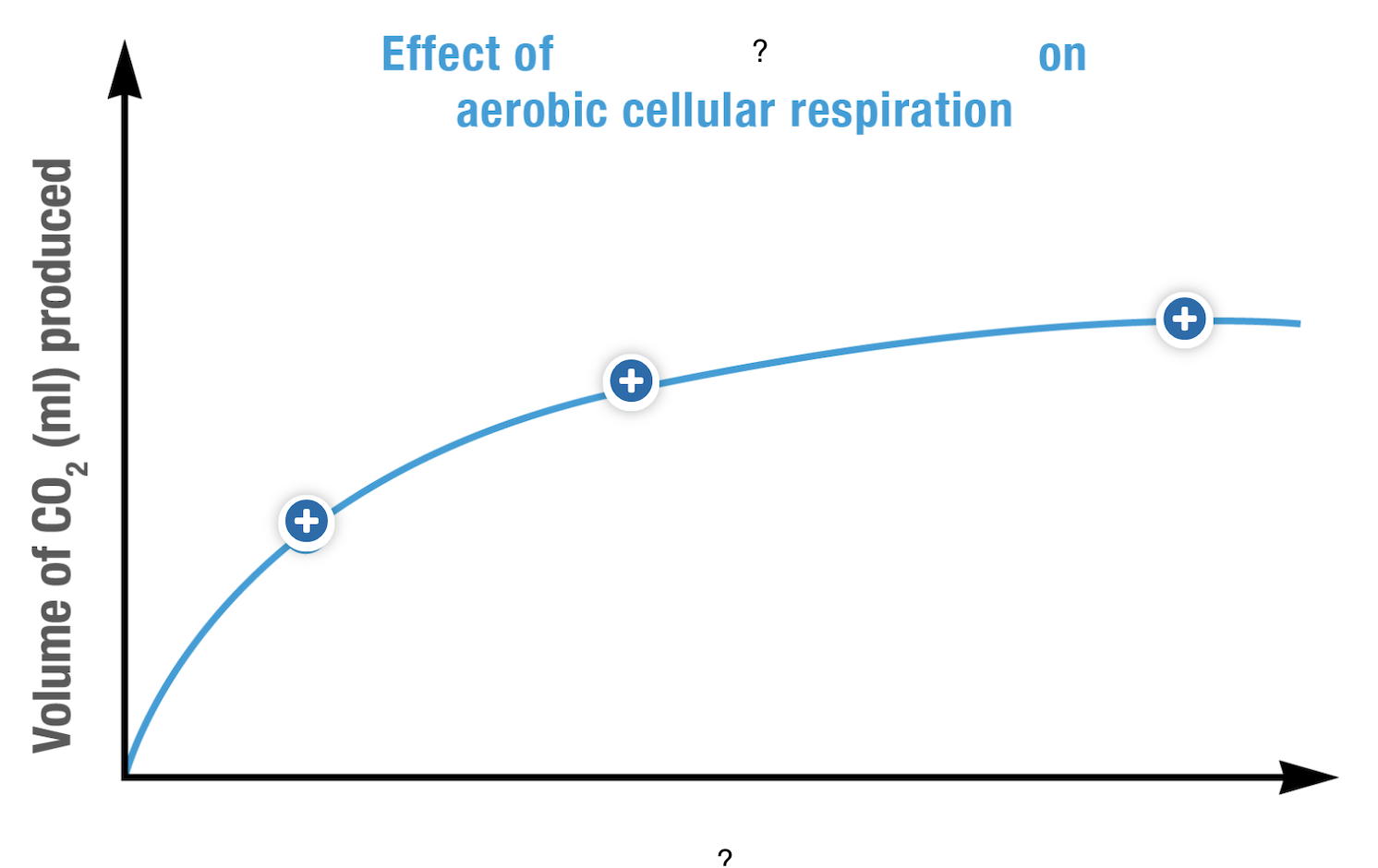
What is the dependent variable represented in this graph?
glucose/oxygen concentration
how does CRISPR works to edit DNA
CRISPR-Cas9 is a gene editing process where genetic engineers target specific genes. The Cas9 enzyme is directed by a sgRNA, which is complementary to the target site. The Cas9 will find the target PAM sequence, check whether the sgRNA aligns with the DNA, and then cut the selected sequence of DNA.
This sgRNA can be engineered to guide the Cas9 to the desired gene, turning it off or changing its function.
key genes regulating these photosynthetic processes can be edited to optimize plant performance
how gene editing can overcome limiting factors of plant value
Enhancing photosynthetic efficiency and crop yield by
enhancing light availability by modifying genes responsible for light capture to improve the absorbance of specific wavelengths of light
transferring certain genes to crop plants to increase biomass
engineering Rubisco to be more efficient at binding to CO₂ by enhancing its specificity to CO₂
Plants switch photosynthesis off when light is too intense, so genes are added from fast-switch plants to slow-switch plants
increased resistance against virus/disease/pests
biomass
the total dry weight of organic material
how is bioethanol produced
Bioethanol is produced by anaerobic fermentation (anaerobic digestion) of biomass.
the fermentation of glucose from the biomass is usually facilitated by yeast
his reaction breaks down the glucose into ethanol and carbon dioxide
Once fermentation is complete, the liquid undergoes distillation to separate ethanol from water and other impurities
issues surrounding the use and production of biofuels
There are ethical considerations of biofuel production as biomass used to make biofuels competes for land use and food crops.
purpose of generating biofuels
renewable and sustainable
readily available
much smaller net output of carbon emissions compared to fossil fuels
biofuel
a fuel that has used biomass as its original material
how is biogas produced
Biogas is gas that is released in the breakdown of organic waste by anaerobic bacteria. Vegetable waste and animal manure are used in these fermenters to produce methane gas.
PEP carboxylase
pep carboxylase is the enzyme used in C4 and CAM plants to convert CO2 into malate