BIOL 112 Final Exam
1/131
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
132 Terms
B
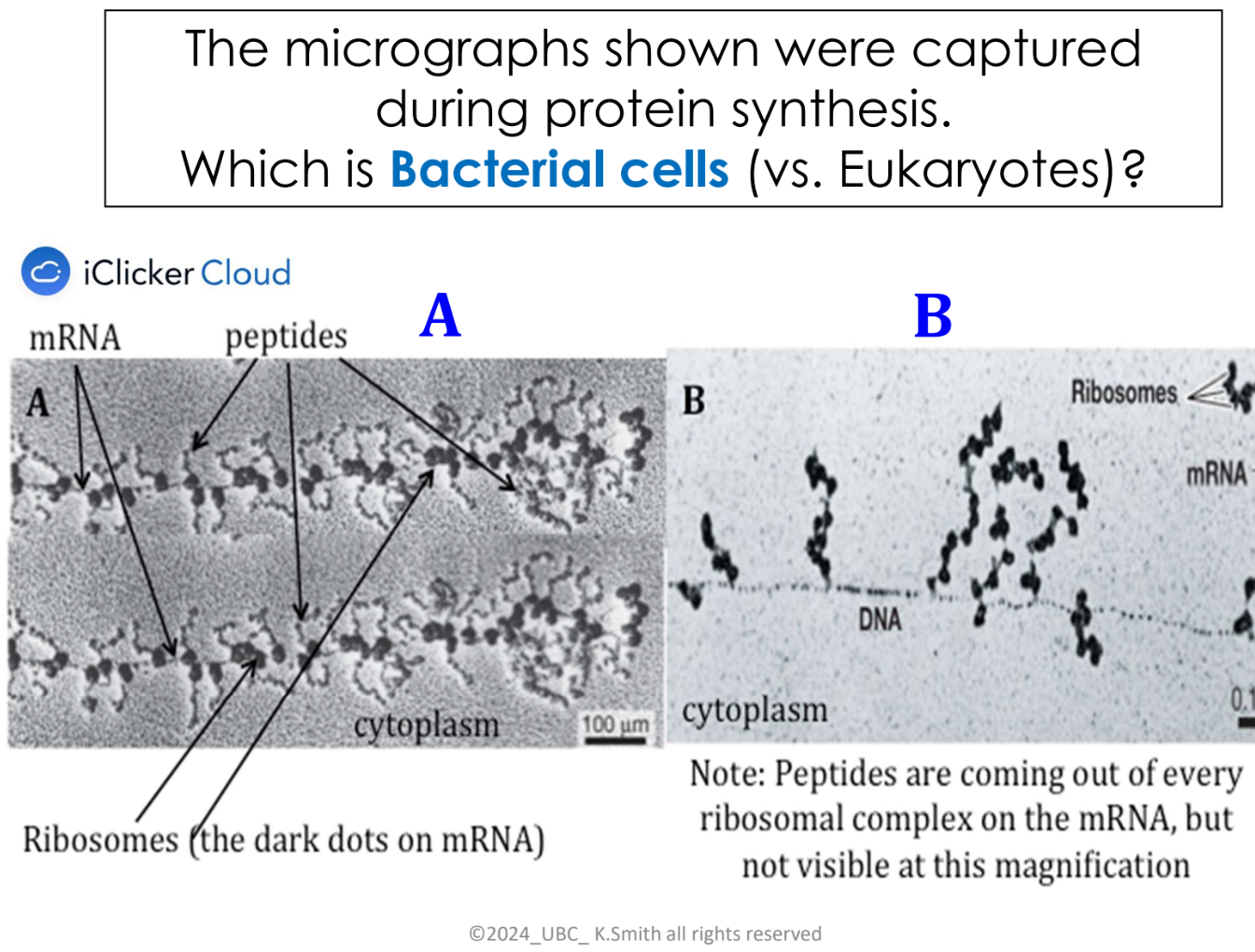
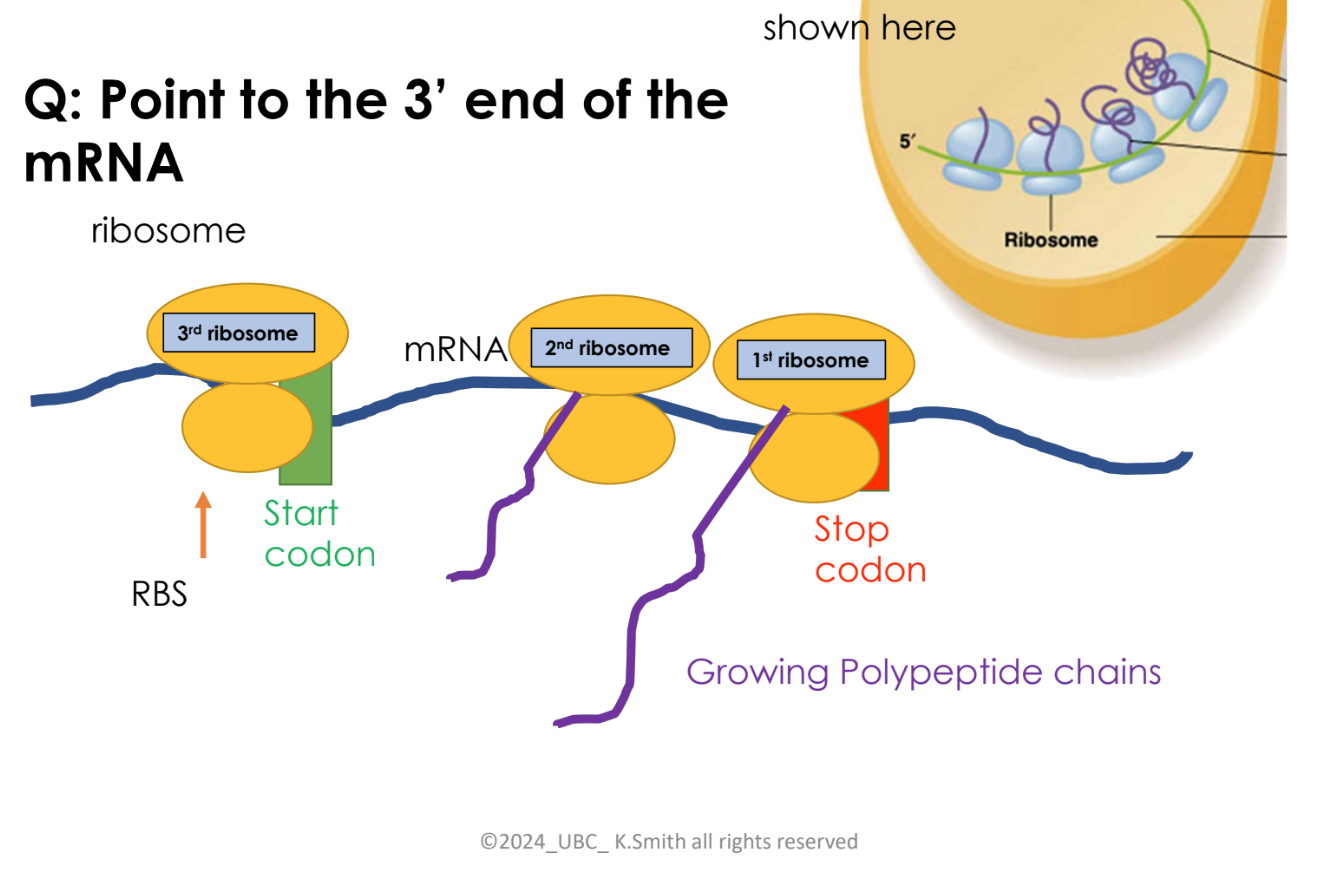
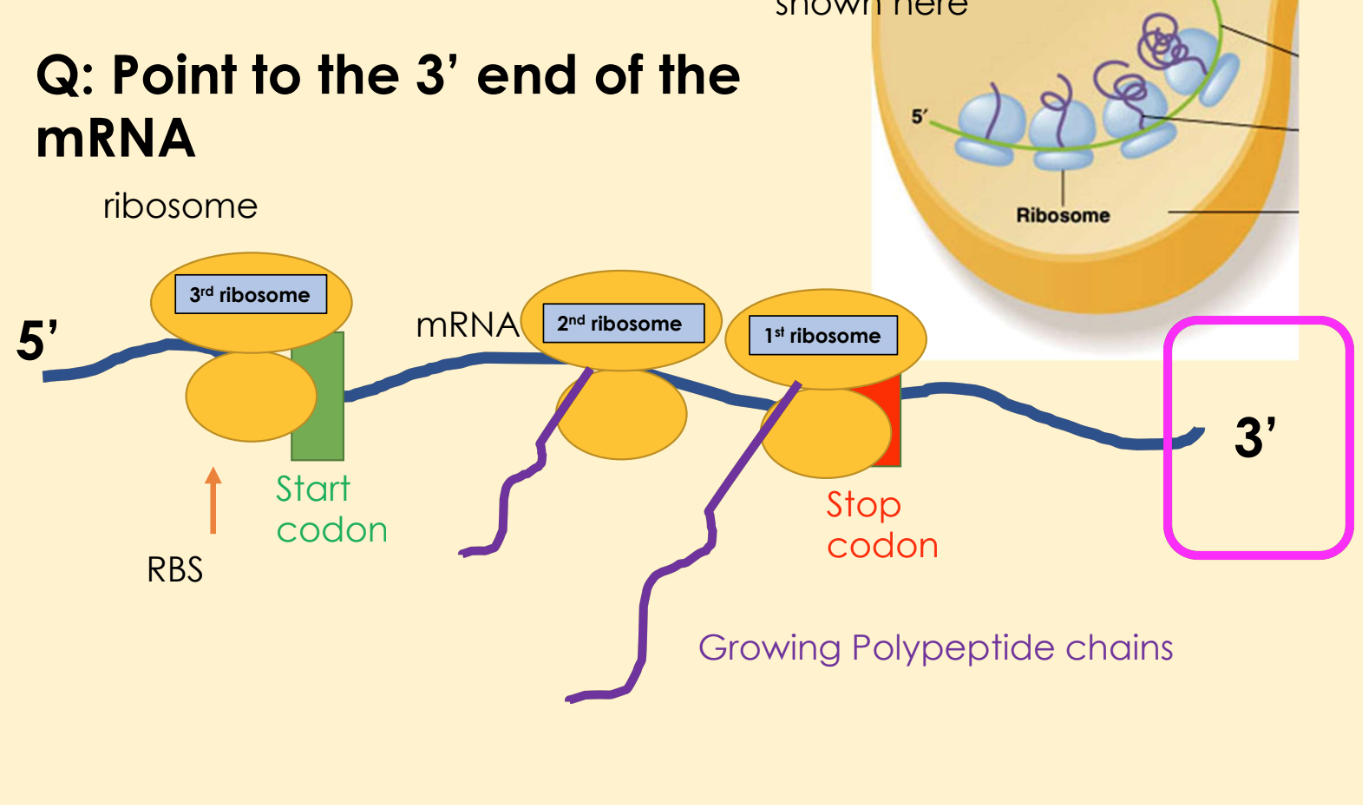
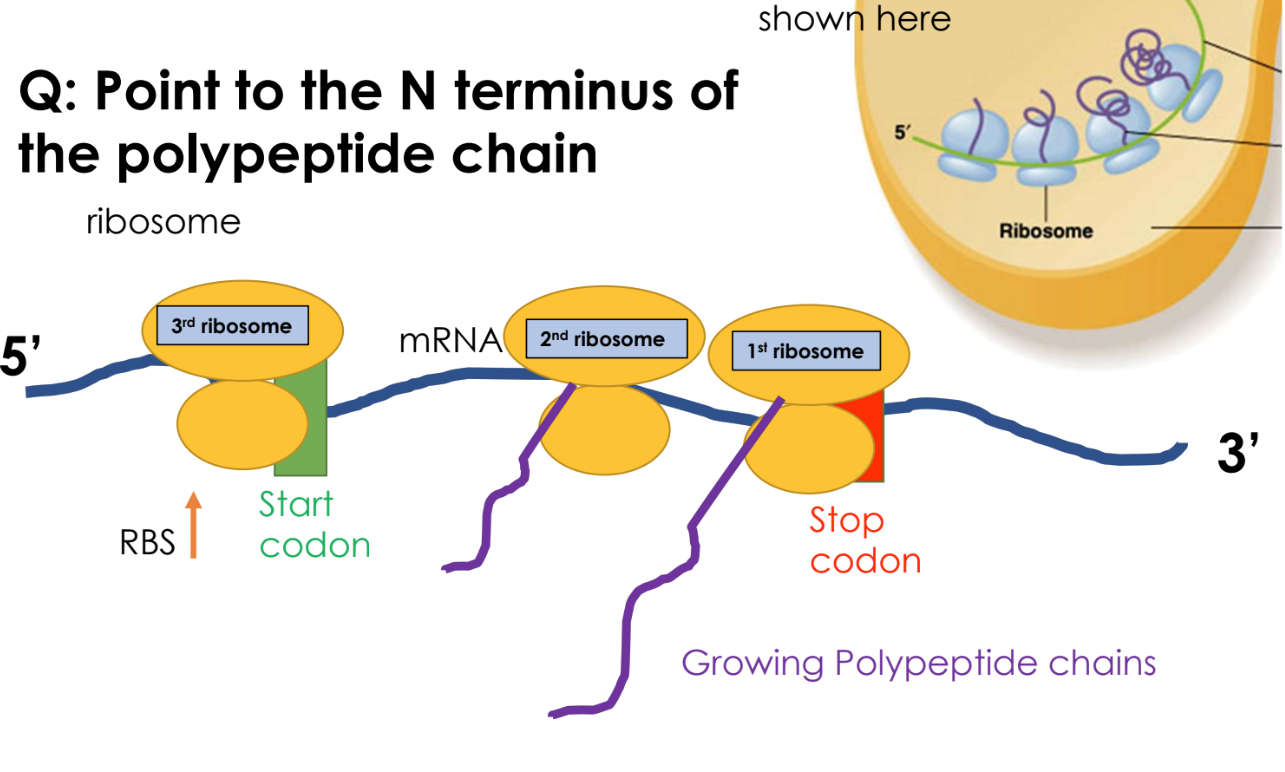
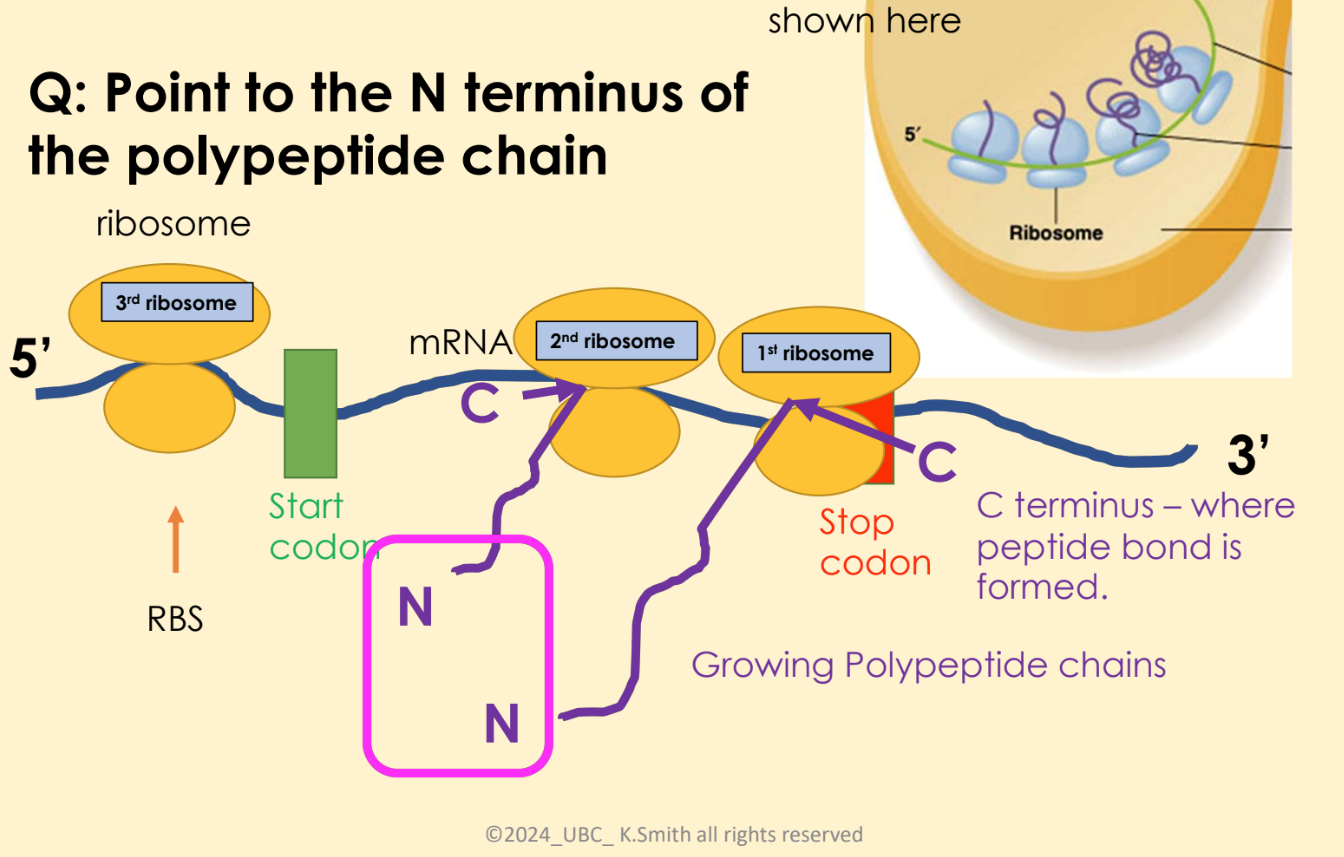
explain the steps of bacterial cells translation (electron micrograph)
First ribosome binds to the mRNA
A second ribosome binds after first ribosome has moved downstream, polypeptide chain is being synthesized by ribosome 1
a third ribosome binds as 1 and 2 are moving along the mRNA downstream
what does CRISPR stand for
Clustered Regularly Interspaced Short Palindromic Repeats
what is the CRISPR/Cas system
a widespread class of immunity system that protect bacteria and archaea against phages and other mobile genetic elements
use repeat/spacer-derived short crRNAs to silence foreign nucleic acids in a sequence specific manner
what are CRISPR arrays
contain all of the viral DNA samples together in a single repeated array
a storage bank that records previous encounters with viruses that infect the bacterial cell
spacers vs repeats
the CRISPR array itself is a permanent part of the bacterial genome and can therefore be passed to offspring cells
each spacer sequence is taken from a different virus
different spacers = different viruses
what is crRNA
the RNA that bacterial cells transcribe from the CRISPR RNA
crRNA is cut into smaller sequences that contain spacer and repeat sequences
what do Cas9 enzymes do
use crRNA to target viral DNA
crRNA binds to Cas9 enzymes
CRISPR-Cas9 complex
multiple copies of Cas9 proteins each with a piece of crRNA
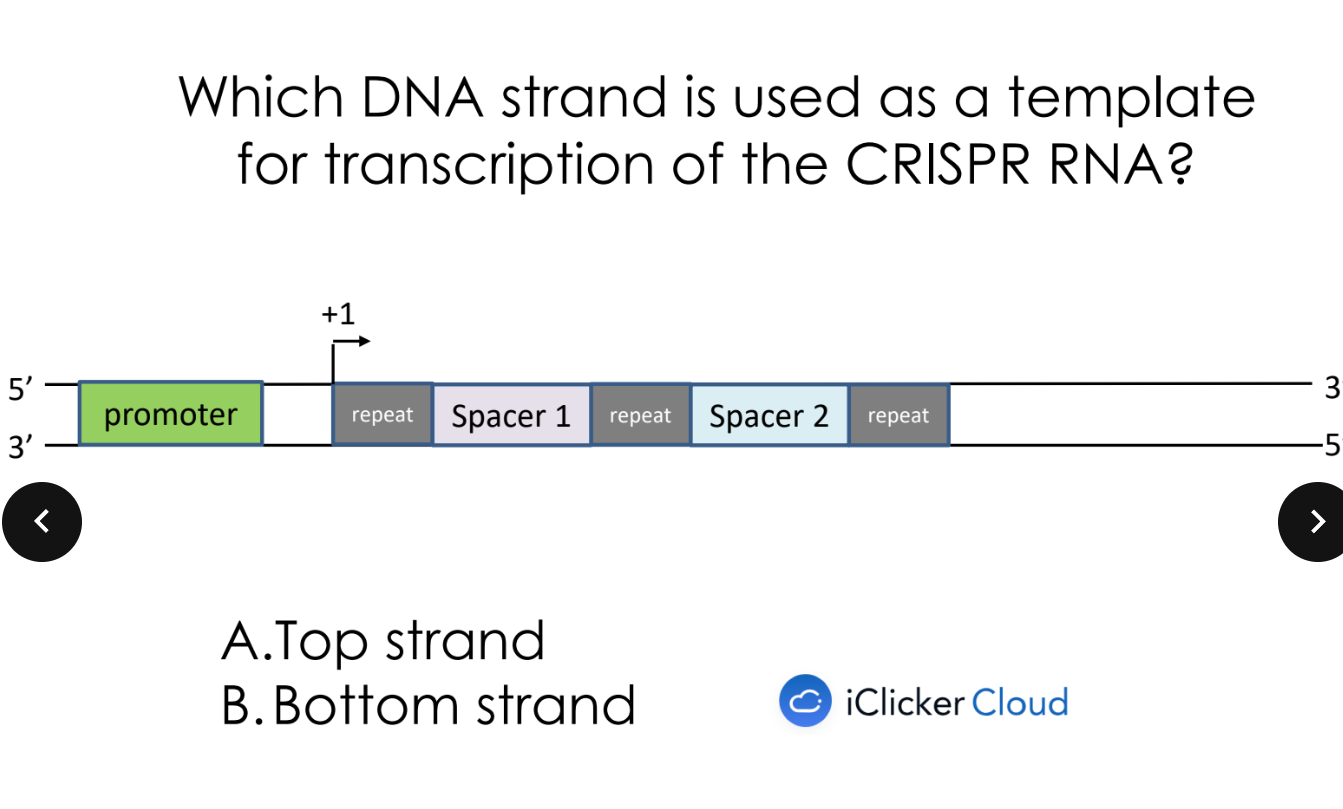
bottom strand
how does bacteria defend itself from new viral infections
finding viral sequences
a new virus infects the cell and inserts the viral DNA into the genome of the bacteria
the CRISPR-Cas9 complex scans the genome of the bacteria to find viral sequences
they use the crRNA as a guide to find a sequences match to the virus genome
matching crRNA with DNA in bacteria
the cas9 opens opens the sequence of viral DNA and checks it against the crRNA samples it is carrying
if it matches crRNA-Cas9 and the genome forms complementary base pairing
the crRNA and DNA match are locked in
cut out viral DNA sequence
when locked in, the CRISPR-Cas9 will proceed to cut out the sequence that matches the spacer (i.e. original viral DNA sequence)
Cas9 cuts up viral DNA
how does cutting/editing defend against viruses
cut in the middle of an important viral gene, killing the virus
other cas9 enzymes maybe carrying crRNA that match other parts of the viral DNA, thus increasing the chances of cutting up and killing the viral DNA before it can hijack the cell
what is gRNA
guide RNA
cut any DNA of interest and then edit it
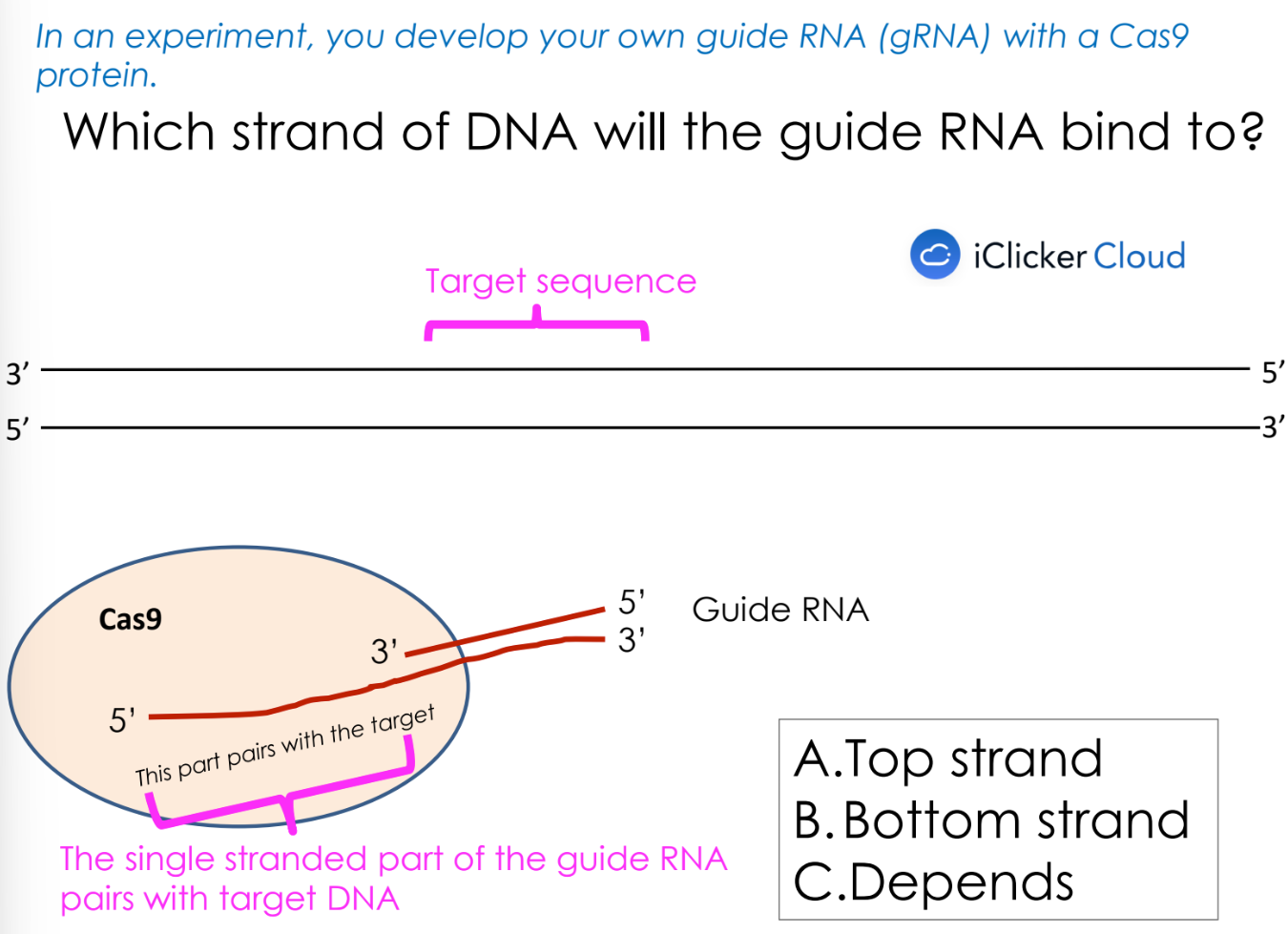

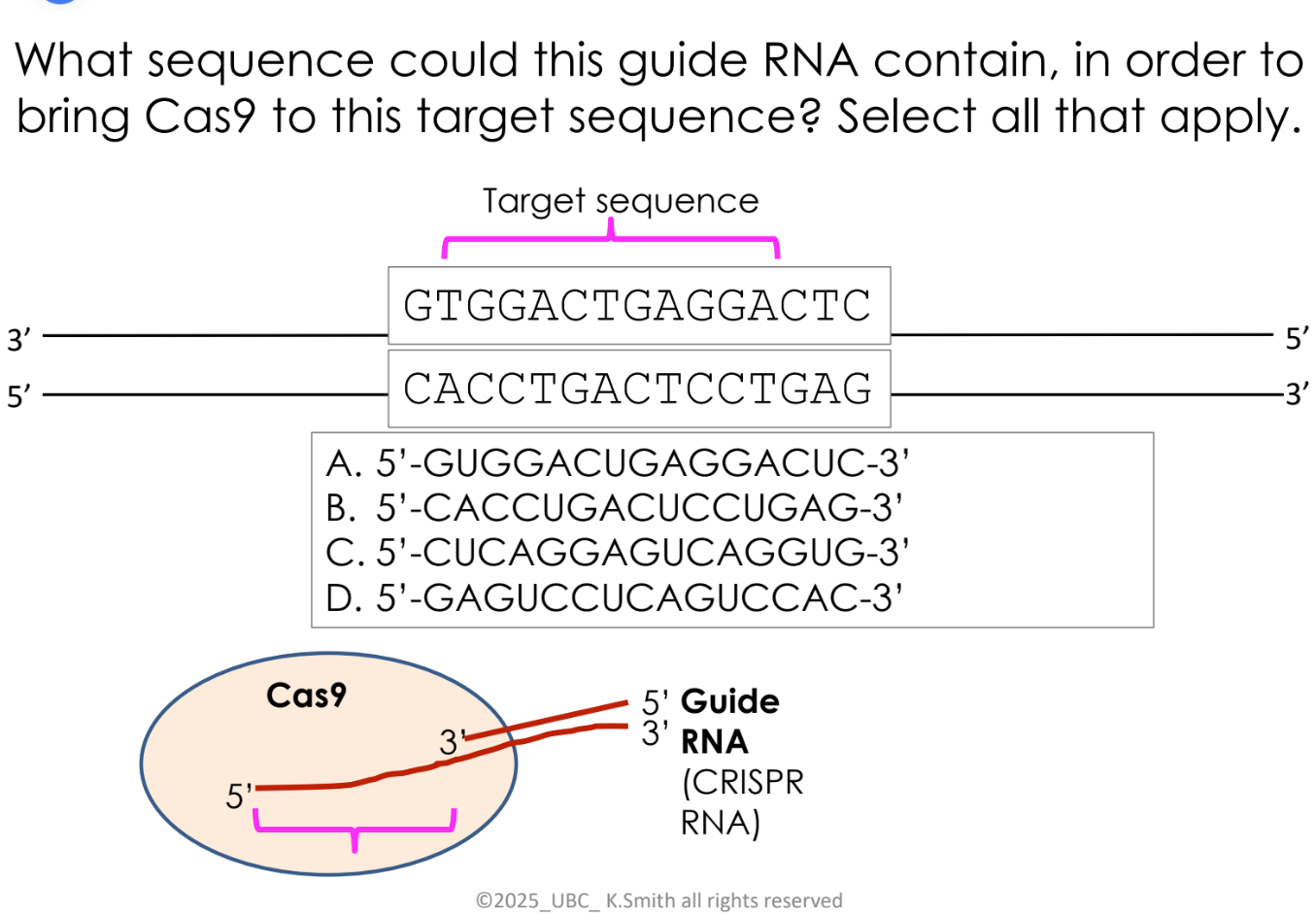
this guide RNA sequence hybridizes to the top strand of this DNA (B sequence)
C sequence hybridizes to the bottom strand of this DNA
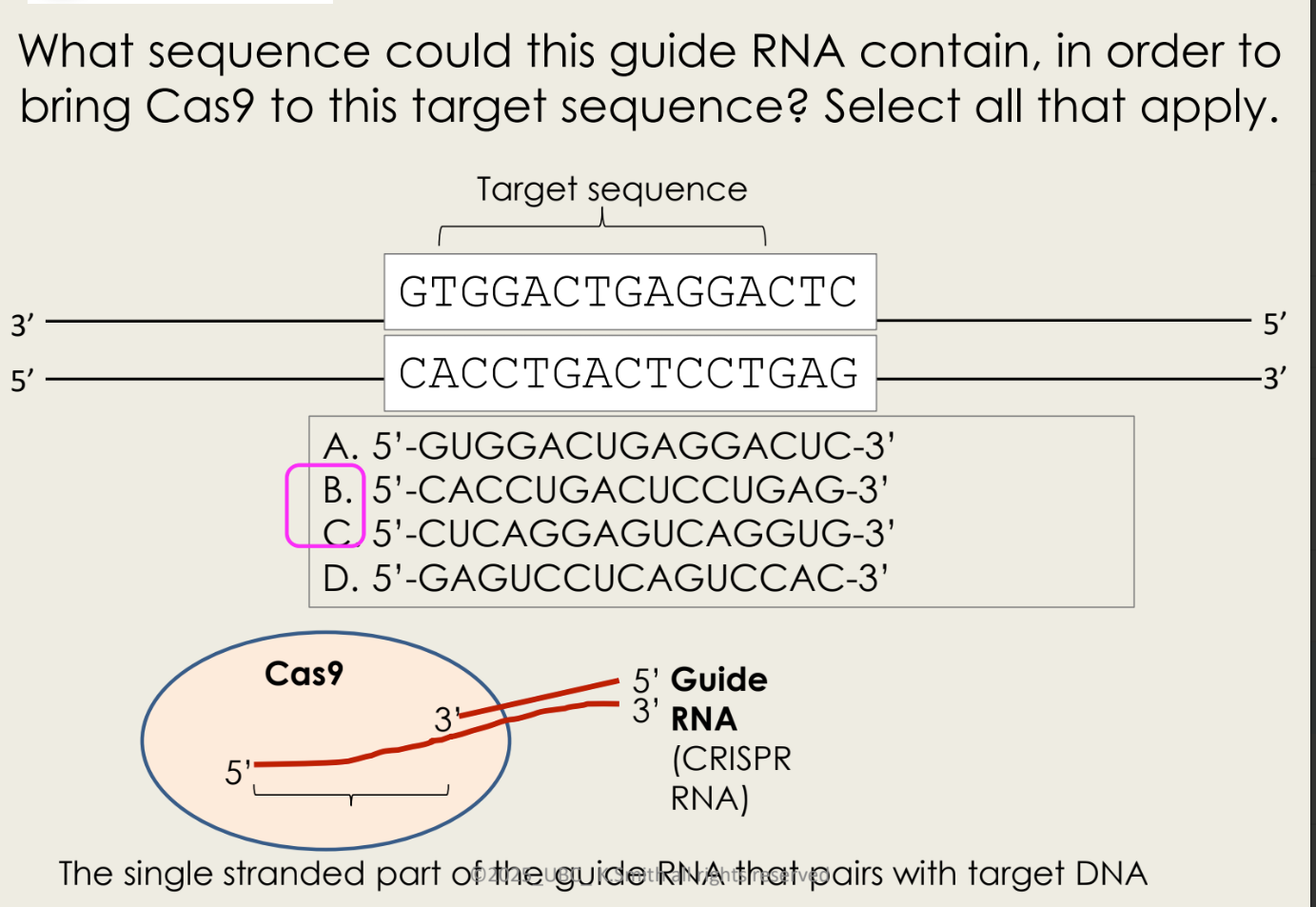
what happens when the DNA is cut
the cell’s DNA repair system are turned on
scientists can then introduce a customized DNA template for the repair system to use, changes DNA sequence
what does gene regulation mean
a cell does not express all of its genes all of the time, very selective about the genes they express
how strongly they are expressed (high vs low)
when they are expressed
a gene is said to be expressed when the gene product is actively being synthesized and used in a cell
what is a constitutively expressed gene
a gene that is expressed all the time because its gene product is needed all the time
eg. rRNA, tRNA, RNA pol, ribosomal proteins, amino acyl tRNA synthetases
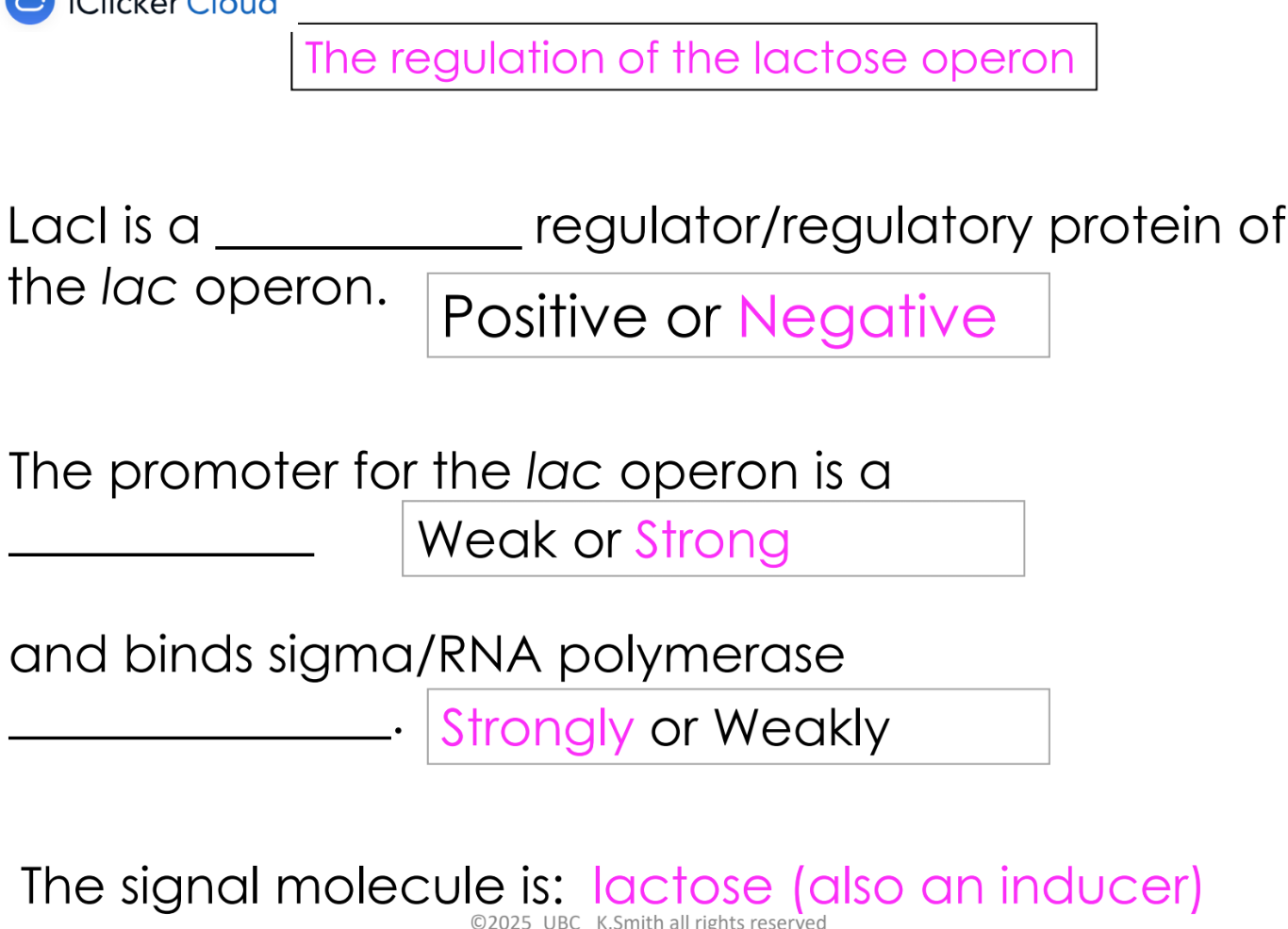
how does promoter strength affect gene expression
determines how frequently transcription is initiated
this determines how many RNA molecules are made
how does mRNA half-life affect affect gene expression
how quickly the mRNA is degraded after it is made
the longer mRNA lasts, the more protein copies can be translated per unit time
only applies to protein encoding genes
tRNA and rRNA are stable RNA so not degraded
transcriptional level of control examples
promoter strength - how strongly RNA pol + transcription factors bind to the promoter
operons - regulatory proteins binding to operator
what is an operon
share a promoter and termination sequence but have multiple coding regions
one mRNA is produced yet multiple proteins are translated
what can be found in operons
a regulatory region called the operator can be found either upstream or downstream (and sometimes overlapping) the promoter
the regulatory protein binds to an operator region
basal definition
means base, minimal
positive regulation
regulatory protein binds a region by the promoter and increases transcription
regulatory protein called an “activator” protein
eg. MalT
negative regulation
regulatory protein binds to a region by the promoter and decreases transcription
regulatory protein called a “repressor” protein
eg. LacI
catabolic operons
lac and mal operons are examples
some cells use lactose or maltose as a source of energy and carbon
the cells only needs to synthesize the proteins in the operons if the sugars are present in the environment
the proteins produced in these operons function to break down sugars
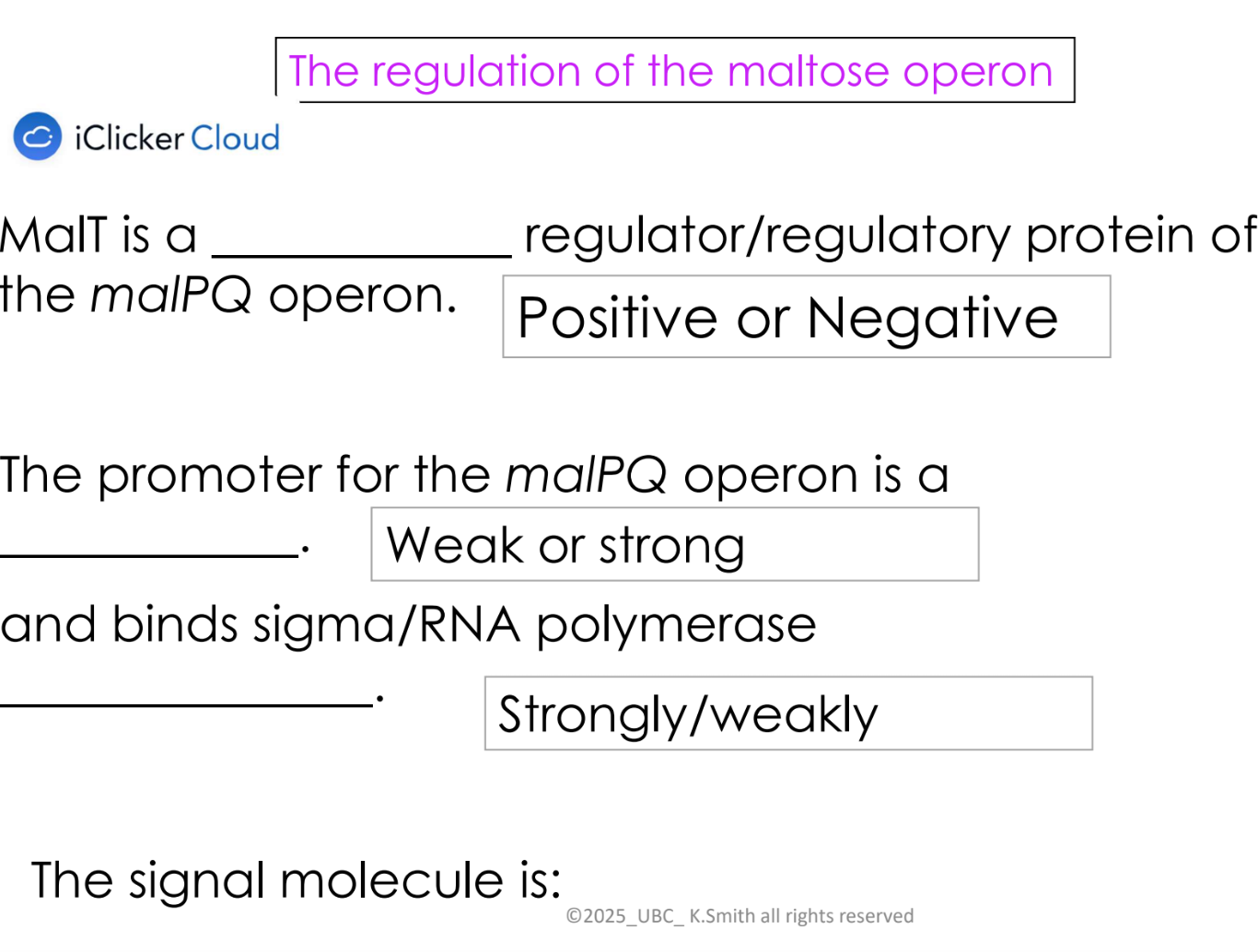
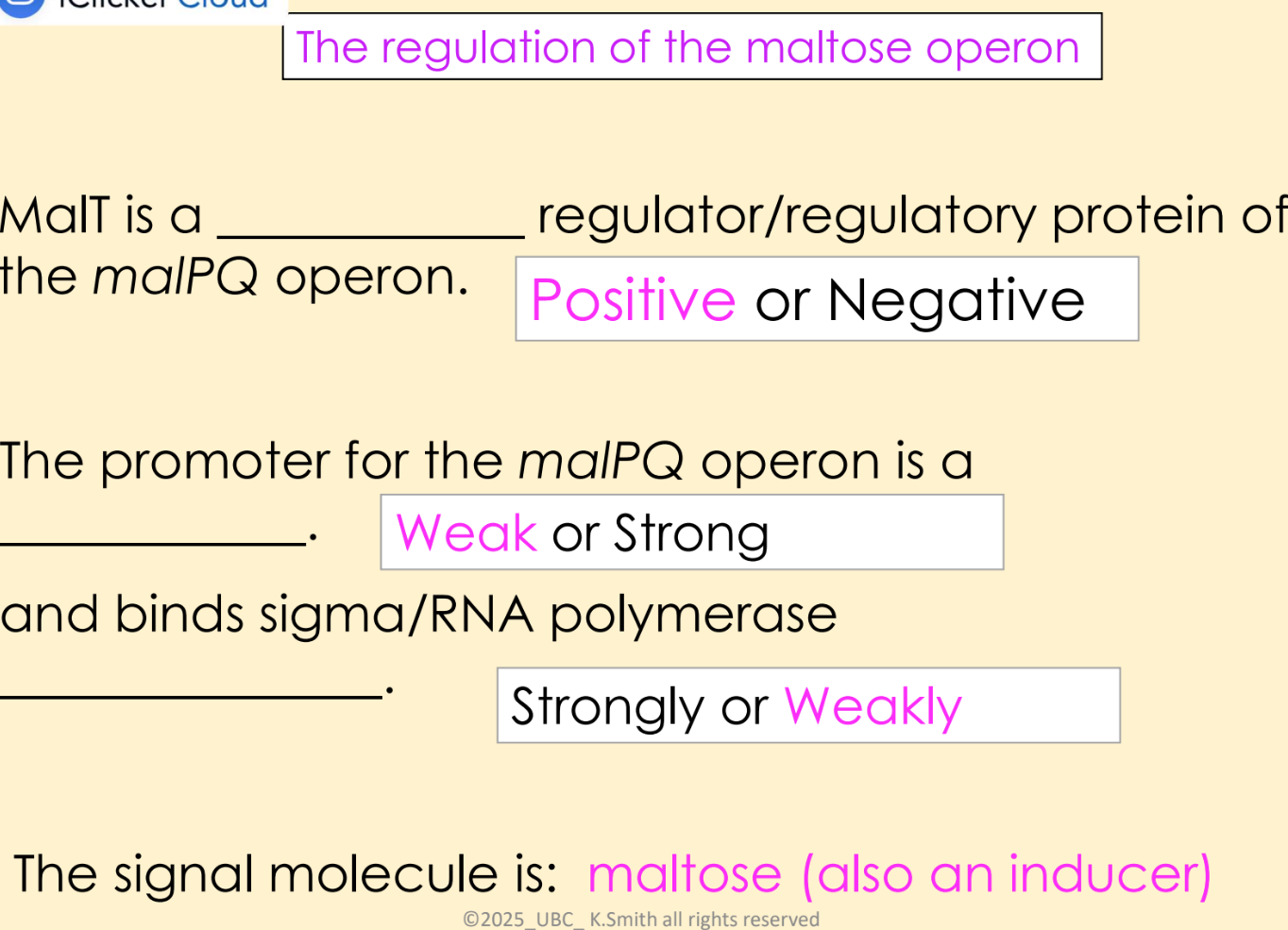
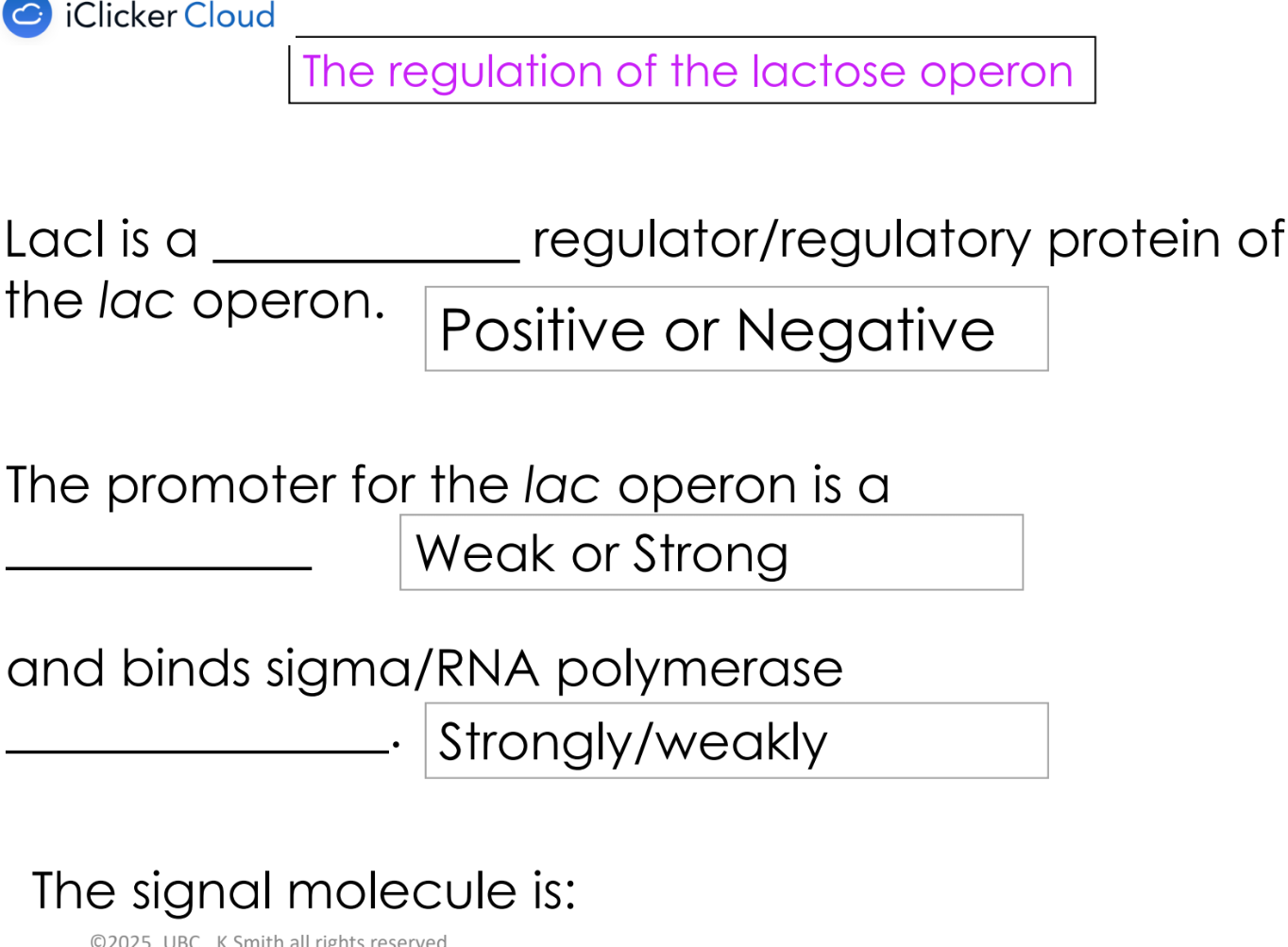
what are inducers
when present, the lac and mal operons are expressed at high levels
what is a co-repressor
if the presence of a signal molecule results in low levels (to zero) of operon expression, this is often referred to as a “co-repressor”
anabolic operons
produce proteins that are involved in the synthesis of the signal molecule
signal molecule (eg. arginine)
the operon are expressed in the absence of the signal molecule, the signal molecule is a co-repressor
i.e. helps the repressor protein, repress the expression of the operon
function of DNA replication
DNA encodes all of the proteins and RNA in the cell to allow for proper cell function
new cells will need a copy of the genome
key properties of DNA polymerase
DNA pol reads the sequence on a template and links nucleotides together, i.e. reads the template 3’ to 5’ and synthesizes nes DNA in the direction 5’ to 3’
DNA pol cannot start on its own, must always start from an existing 3’OH end of an existing template
in the cell, the primer is RNA
what are requirements for polymerization of RNA/DNA
requires an enzyme (DNA or RNA polymerase)
requires a form of energy
this energy form is a monomer with 2 or more phosphates, most common forms = triphosphates
polymerization of DNA or RNA
the 5’ carbon of a new monomer is added to the 3’ carbon of the existing strand (or 1st monomer)
why are monomers shown as a triphosphate?
binding of three phosphates creates an unfavourable state (3 negative ions in close proximity)
removing the outermost phosphates release energy
the energy can be used to link the monomer to the polymer
explain why DNA replication is semi-conservative
each cell will receive an original template strand that is base paired with a newly synthesized strand
what is the rate of replication
~1000 base pairs/second
how does linear DNA replicate
needs more than one origin of replication (OriR)
how does circular DNA replicate
bacterial cells can replicate in a circle (both directions), ends will meet up
usually has 1 origin of replication (OriC)
what are three problems the cell has to solve to replicate DNA
separate the DNA strands only a little at a time, can’t separate an entire genome
make primers for DNA polymerase
can’t make a DNA primer without a DNA primer
cell makes an RNA primer - can start without a 3’ OH
allow synthesis to happen simultaneously from the two template strands
DNA strands are anti-parallel, yet DNA polymerization has to occur in the 5’-3’ direction
synthesis has to happen in opposite directions
solutions to the problems (DNA replication)
helicase unwinds DNA at replication fork
RNA primase → RNA polymerase (RNA primase) makes RNA primers, DNA pol can add dNTPs to the 3’ OH end of this short RNA sequence
from the replication fork - synthesis occurs in the opposite directions (i. e. in one direction on one strand and in the other direction on the other strand)
but overall, replication must progress on both strands in the direction of the growing replication fork
what do primers do
direct the start of polymerization
DNA pol cannot start on its own, must always start from an existing 3’ OH end of an existing template. in the cell, the primer is RNA
primase is an enzyme that creates the primer
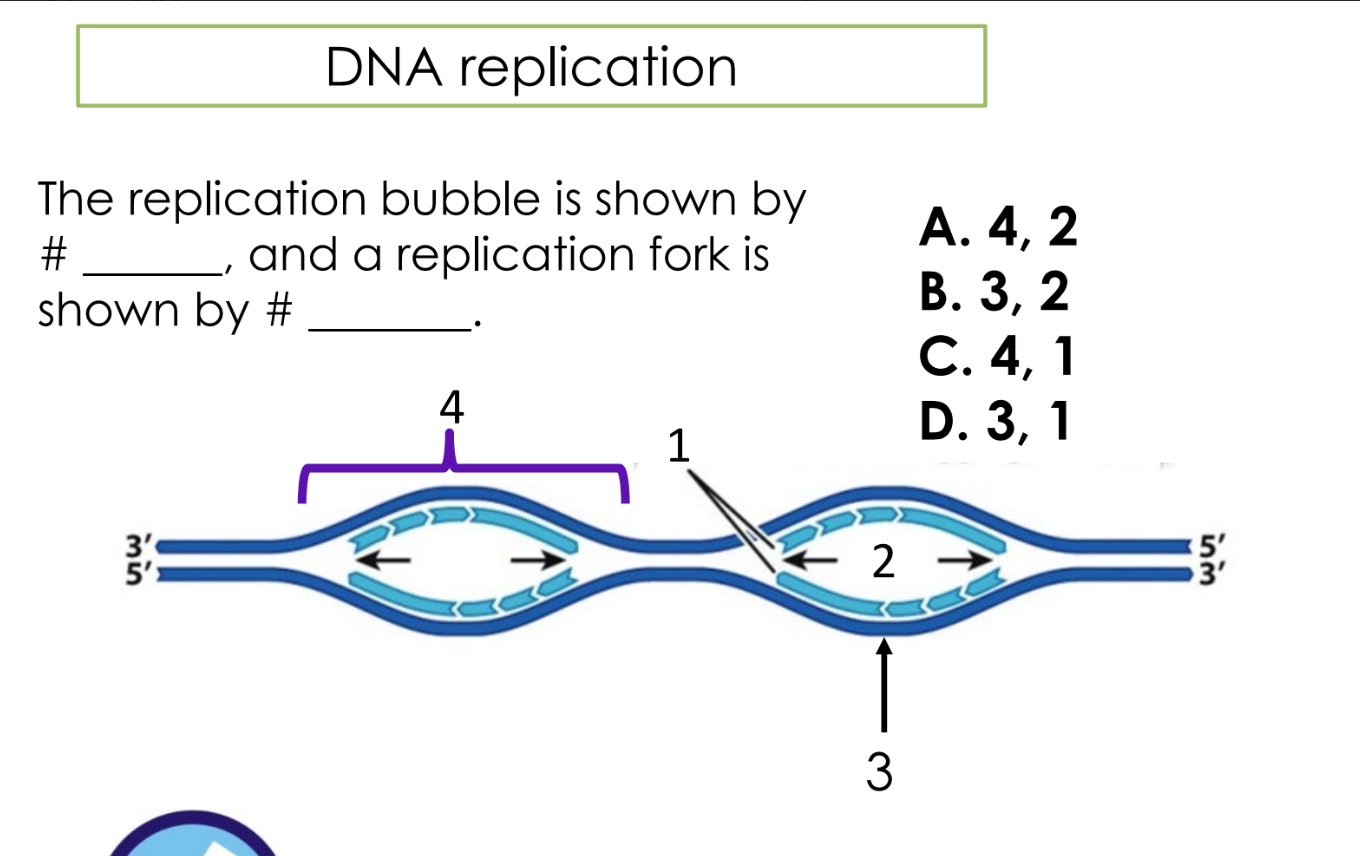
A
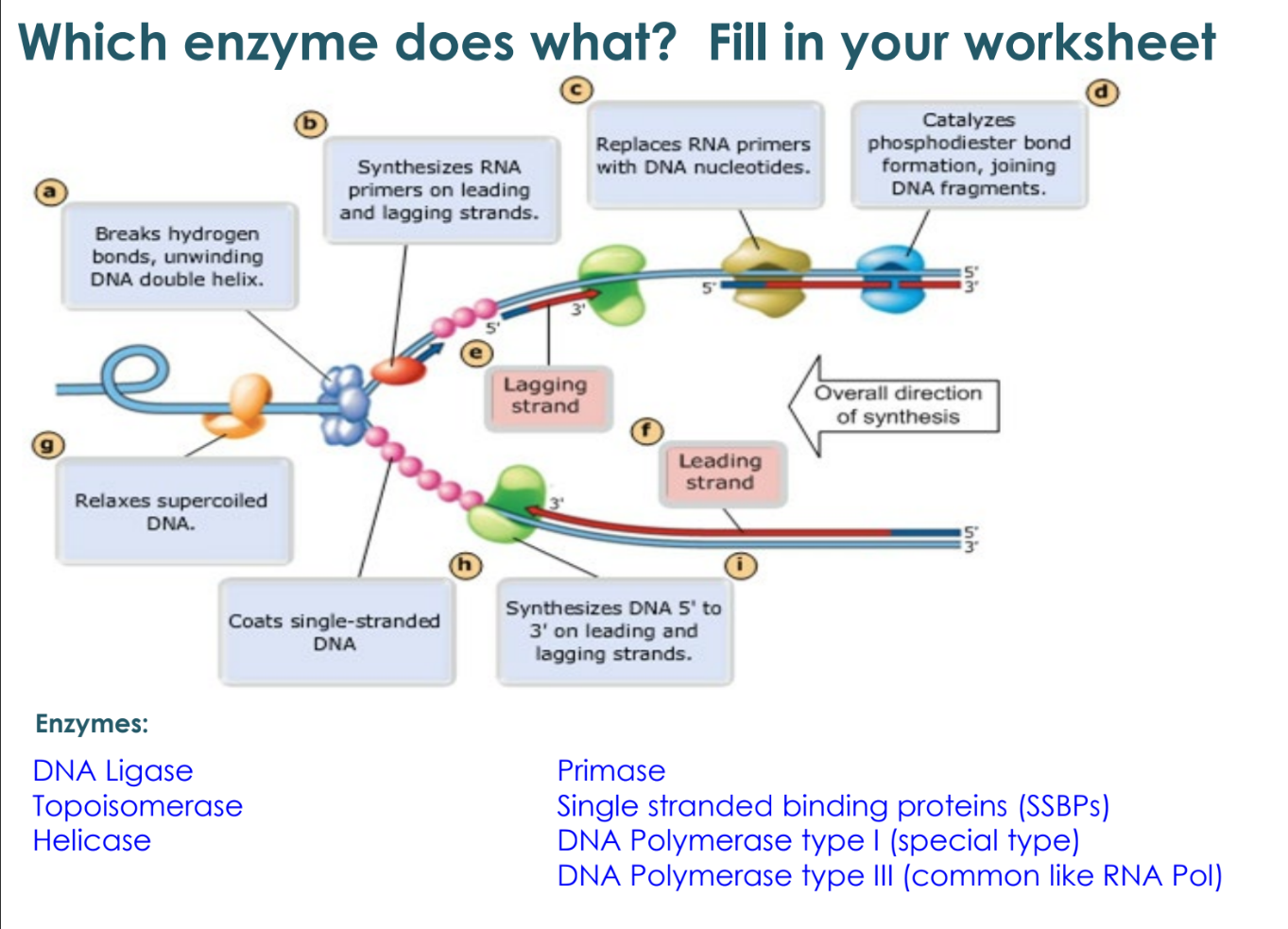
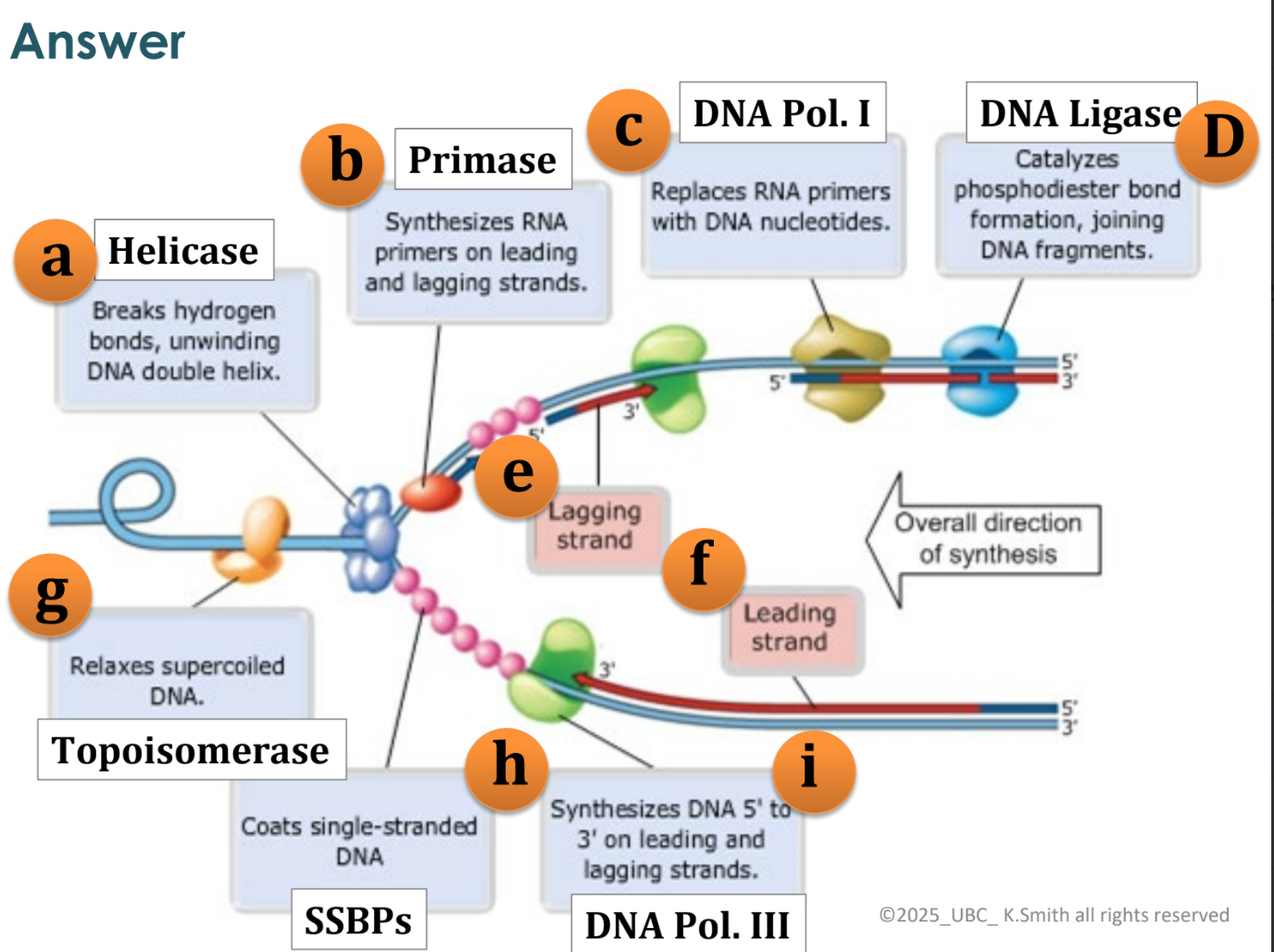
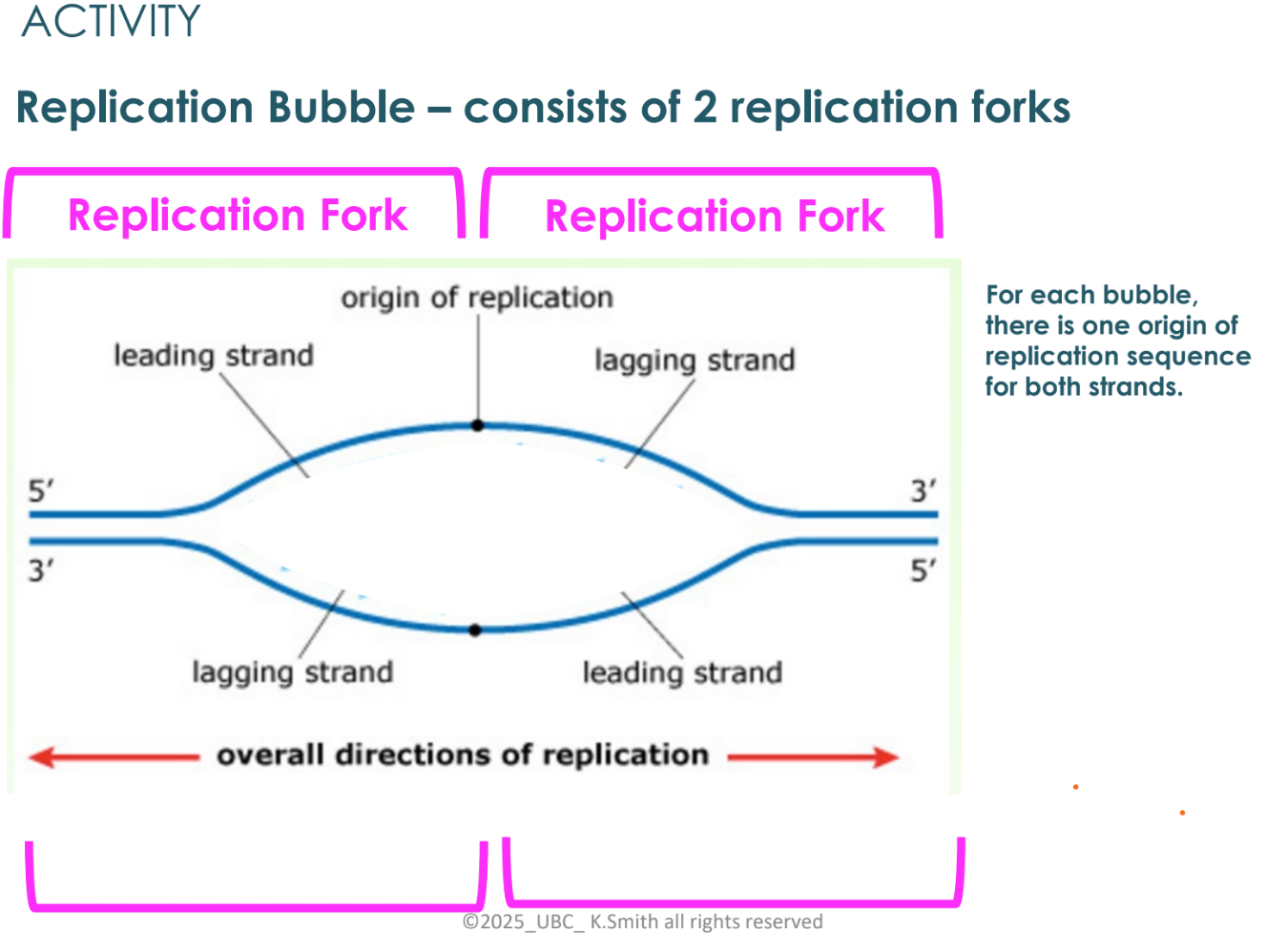
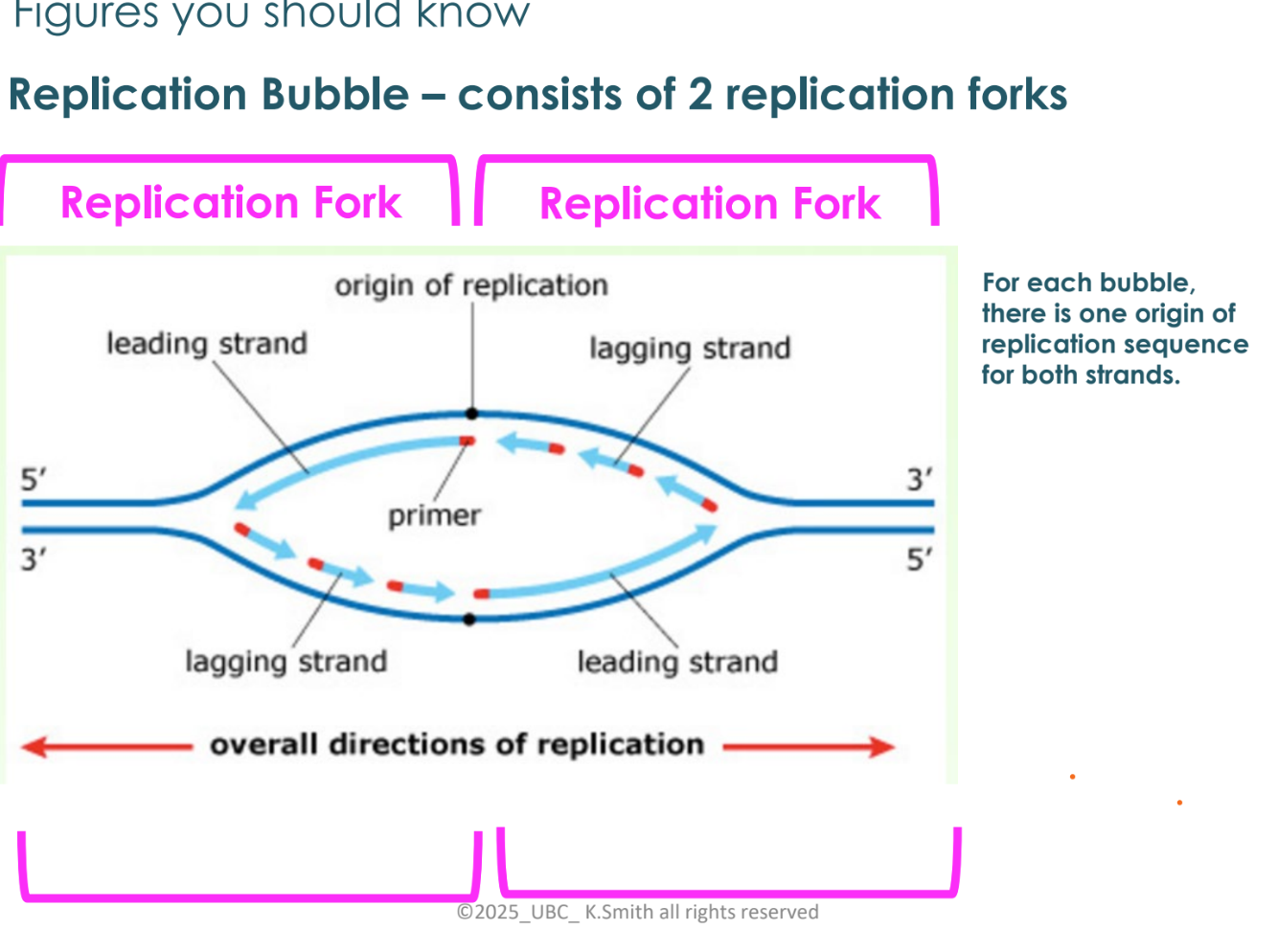
what happens as the replication fork opens?
at first, both strands (leading and lagging) are synthesized as the replication fork
the replication fork has opened further to the left so need to start a new Okazaki fragment
as replication fork opens further, the top strand has DNA pol moving to the right and leaving a gap at the fork, so another Okazaki fragment is added to join the first fragment
discontinuous vs continuous synthesis
leading strands are referred to as continuous synthesis
lagging strands are referred to as discontinuous synthesis
DNA polymerase I function
removes the RNA primer (red) and replaces it with DNA
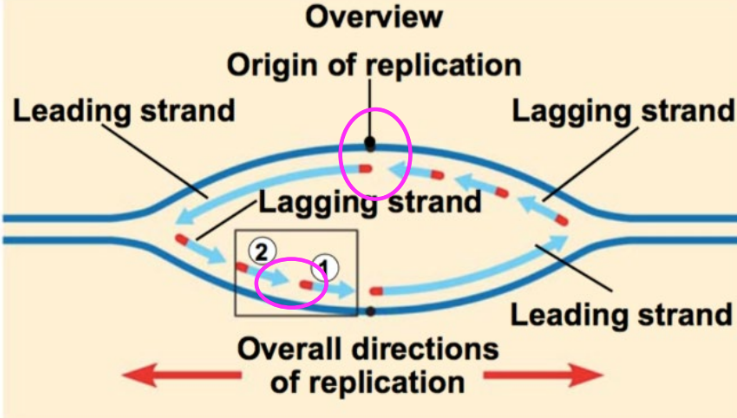
DNA ligase function
joins all the fragments together
joins all Okazaki fragments
joins leading and lagging strands at origin of replication (pink circles)
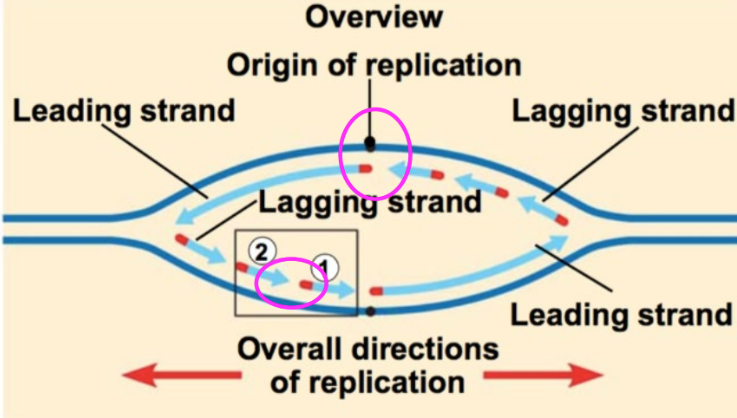
topoisomerase function
relieves stress on winding helix
single-stranded binding proteins
keep the single stranded DNA apart
metabolism definition
the set of biochemical reactions that transforms biomolecules and transfers energy
catabolism
energy released from “breaking down” or degrading molecules can be converted into useable forms of energy for the cell
note: ATP synthesis is coupled to substrate catalysis
anabolism
“making” or building molecules and structures in the cell uses energy
note: ATP catalysis is coupled to substrate anabolism
what are nutrients
substances taken from the environment by organisms for their growth, development and to sustain them
what is a bio element
elements found in cells and that are required for cellular function
autotrophs
organisms that can convert CO2 into organic carbon
heterotrophs
organisms that get organic carbon from other organisms
organic compounds
glucose (C6H12O2)
glycerol
inorganic compounds
water
nitrite
hydrogen sulfide
ammonia
what is cellular respiration
oxidation of glucose to carbon dioxide
glucose donates electrons to O2
the carbons in glucose end up as the carbons in CO2
4 stages
1. glycolysis
2. pyruvate oxidatiaon
3. citric acid cycle
4. oxidative phosphorylation
location of cellular respiration in eukaryotes
step 1 = cytoplasm (glycolysis)
step 2 + 3 = mitochondria (pyruvate processing + citric acid cycle)
step 4 = mitochondria (oxidative phosphorylation)
location of cellular respiration in bacteria
steps 1-3 = cytoplasm/cytosol (glycolysis + pyruvate processing + citric acid cycle)
step 4 = oxidative phosphorylation (cell membrane)
what are some high energy intermediates
ATP, NADH, FADH2
these are usable forms of energy for the cell
where does a cell get the “energy” from
from nutrients
energy is captured by oxidation and reduction reactions
nutrients are electron donors
the electron acceptor molecules capture some of the energy loss from the electron donors
oxidation
loss of electrons by an atom
and usually gains O atoms or loses H
reduction
gain of electrons by an atom
usually also gains H atoms or loses O
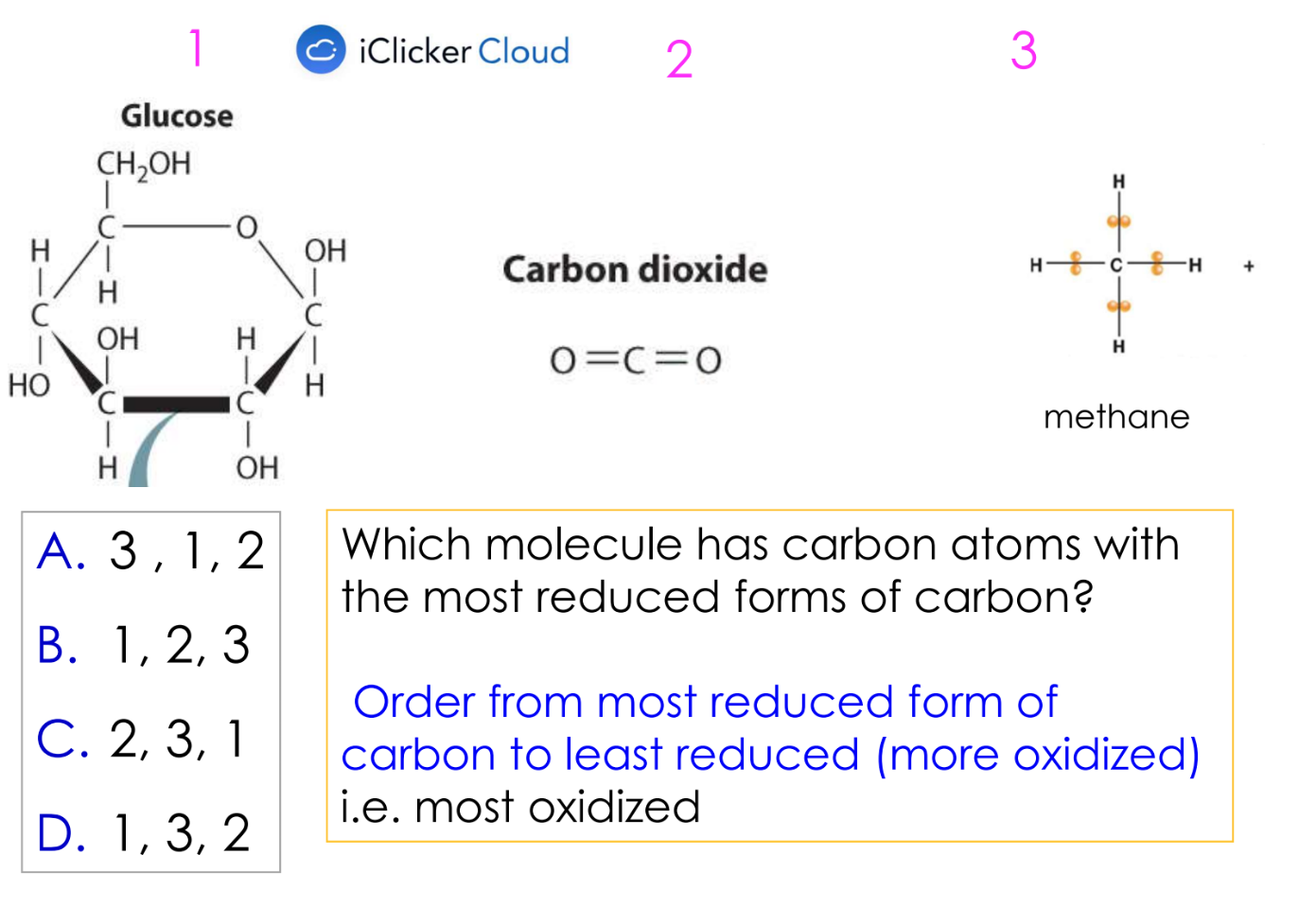
A
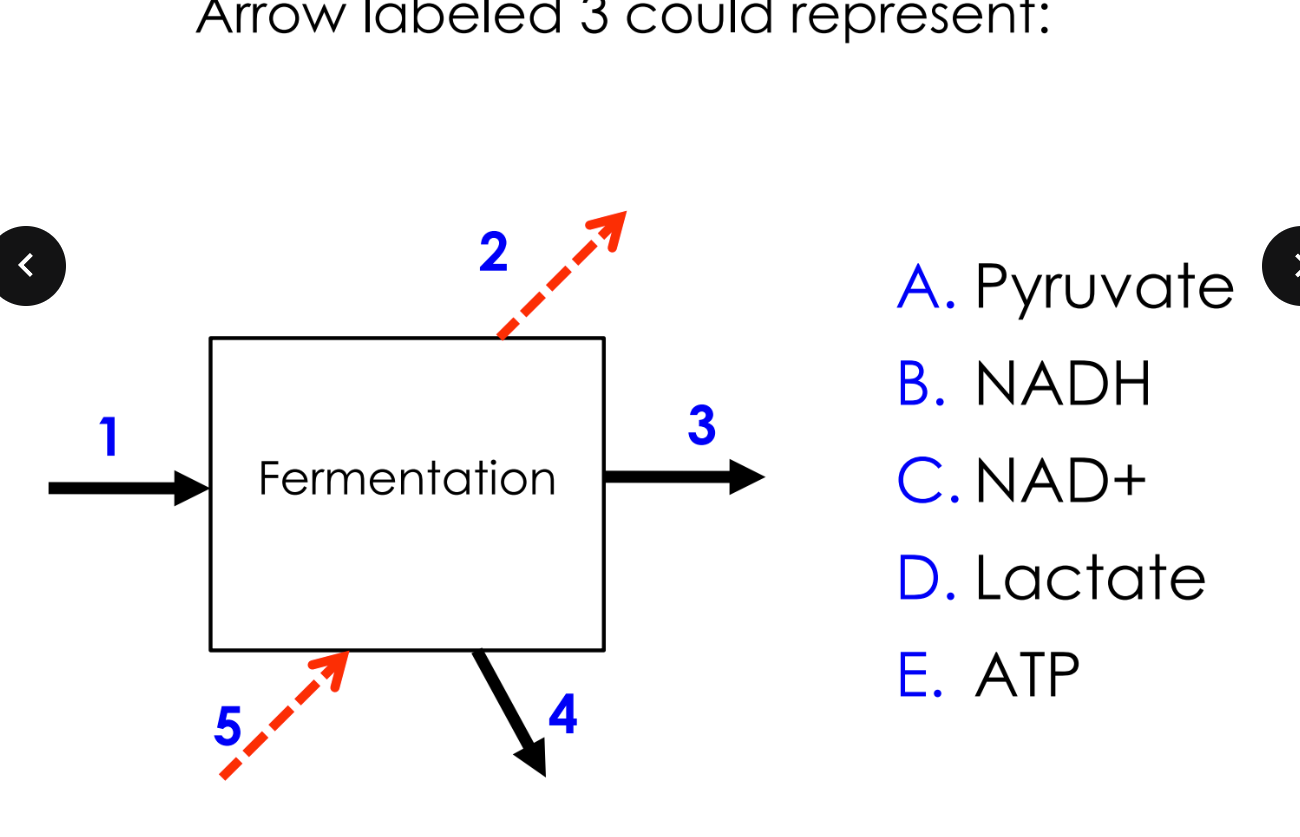
what is fermentation
a metabolic process that converts sugars to acids, gases or alcohols
fermentation is an alternate pathway after stage 1, not all cells can ferment
no terminal electron acceptor
if glucose is available, the cell will undergo glycolysis and produce ATP and NADH but ends there
respiration vs fermentation
respiration: ATP made in stage 1, 3, 4 i.e. cell makes most of its ATP by oxidative phosphorylation
all 6 carbon atoms in glucose are completely oxidized to CO2
oxidized C atoms discarded as CO2 waste
more ATP can be synthesized if the cell can complete all 4 stages of cellular respiration
fermentation: cell makes all of its ATP by substrate level phosphorylation (in glycolysis)
no net oxidation of C
electrons removed from some C atoms and returned to others
the products of fermentation are waste products
much less ATP if a cell can only complete stage 1 (glycolysis) and fermentation
when cells respire, what element do they use as a terminal electron acceptor
O2
why can’t pyruvate continue in cellular respiration
most likely reason is lack of terminal electron acceptor O2
what are the two types of fermentation
ethanol fermentation
lactic acid fermentation
can pyruvate undergo fermentation?
if the cell has the capacity to do so

C
slp = substrate level phosphorylation
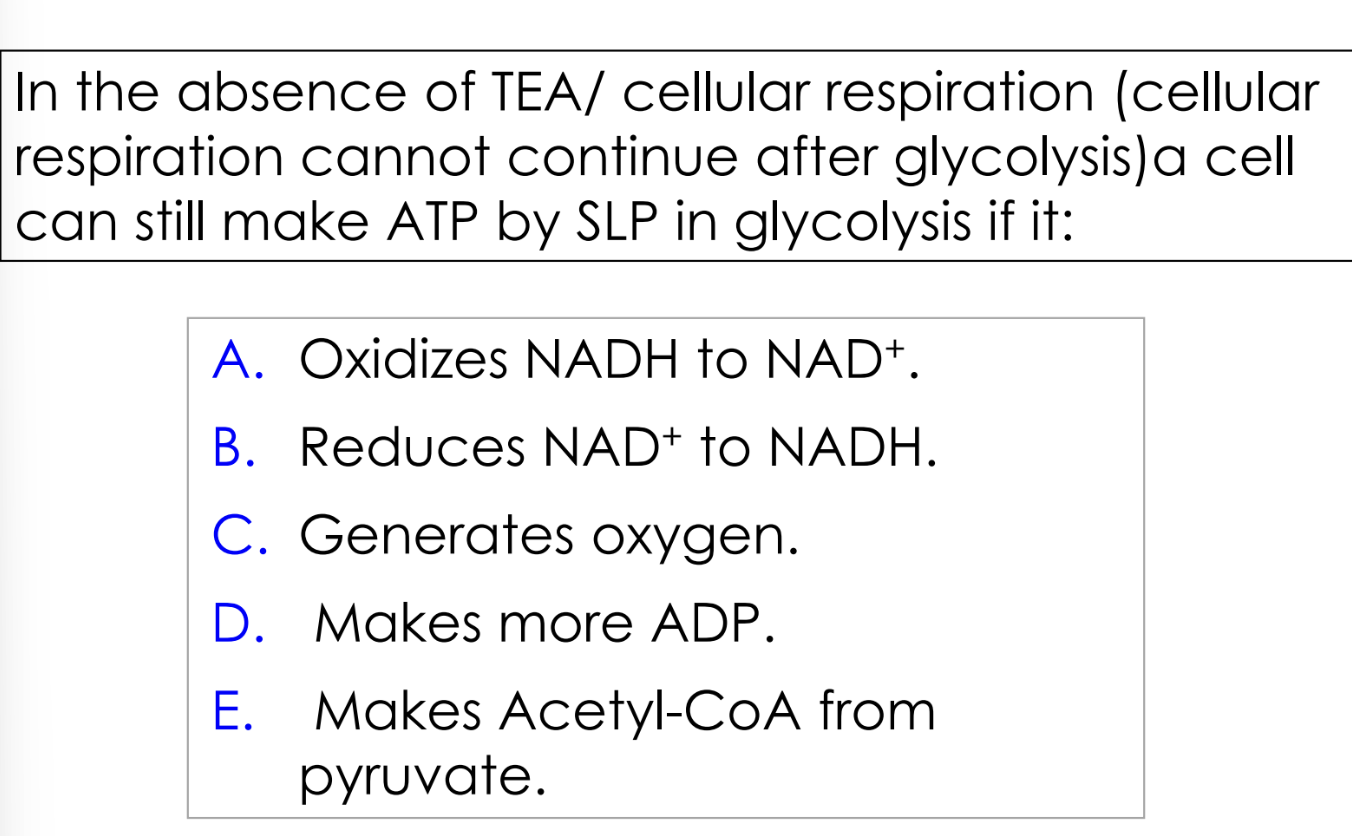
A
fermentation if there is no terminal electron acceptor
pyruvate will undergo fermentation
NADH will deliver electrons to fermentation intermediates
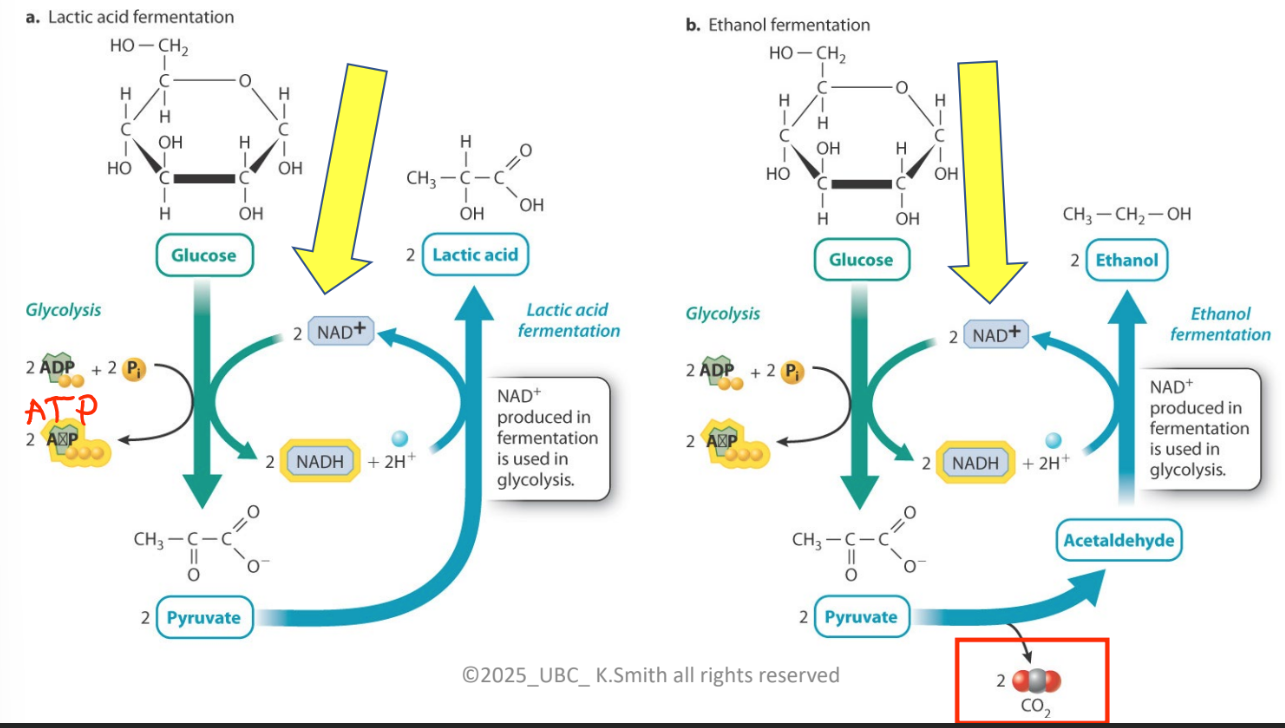
what is being described in this picture
NADH donates electrons to re-generate NAD+
Now, NAD+ can return to glycolysis to pick up more electrons
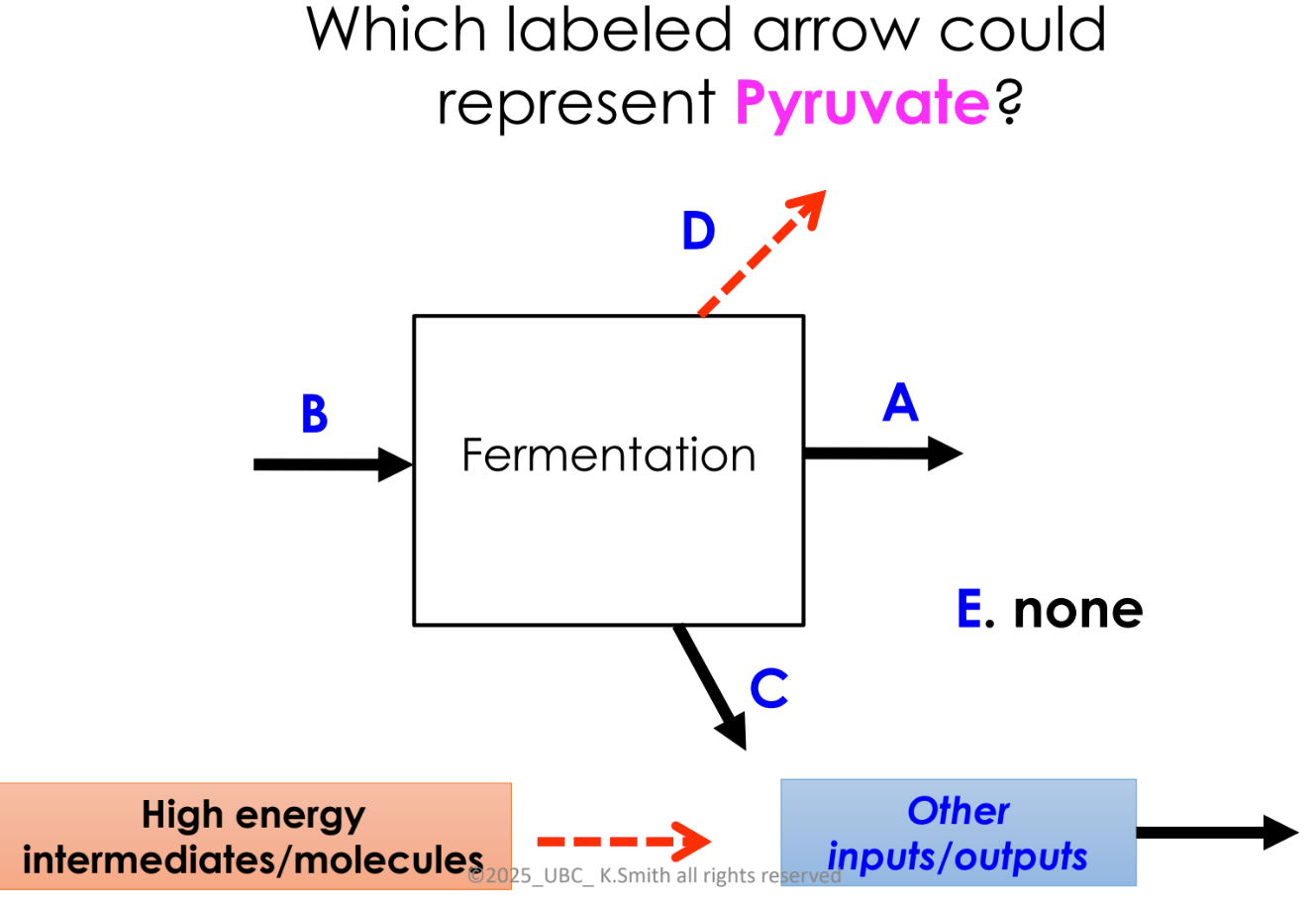
B
D (lactic acid)

E
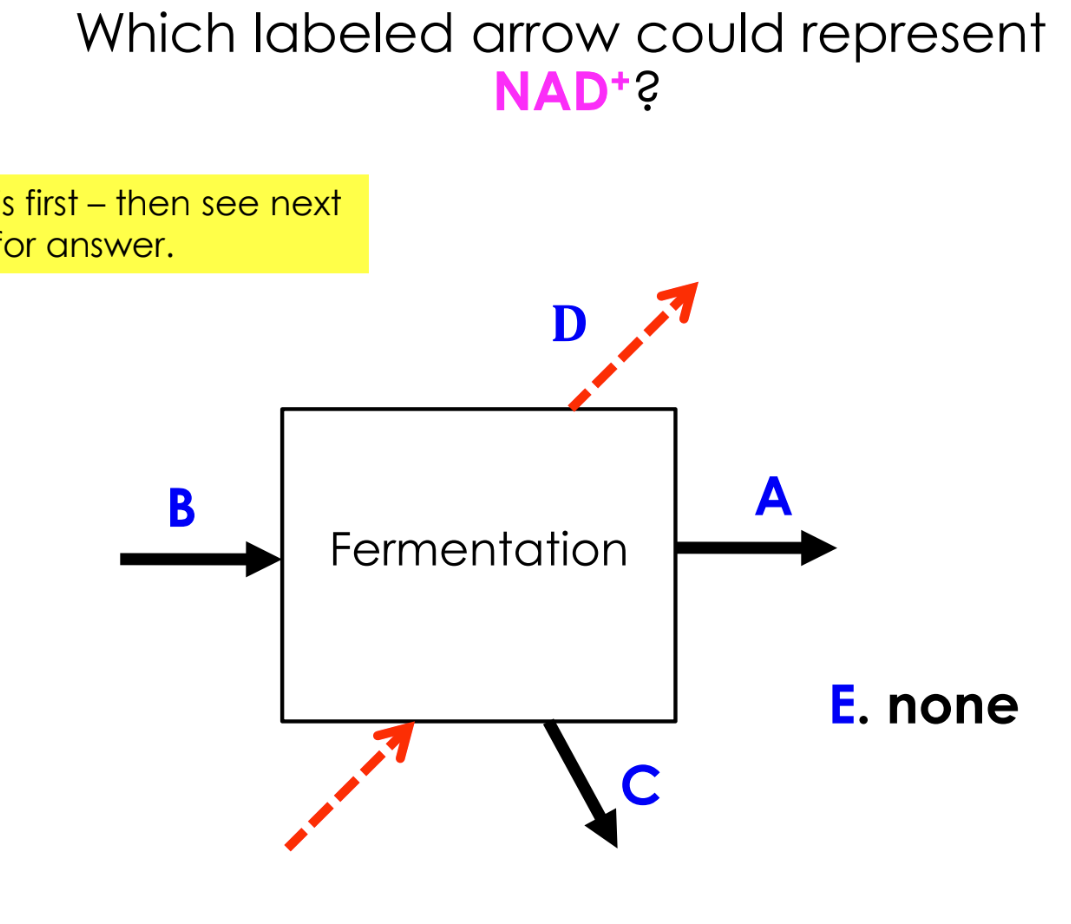
D
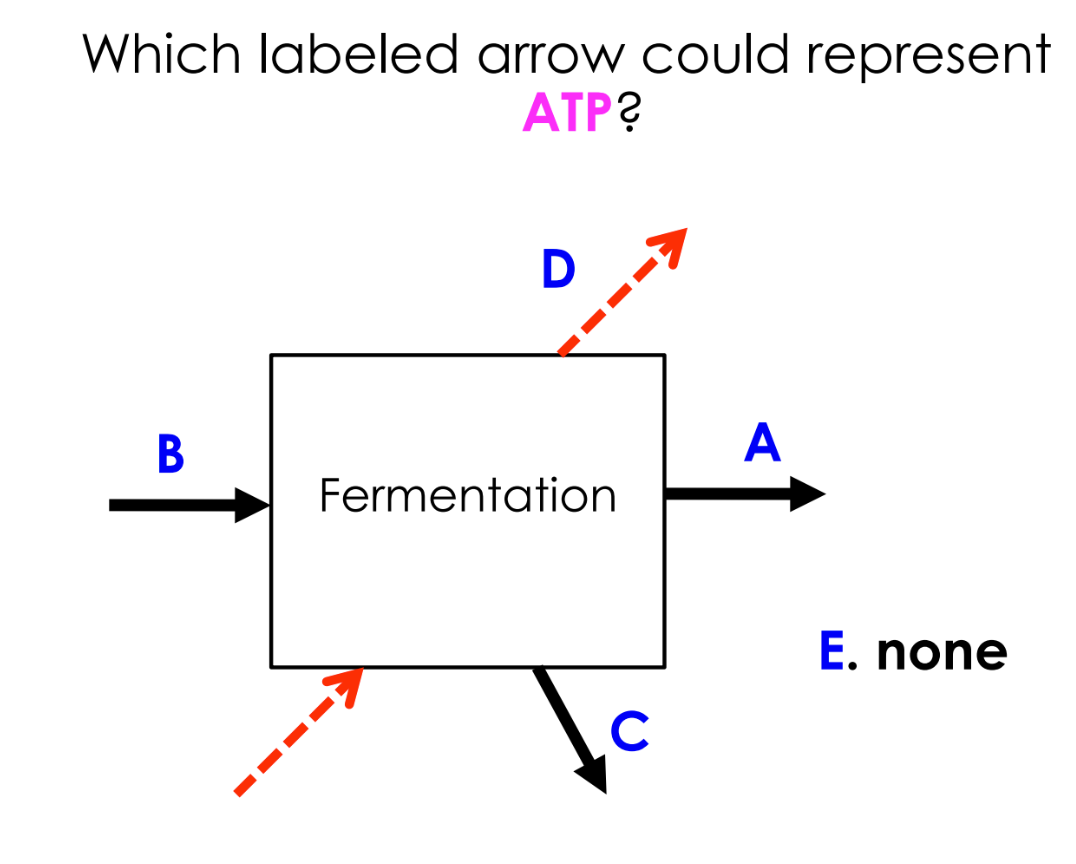
E
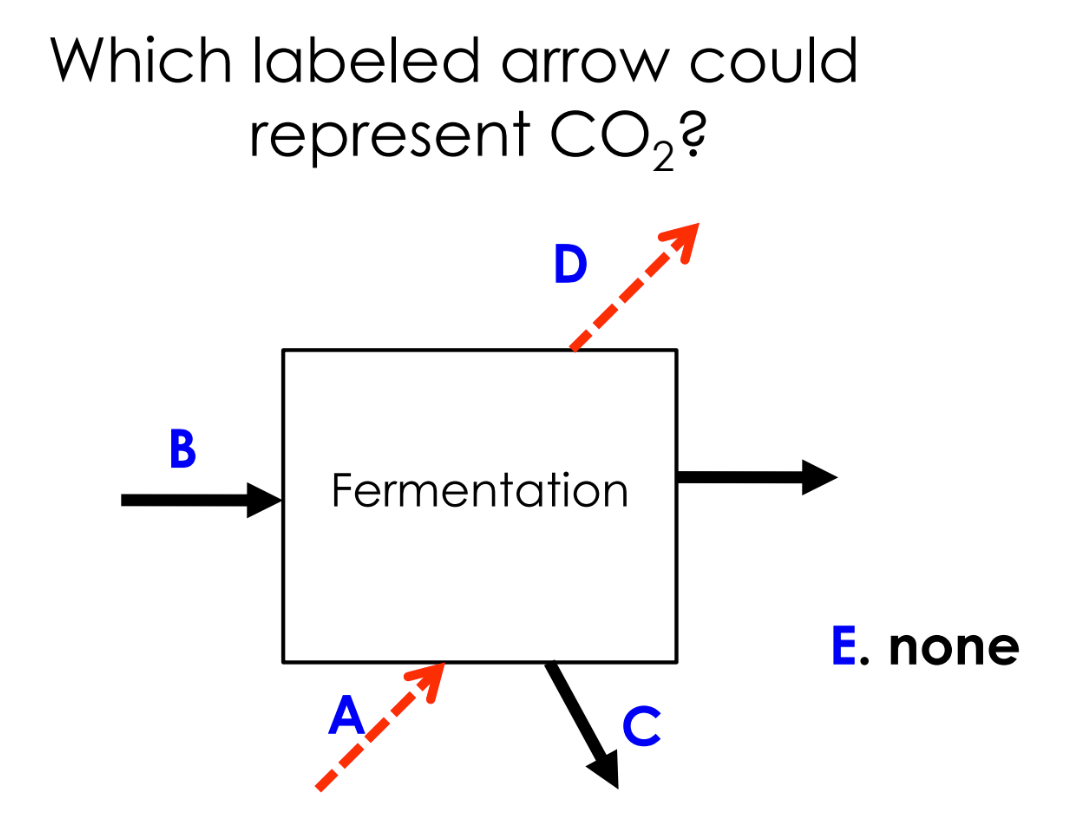
C
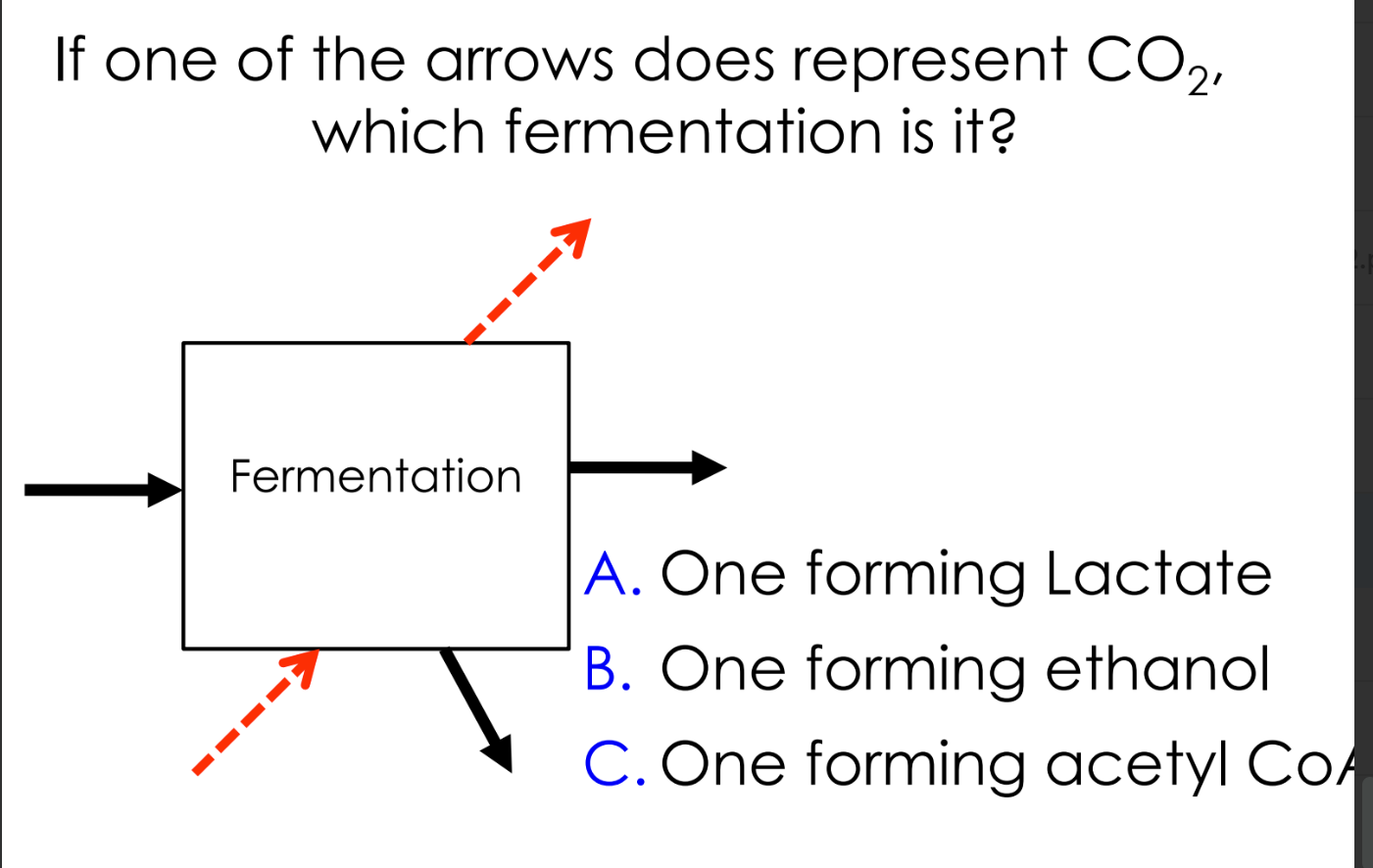
B
glycolysis
glucose is broken down to pyruvate
ATP and other high energy intermediates are produced
occurs in cytosol
goal is to degrade glucose, extract energy from the reduced form of carbon (glucose) in several steps, synthesize high ATP and NADH (electron carrier), create pyruvate - a molecule that can be used in different pathways
uses 2 ATP and synthesizes 4 ATP
synthesizes 2 NADH (4 electrons - 2 to each NAD+)
explain phase 1 of glycolysis
energy investment phase
2 ATP used to phosphorylate a 6C sugar with 2 negatively charged phosphate groups
explain glycolysis phase II
energy payoff phase
4 ATP made by substrate level phosphorylation
4 electrons removed (from G3P) to reduce 2 NAD+ to 2NADH
oxidized form of NAD
NAD+
can take up/accept 2 electrons
reduced form of NAD
NADH
can donate 2 electrons
what is substrate level phosphorylation
enzyme catalyzes transfer of phosphate from a phosphorylated molecule to ADP
location of substrate level phosphorylation
euk cytosol or mitochondrial matrix
cytosol for bacteria
SLP vs oxphos
SLP involves enzymes and substrates that phosphorylate ATP
oxphos involves membrane-bound enzymes and H+ gradient drives ATP phosphorylation
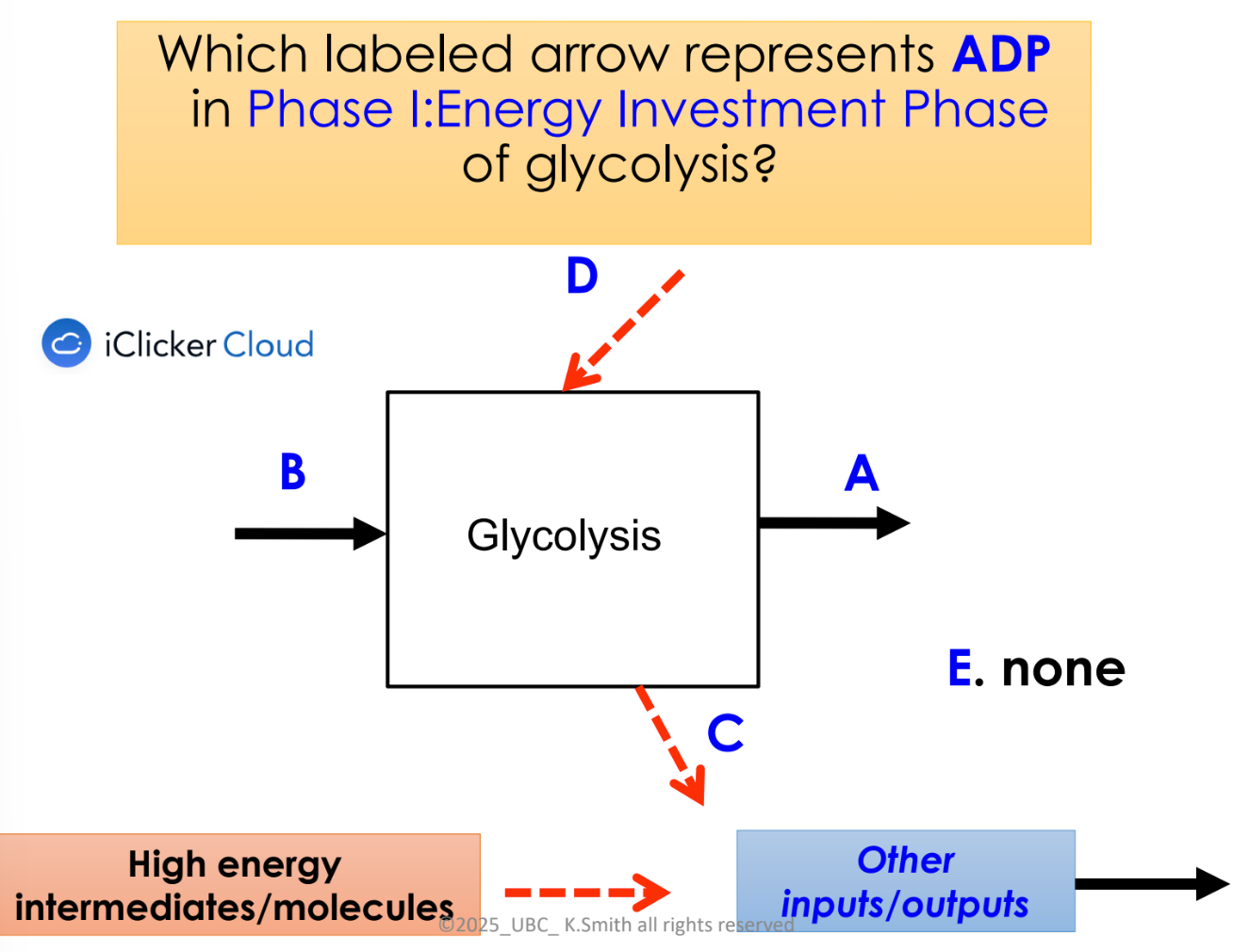
C
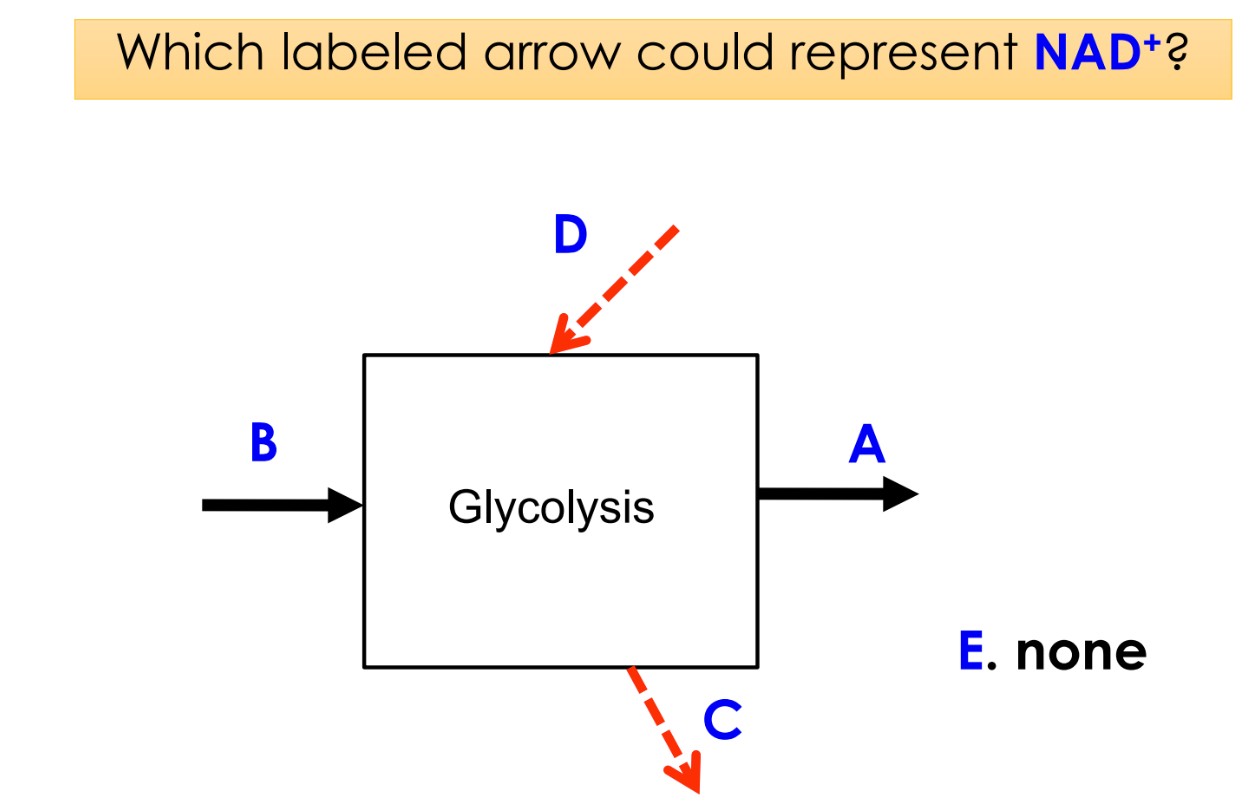
D