Comparative human evolution
1/47
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
48 Terms
two phenotypic sources of evolution
morphological and behavioral

large vs small scale sources for evolution
gene deletions/duplications vs. single-nucleotide polymorphism

some phenotypic traits listed as diff. from great apes

Early evolved traits in Hominid Evolutions

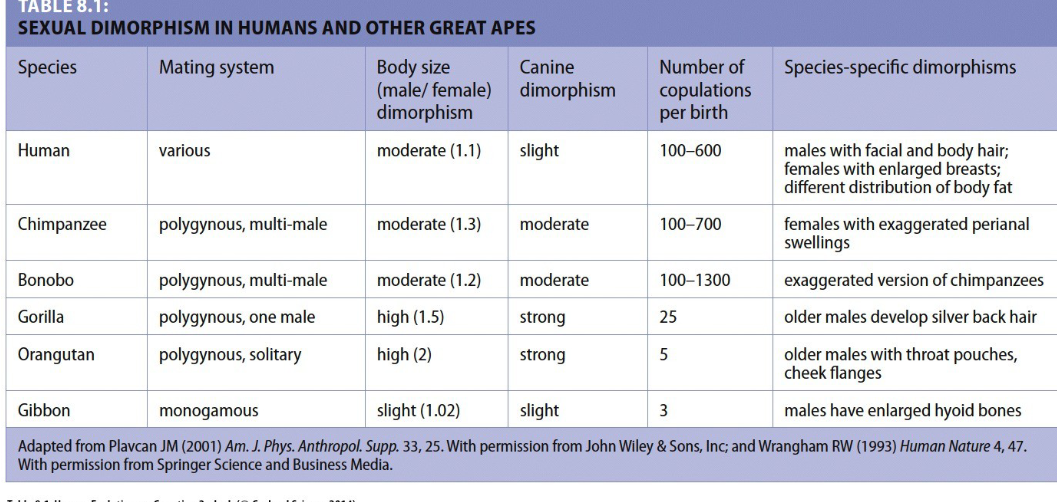
extensive sexual dimorphism in early hominins/australopithicines of body size due to (CMD)
change in env., mating practices, diet changes

humans sometimes thought of as
netonic apes; meaning they retain juvenile characteristics of their primate ancestors longer than other primates

cranial shape of human infant similar to that of infant chimp but human baby is twice as large than gorilla and chimp baby overall due to
fat deposits
human newborns differences in growth, fat reserves, and weaning reasons
longer growth for bigger brain, fat reserves for coping with bodily stress, and weaning time shorter to decrease energy costs for mother all adaptations were likely helpful for african migration

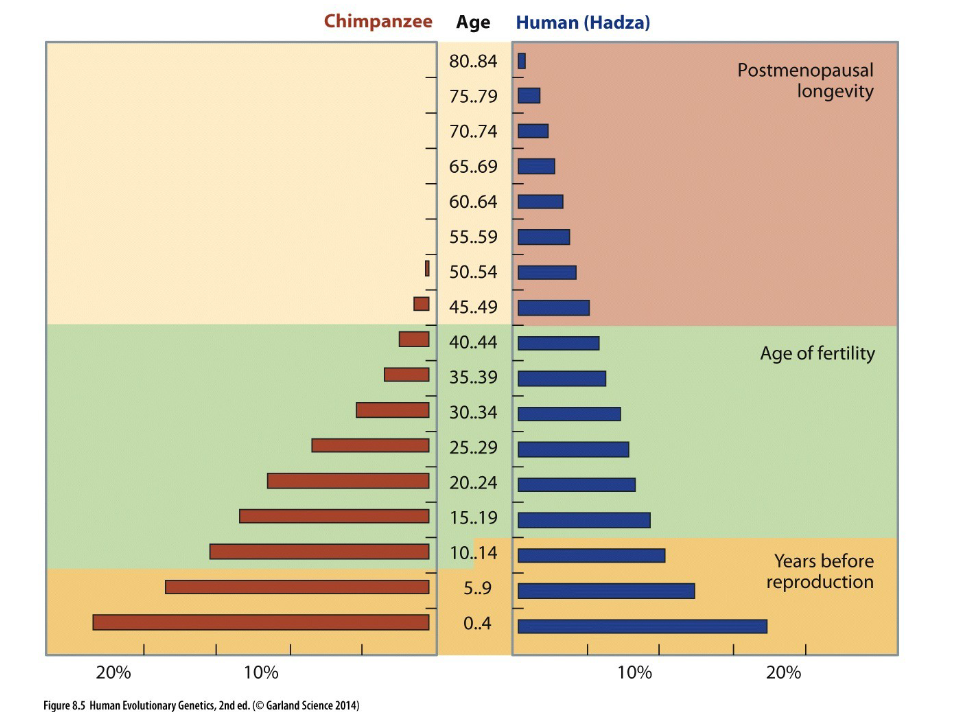
Slower growth in childhood is compensated for by a
growth spurt in adolescence; not seen in other mammals
why are humans good at long distance running compared to other mammals

handedness in chimps vs. humans and neanderthals

hypothesis for probable speech of ancient homos with evidence (CHFTL)
complex tool usage, hyoid bone, facial expression development, tears, and lower larynx

clavicle diff. in neanderthal vs modern human
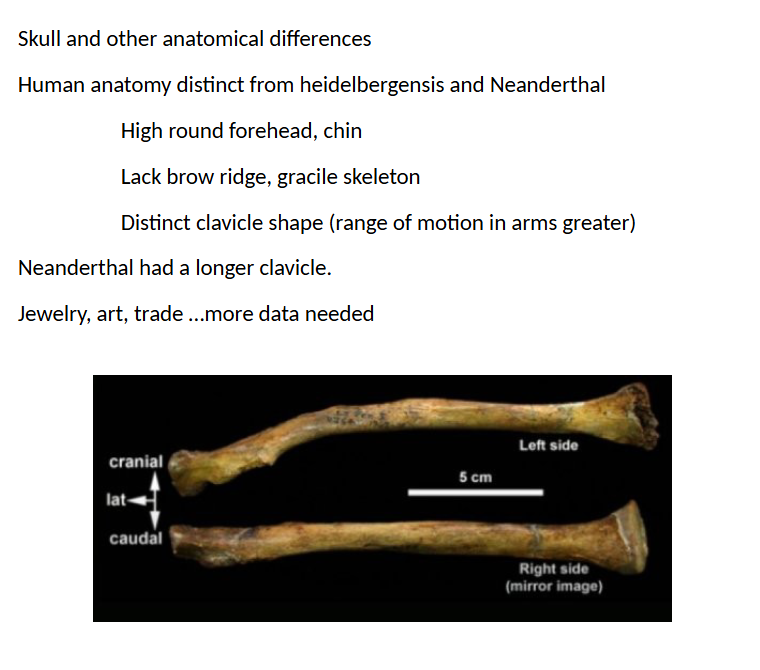
skull diff. in neanderthal and modern human
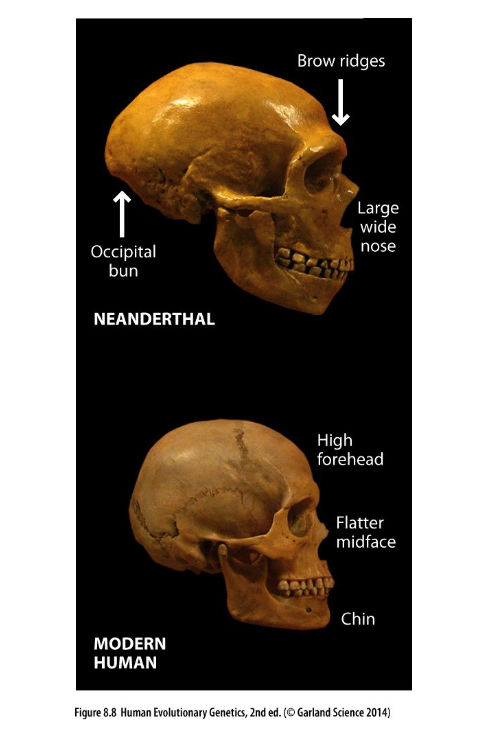
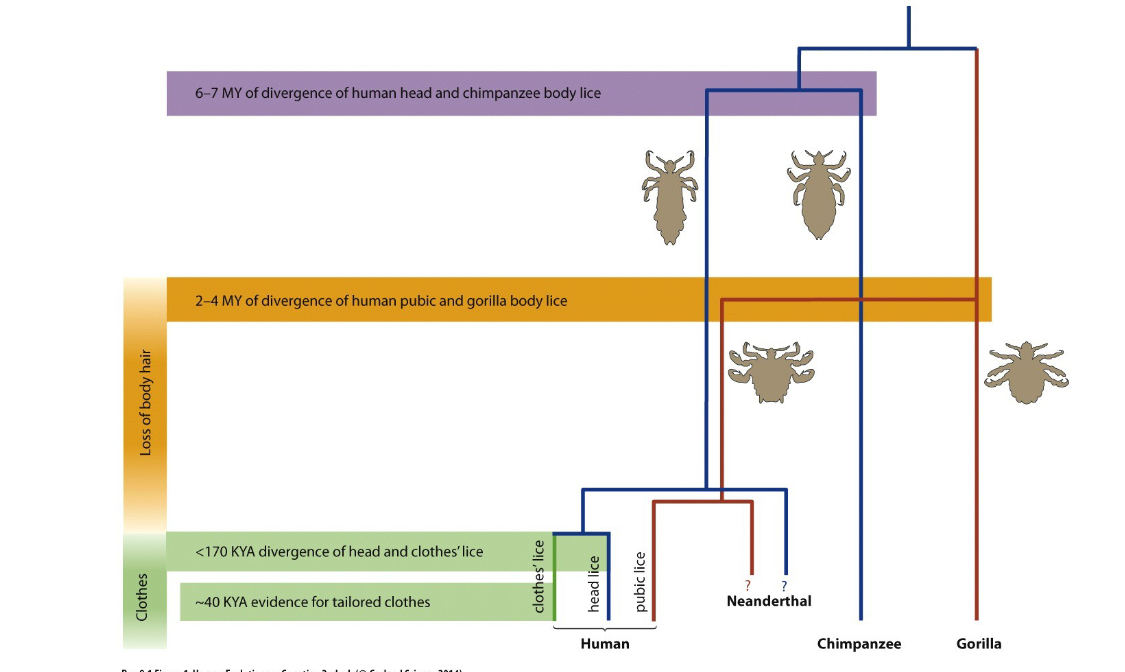
what is this saying
shows evolution of lice when humans began to lose body hair some stayed on the head, others moved to the pube, then others evolved on clothes
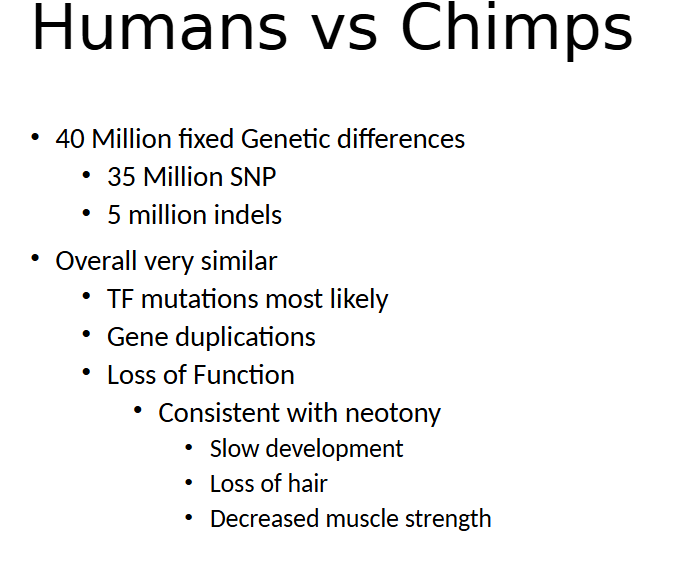
how many fixed genetic differences in chimps and humans
40 mil
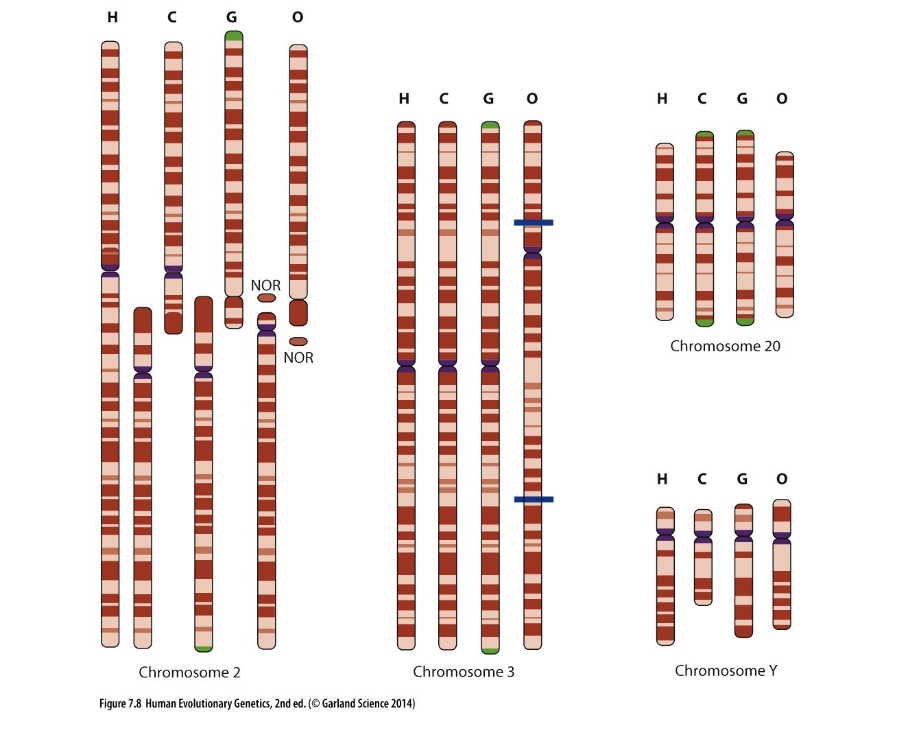
chromosome num. in great apes vs. humans
humans have 46 great apes have 48

why do humans have a large chromosome #2
Humans have a large chromosome 2 that actually formed from the fusion of two ancestral ape chromosomes
does non coding genes influence phenotypic diff. between humans and great apes
Non-coding DNA usually doesn’t cause obvious traits, but some jumping genes (like retrotransposons) might mess with gene function. This is hard to study
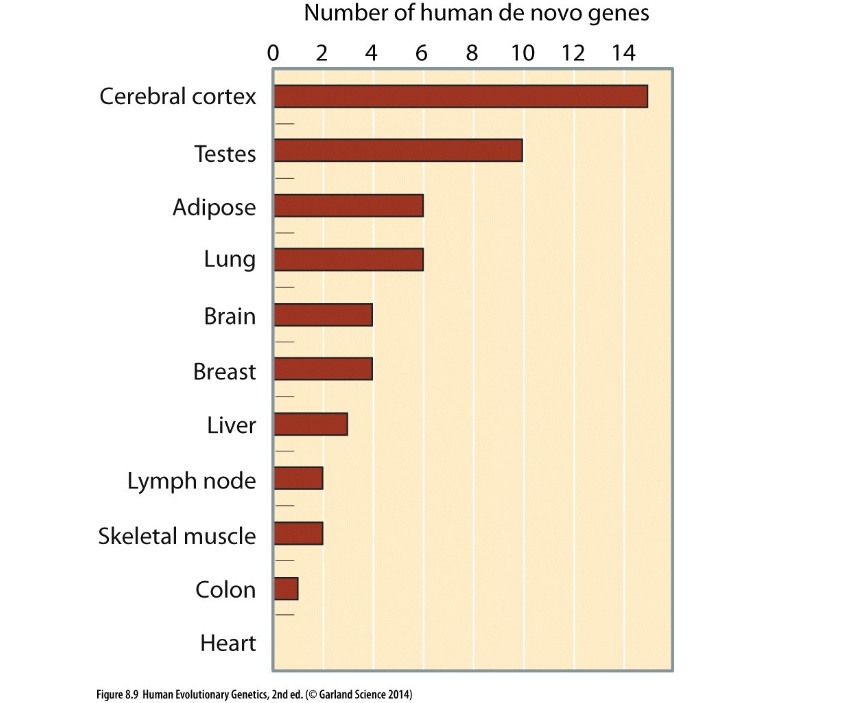
compared to great apes, humans have how many denovo genes that didn’t evolve in diff. species from common ancestor (orthologs)

why do humans have 60+ denovo genes compared to great apes
Likely arose from previously non-coding DNA that got turned into functional genes.
what are the deletions and duplications in genes in humans most likely involved in (RIC)
reproduction, immune response, and chemosensory activity
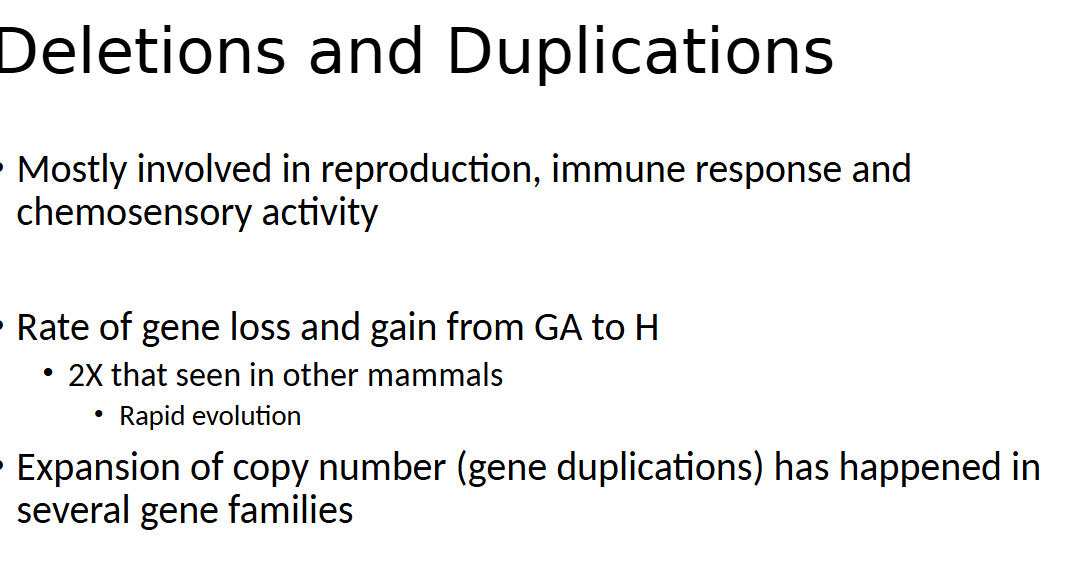
humans gained and loss genes at a rate x2 faster than other mammals which points to
rapid evolution
gene deletions in humans vs other mammals
510 regions deleted in humans compared to other mammals
GADD45G-loss of function correlated with
expansion of specific brain regions (loss means brain expanded to greater extent)
how many snp differences betw humans and chimps
35 million


what dis slide say
Fatty acid metabolism and G-protein signaling genes show signs of positive selection, with 10–20% of changes fixed adaptively based on MK test results. Adaptive changes are common in genes related to sensory perception, immunity, apoptosis, and reproduction, but are rare in brain-expressed genes. Many of these changes may occur outside coding regions, highlighting the role of non-coding DNA in human evolution.


what dis slide say
Insertions/deletions (indels) and SNPs that affect alternative splicing are a key mechanism for increasing transcriptome and proteome diversity in mammals. These splicing changes impact over 90% of genes


The gene KLK8 (kallikrein-related peptidase 8) has a fixed human-specific splice variant. In human populations, variation in this gene is linked to
learning and memory. This splice form hasn't been found in other primates, suggesting a potential human-specific adaptation


what dis slide say
Only 2% of the genome codes for proteins, but 5.5% is conserved and functional across mammals. To understand what makes us human, scientists study Human Accelerated Regions (HARs), which often target functional RNA genes. Among 49 identified HARs, two map to known genes, and one is linked to a novel RNA expressed during fetal brain development.


what dis slide say
HARs include cis-regulatory elements that increase expression of genes related to neural function and glucose metabolism. A notable example is the amylase gene, which has up to 8 copies in humans but only 1 in great apes (GA)—a difference likely driven by dietary shifts and adaptive selection


what dis slide say
miR-1304 is not repressed in humans, unlike in great apes, and it plays a role in tooth formation. This may contribute to differences in dentition between humans and great apes


what dis slide say
Gene expression differs by 30% in testes and 10% in the brain, with brain gene expression being more stable within species. Brain regions in humans and chimps show similar divergence patterns, but brain-expressed genes evolved rapidly in humans. Human brain gene expression is generally higher than in chimps, and gorillas show the most similar patterns to humans. Additionally, 259 genes are upregulated in humans, especially those involved in neuron insulation and motor neuron development.

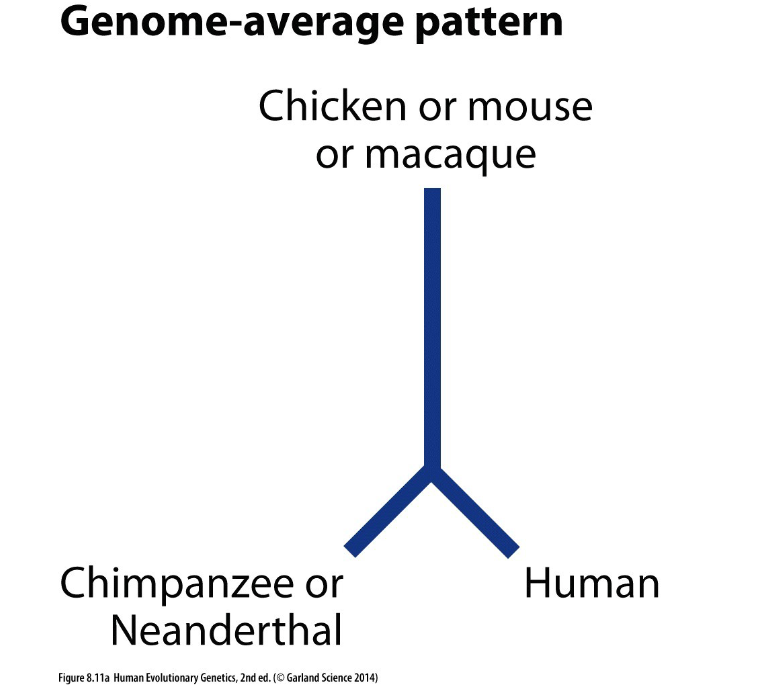
what dis sayin
the human genome is more closely related to chimpanzees or Neanderthals than to more distantly related species like chickens, mice, or macaques
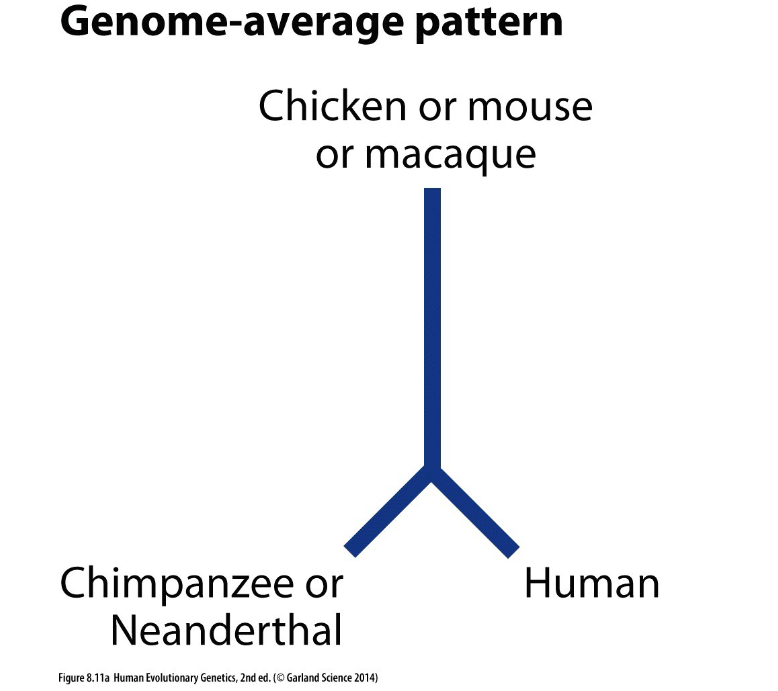
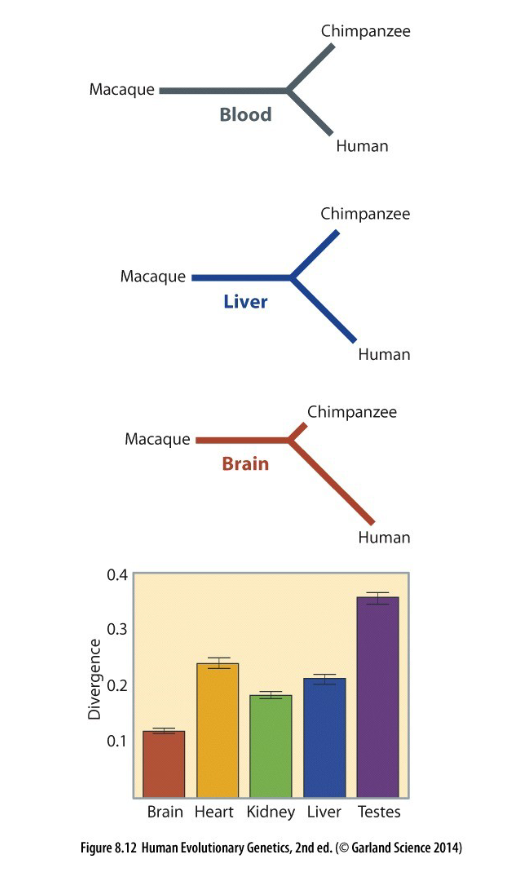
what dis sayin
humans most close to chimps and bottom measures divergence in traits

what dis sayin
Humans and Neanderthals differ by fixed amino acid changes in 78 genes. Only 6 of these genes have more than one change. Interestingly, 3 of those 6 genes are expressed in the skin, though it's currently unknown whether these changes are biologically significant
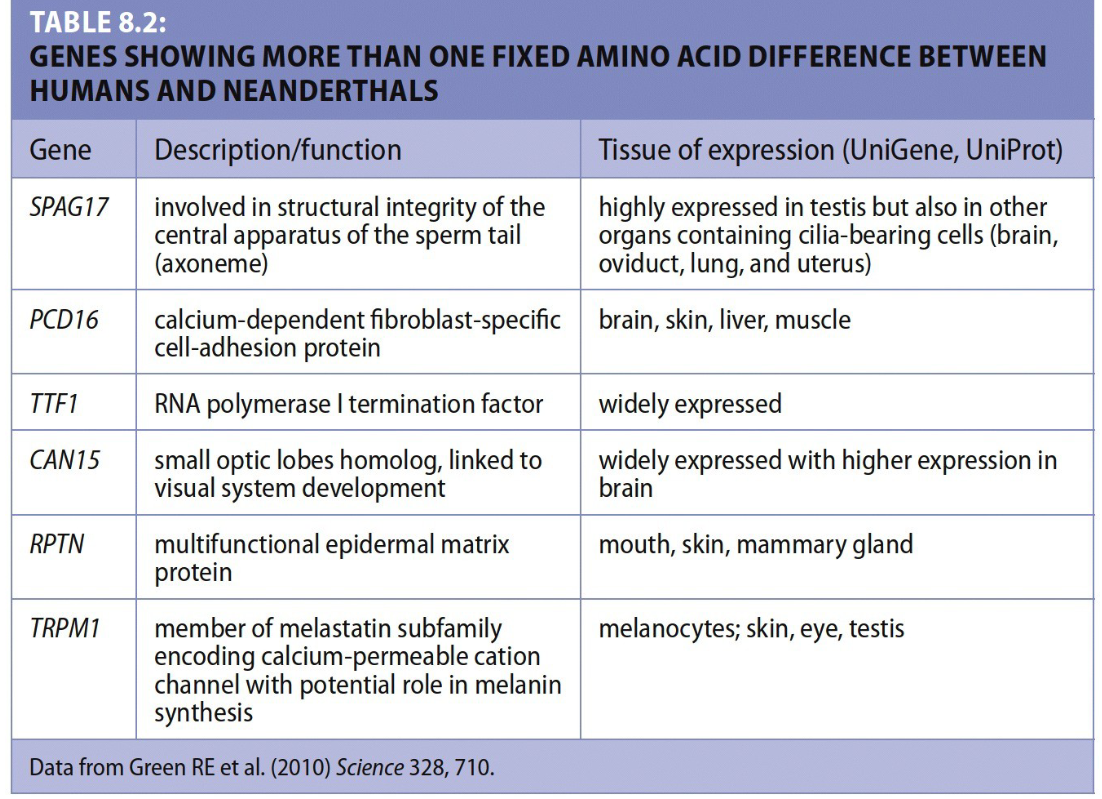
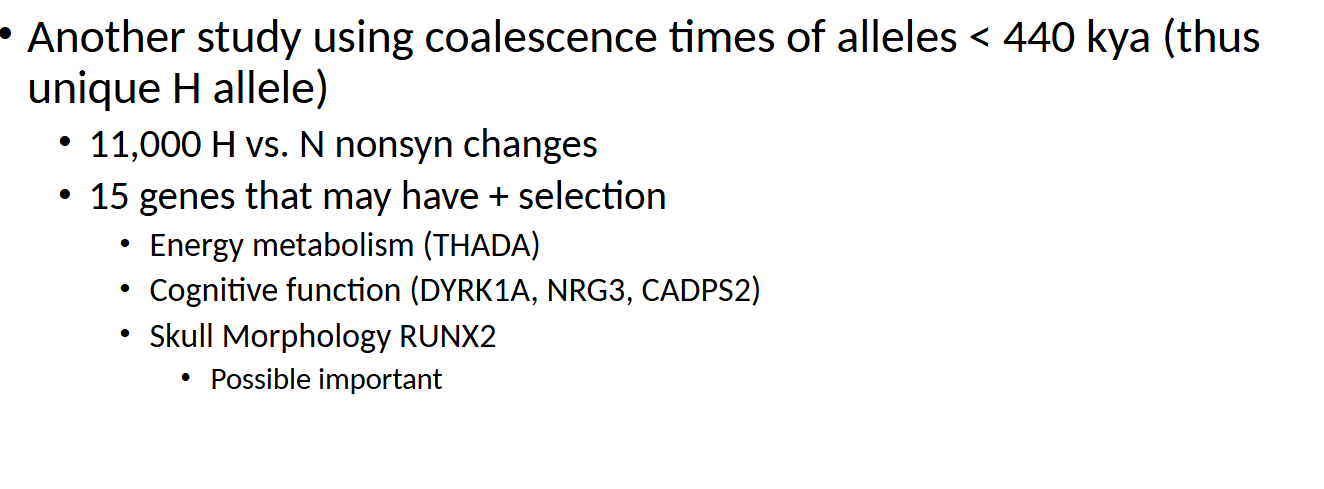
what dis sayin
study that looked at alleles unique to humans by analyzing coalescence times under 440,000 years ago. It identified 11,000 nonsynonymous (protein-changing) differences between humans and Neanderthals. Of these, 15 genes may have undergone positive selection, including genes related to energy metabolism (THADA), cognitive function (DYRK1A, NRG3, CADPS2), and skull morphology (RUNX2)

human and Denisovans differences in amino acids
Humans and Denisovans differ by 129 amino acid substitutions and 14 indels, with humans carrying the derived versions. Two of these fixed differences are linked to skin disease, although the specific condition is unknown

some unique traits for humans

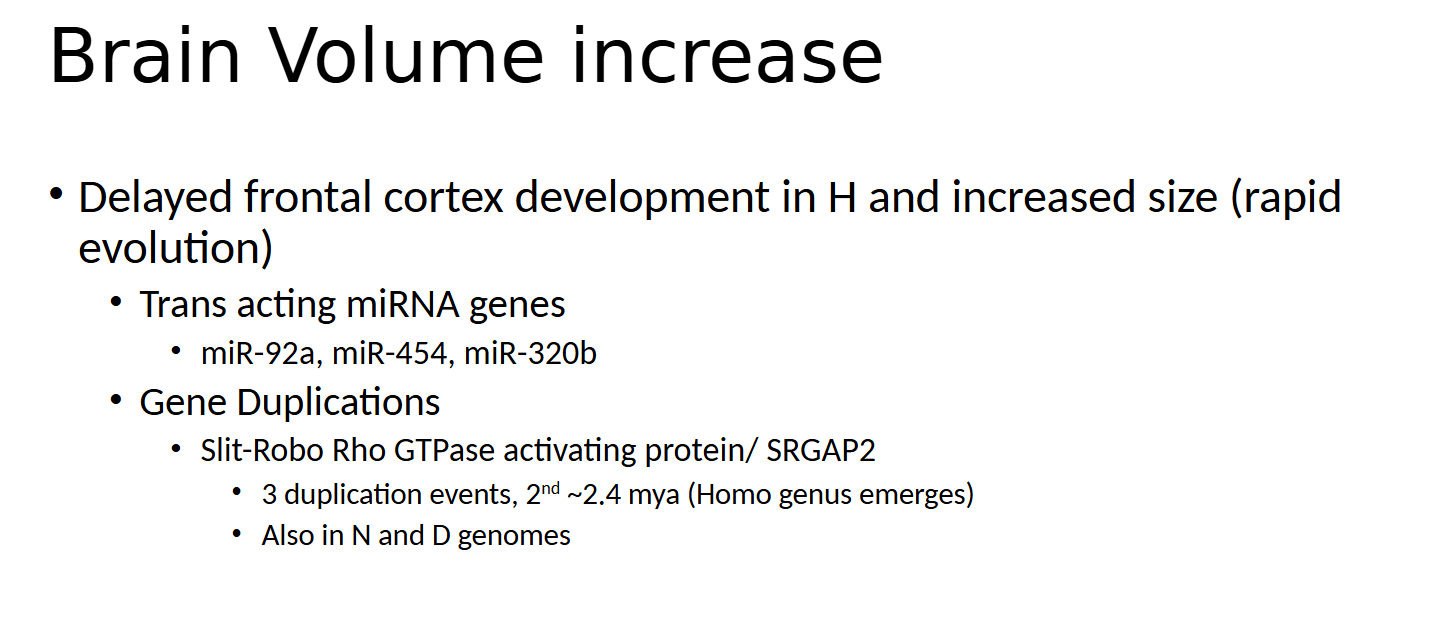
what dis say
humans experienced a delay in frontal cortex development alongside a rapid increase in brain size, suggesting fast evolutionary changes

Paralog means genes within the same species that arose from gene duplication. These genes are expressed in both developing and adult brains, suggesting a role throughout brain growth and function. In mice, the SRGAP2 ortholog promotes the maturation of dendritic spines

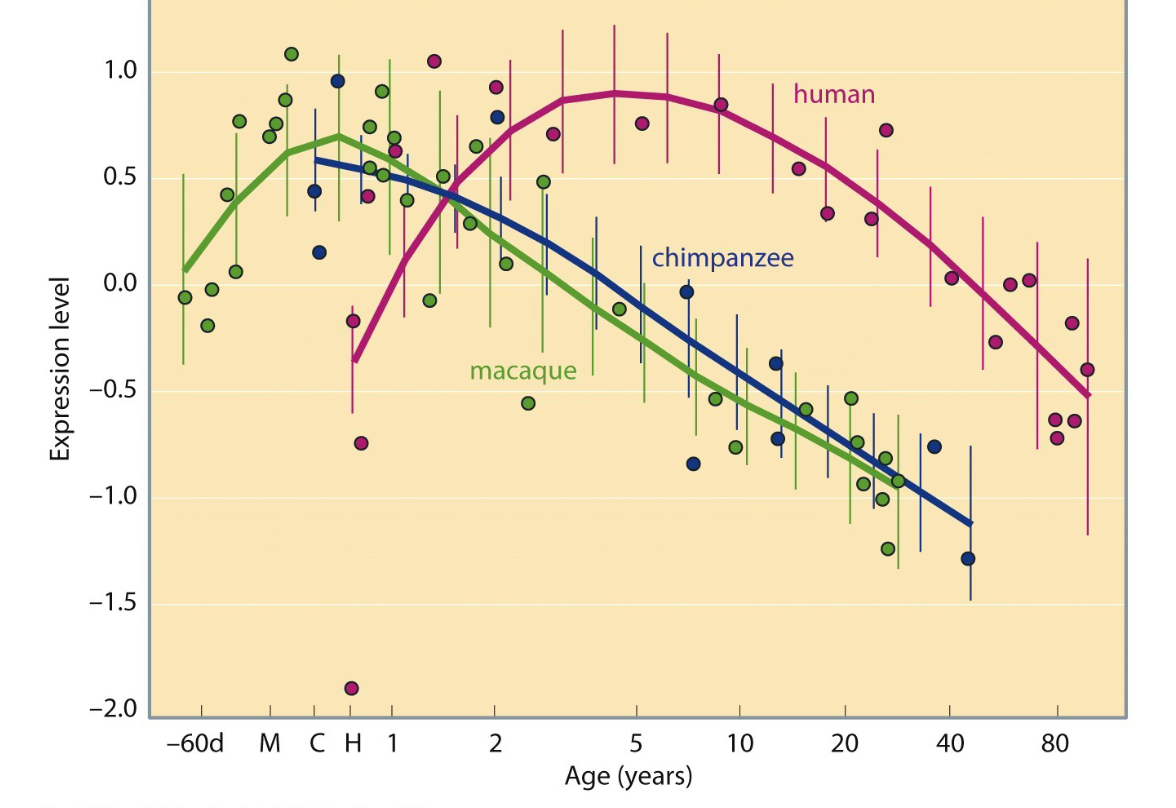
what this portray fr
Humans exhibit a delayed and extended peak in gene expression in the prefrontal cortex compared to chimpanzees and macaques.
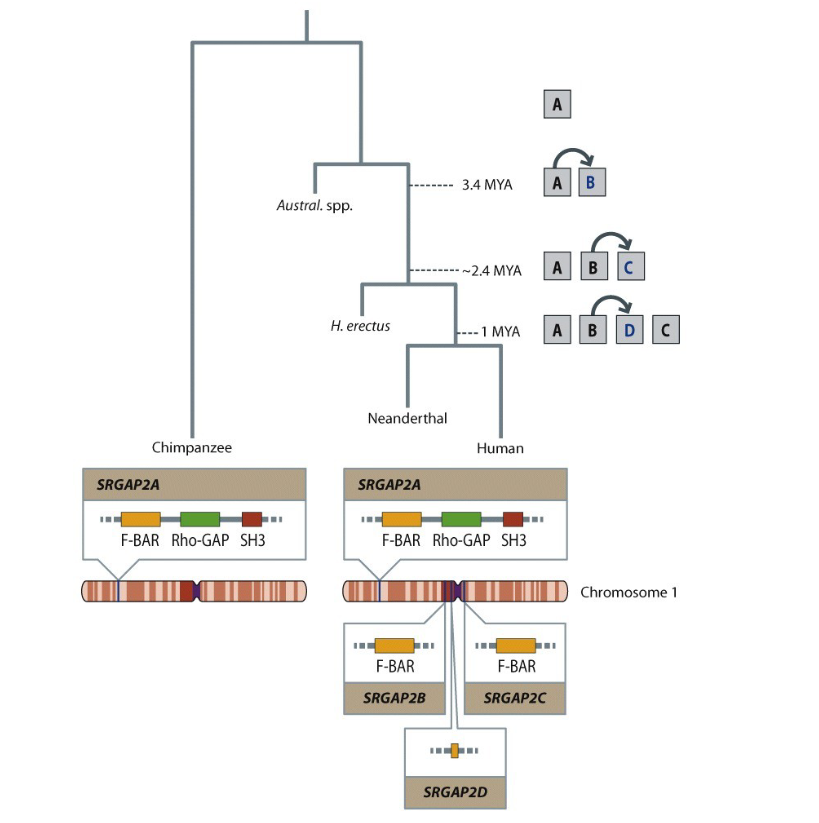
what
The SRGAP2 gene underwent multiple duplications during human evolution. The original gene, SRGAP2A, is present in chimpanzees and early hominins. Around 3.4 million years ago, SRGAP2B emerged, followed by SRGAP2C around 2.4 million years ago—around the time the Homo genus appeared. A final duplication, producing SRGAP2D, occurred about 1 million years ago
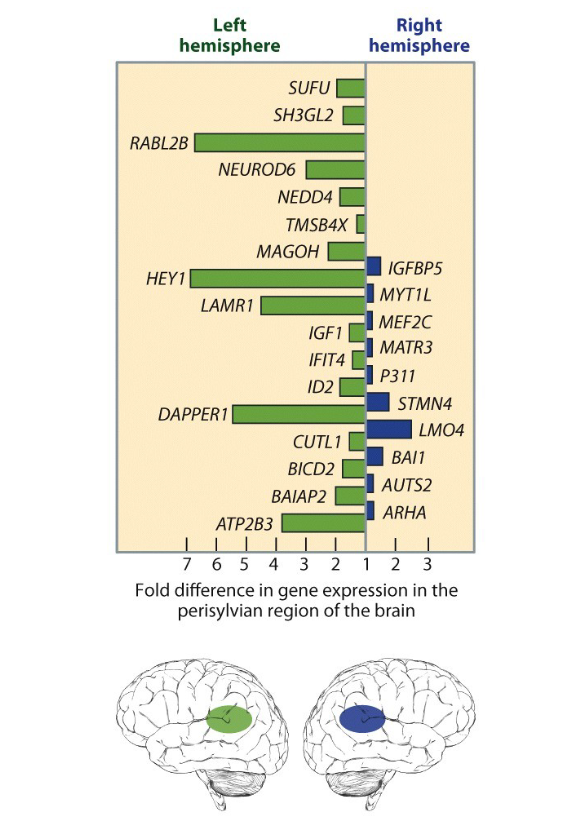
explain

broca vs wernickes brain areas control
word formation vs. recognition


what
The FOXP2 gene, associated with language, is identical in humans and Neanderthals. However, both species have two nonsynonymous substitutions that differ from those found in chimpanzees and gorillas. These changes are likely related to learning, cognition, and speech
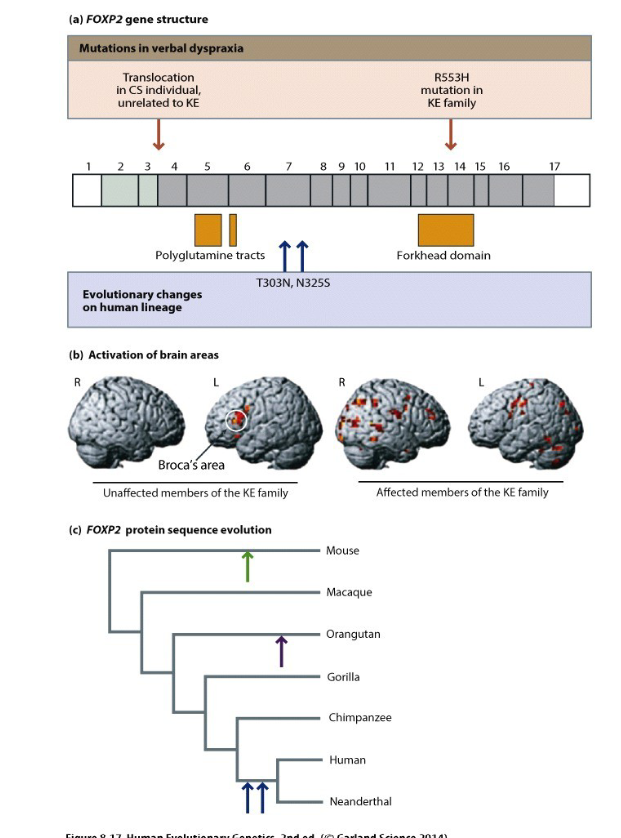
The FOXP2 gene includes 17 exons and contains important regions like polyglutamine tracts and a forkhead domain. Mutations in this gene, such as the R553H mutation in the KE family, are linked to verbal dyspraxia, a speech disorder. Brain scans show affected broca area
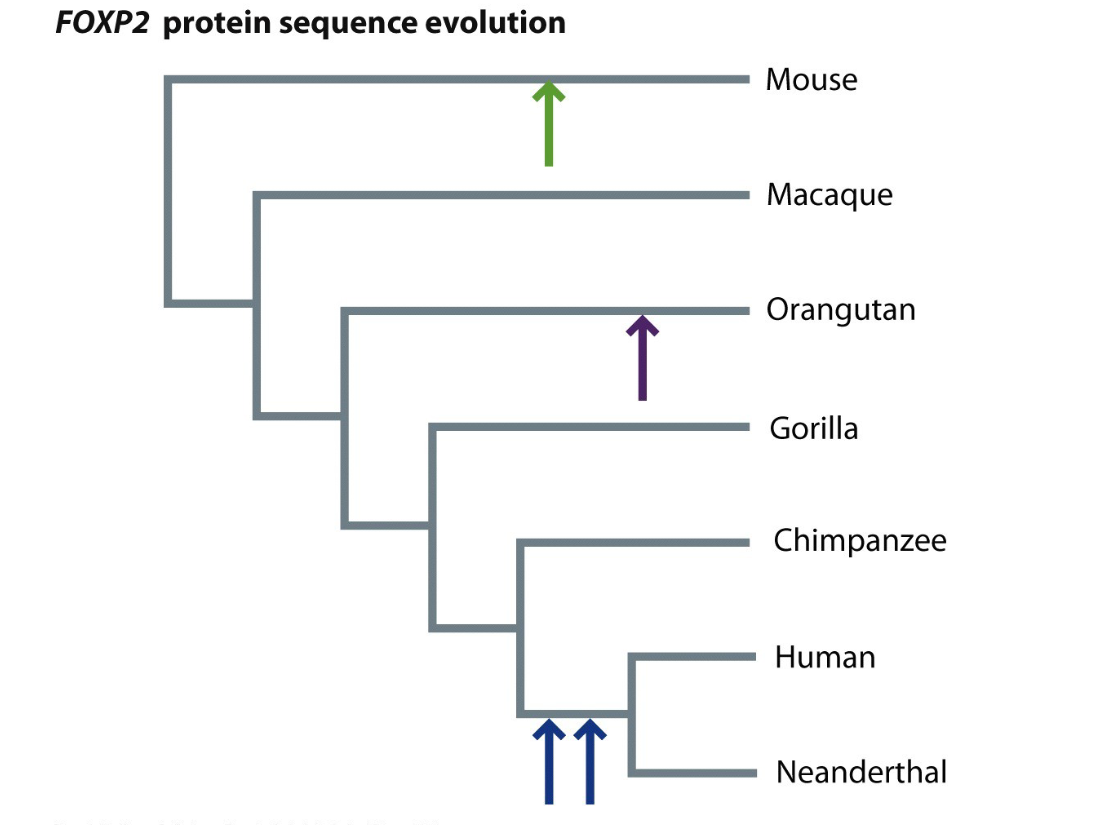
The arrows indicate where amino acid changes occurred. Two changes happened specifically on the human lineage, shared with Neanderthals but not seen in chimpanzees or other primates. It could be linked to language since it’s related to FOXP2