BIS104 MT1
1/92
Earn XP
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
93 Terms
How are coronaviruses (CoV) named?
Named for the crown-like spikes on the surface and belong to the family Coronaviridae within the order Nidovirales
What do coronaviruses infect?
vertebrates such as humans, birds, bats, etc
SARS-CoV-2 (COVID) Genome
single stranded; positive sense (can be directly translated into proteins)
Flu/Influenza virus
ss negative sense RNA virus (needs RdRp, cannot be directly translated)
What did Robert Hooke discover?
Termed pores inside of cork as cells bc they reminded him of the cells inhabited by monks. Were actually empty cell walls made of dead plant tissue
What did Antonie van Leeuwenhoek discover?
Examined pond water and observed microscopic “animalcules”
Saw bacteria from peppercorn water and dental plaque
Credit for discovering living cells, first to see bacteria
Cell Theory; who was it articulated by and what was added later
Articulated in mid-1800s by Matthias Schleiden, Theodor Schwann, Rudolf Virchow
All organisms are composed of one or more cells
The cell is the structural unit of life
Cells arise only by division from a pre-existing cell
Added since:
Cells contain genetic information (DNA) passed to next cell generation
Prokaryotic vs Eukaryotic Cell Differences
Prokaryotic
bacteria
Genetic material in nucleoid
single circular chromosome
cytoplasm is devoid of membranous structures
Eukaryotic
Plants, animals, protists, fungi
Genetic material is membrane bound (nucleus)
single linear molecule of DNA
More complex (structurally and functionally)
Prokaryotic vs Eukaryotic Cell Similarities
Share an identical genetic language, a common set of metabolic pathways, and many common structural features
Both may be surrounded by a rigid cell wall that protects the cell
Both bounded by plasma membranes of similar construction, serving as selectively permeable barrier
Lecture 3 04/04
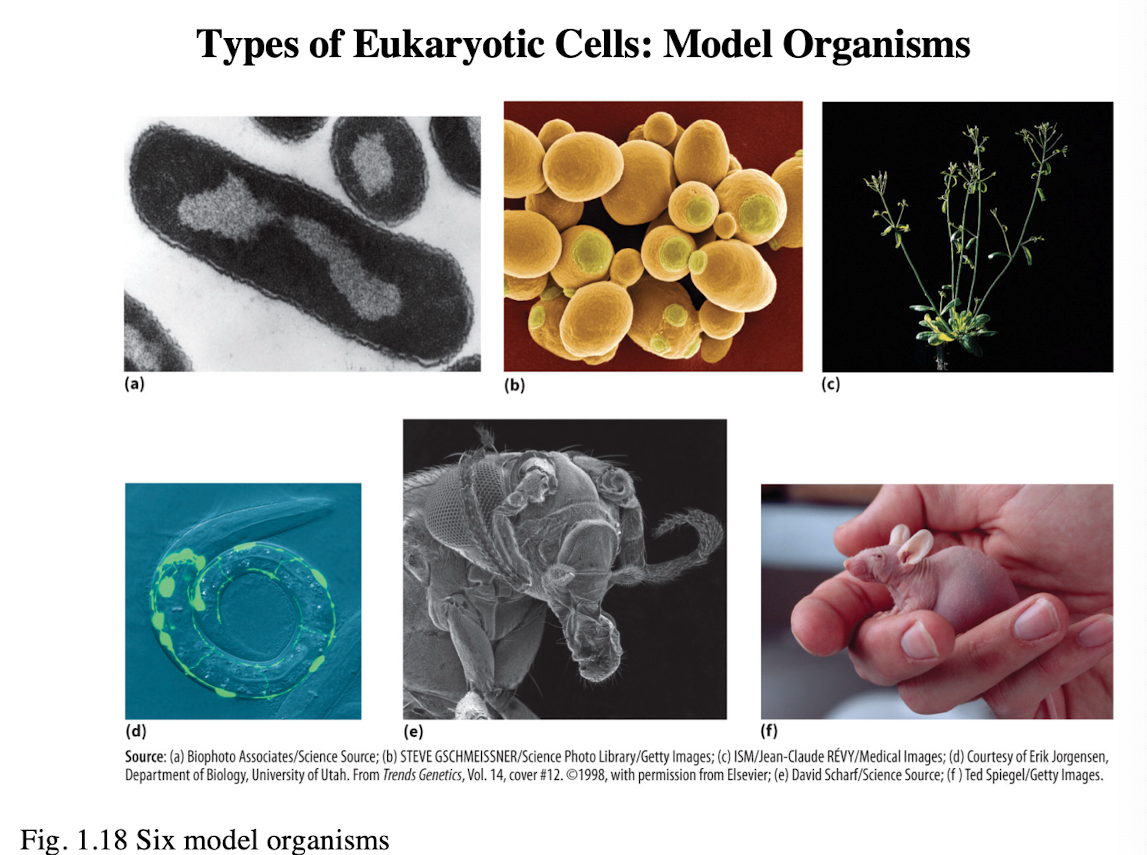
What are the six model organisms for eukaryotic cells
a. Escherichia coli (E. coli/ bacteria)
b. Saccharomyces cerevisiae (Brewer’s yeast)
c. Arabidopsis thaliana (thale cress/ flowering plant)
d. Caenorhabditis elegans (roundworm/ nematode)
e. Drosophila melanogaster (fruit fly)
f. Mus musculus (mouse)
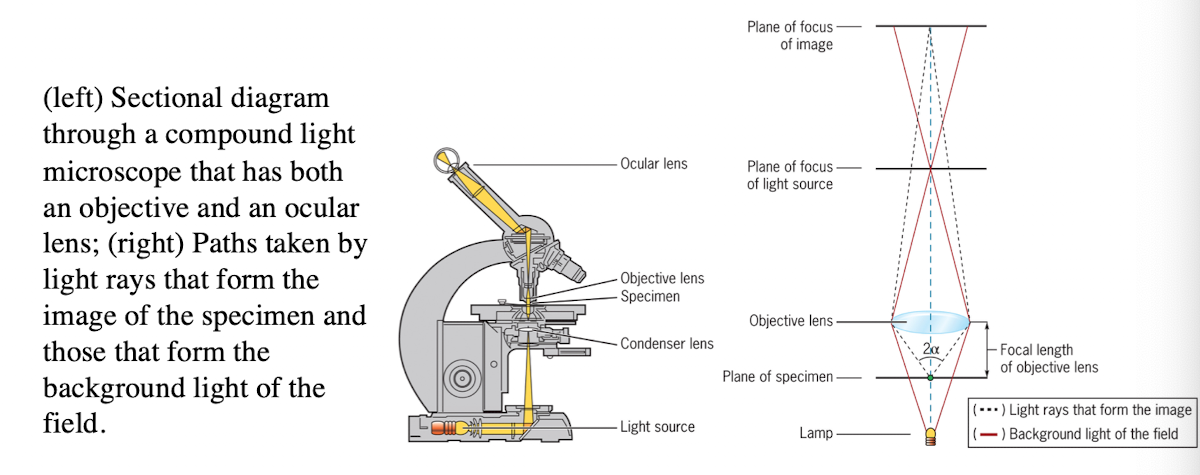
Light microscope
uses refraction of light rays to magnify an object
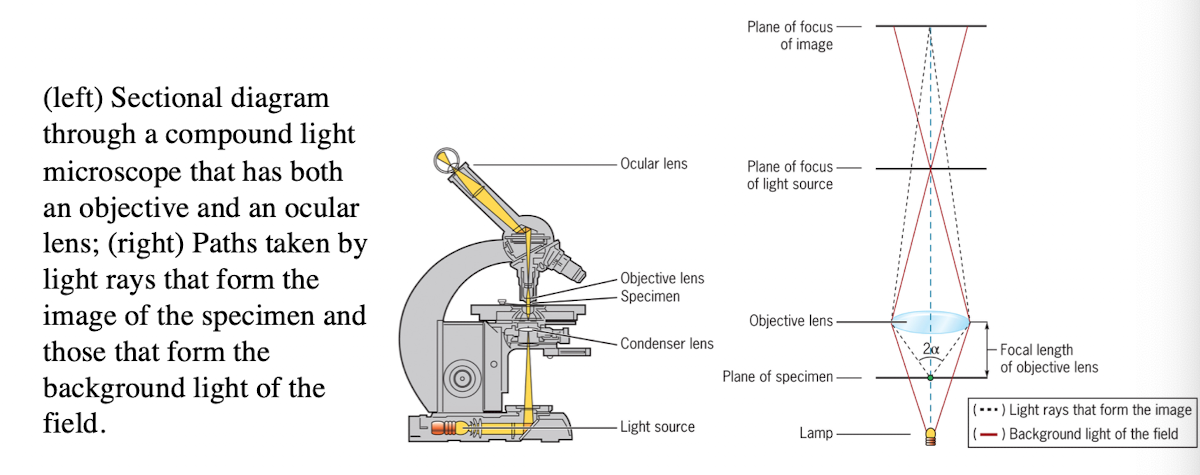
Light Microscope: Condenser lens
Directs light toward the specimen
(more technical description - gathers the diffuse rays from the light source and illuminates the specimen with a small cone of bright light that allows very small parts of the specimen to be seen after magnification)
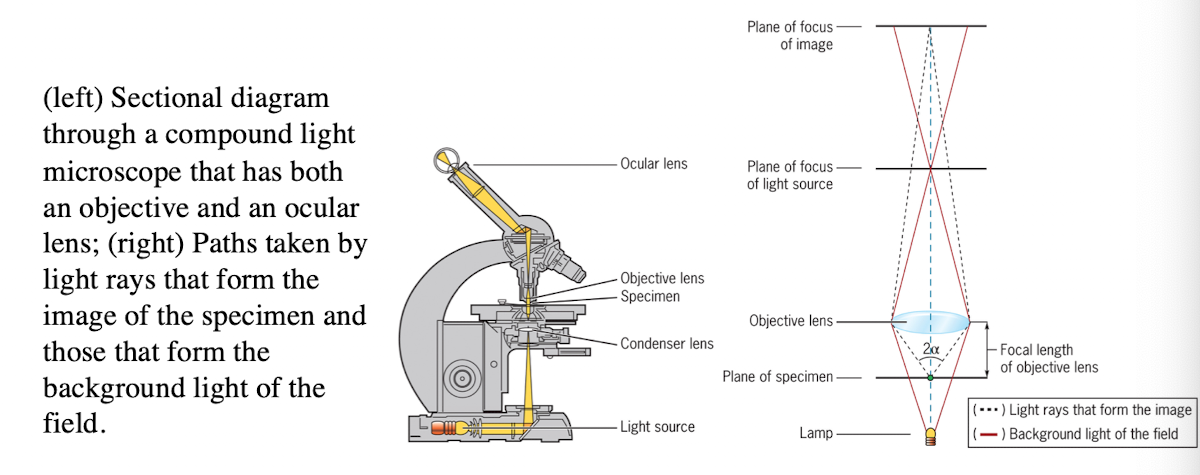
Light Microscope: Objective lens
collects light from the specimen
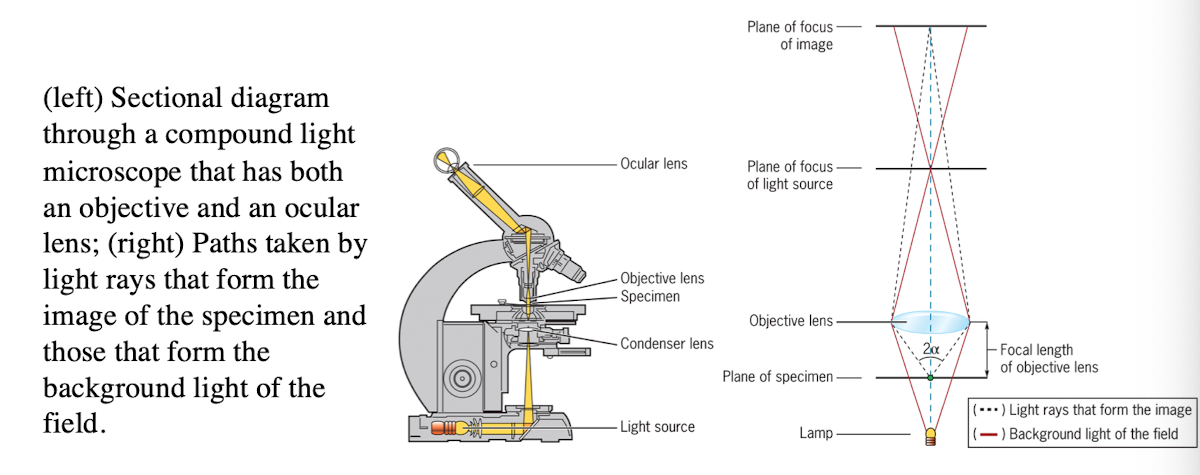
Light Microscope: Ocular Lens
Forms an enlarged, virtual image
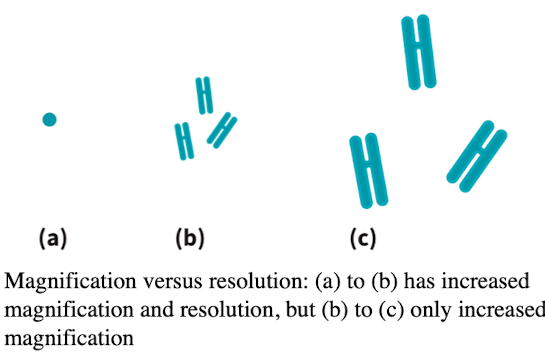
Light microscope: Resolution
Ability to distinguish two points:
Numerical aperture measures the lens light-gathering qualities
Limit of resolution depends on the wavelength of light
Light microscope: Visibility
requires specimen and background to have different refractive indexes
Stains add color and contrast
Suitable for tissue slices and non-living cells
3 ways to study cells
microscope
Genetics: Break the function of genes
difficult to conclude sufficiency
redundancy
Biochemistry
What does it mean when a gene is necessary for a function?
The function cannot happen without that gene. If you knock it out and the function is lost, it’s necessary
How do you experimentally test if a gene is necessary?
Use a knockout experiment by removing or disabling the gene and observe if the function is lost
What does it mean when a gene is sufficient for a function?
the gene alone can cause the function to happen, even in a system where it usually doesn’t occur
How do you experimentally test if a gene is sufficient?
Add the gene or protein to a new system that normally doesn't do the function and it now does, the gene is sufficient
Limit of Resolution Equation
D = (0.61*λ) / (n*sinø)
n*sinø = numerical aperture (N.A)
sinø = maximum amount of light collected (1.0)
λ = wavelength
n = refractive index (R.I)
air =1.0
oil = 1.5
H2O = 1.3

Limit of Resolution for:
Naked Eye
Light Microscopy
TEM
Fluorescence Microscopy
Naked eye = ~200 µm
Light microscopy = 0.2 µm
TEM = 2nm
FM = 0.2 µm (still a light microscopy)
Lecture 4 04/07
Transmission Electron Microscopy (TEM)
Uses electrons instead of light → very short wavelength → higher resolution (2nm)
Can see membranes and very fine structures
TEM limitations
requires extensive sample prep
not commonly used unless necessary
needs training; typically done through collaboration
Transmission Electron Microscopy:
Limit of resolution
Magnification
What helps provide contrast
What kind of images may be taken
Limit of resolution = 10-15 Å
Magnification enhanced 105 times due to increased resolution
heavy metals stains provide contrast
photographic emulsion or digital images may be taken
Fluorescence Microscopy
Useful for in live-cell imaging
Uses fluorophores that absorb light (excitation) and emit longer wavelength light (emission)
Works best with flat or thin cells/tissues
Limitation: blurred images in thick staples due to multiple focal planes and out-of-focus light
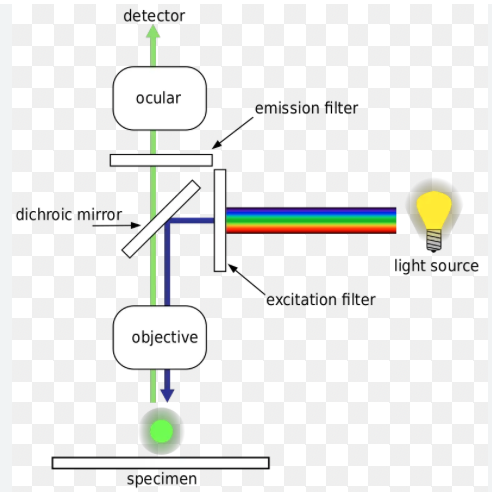
Fluorescence Microscopy 3 parts to setup
First barrier filter
Beam-splitting mirror
Second barrier filter
Fluorescence Microscopy: 1) First Barrier Filter (light & wavelength)
lets through only blue light with a wavelength between 450-490nm
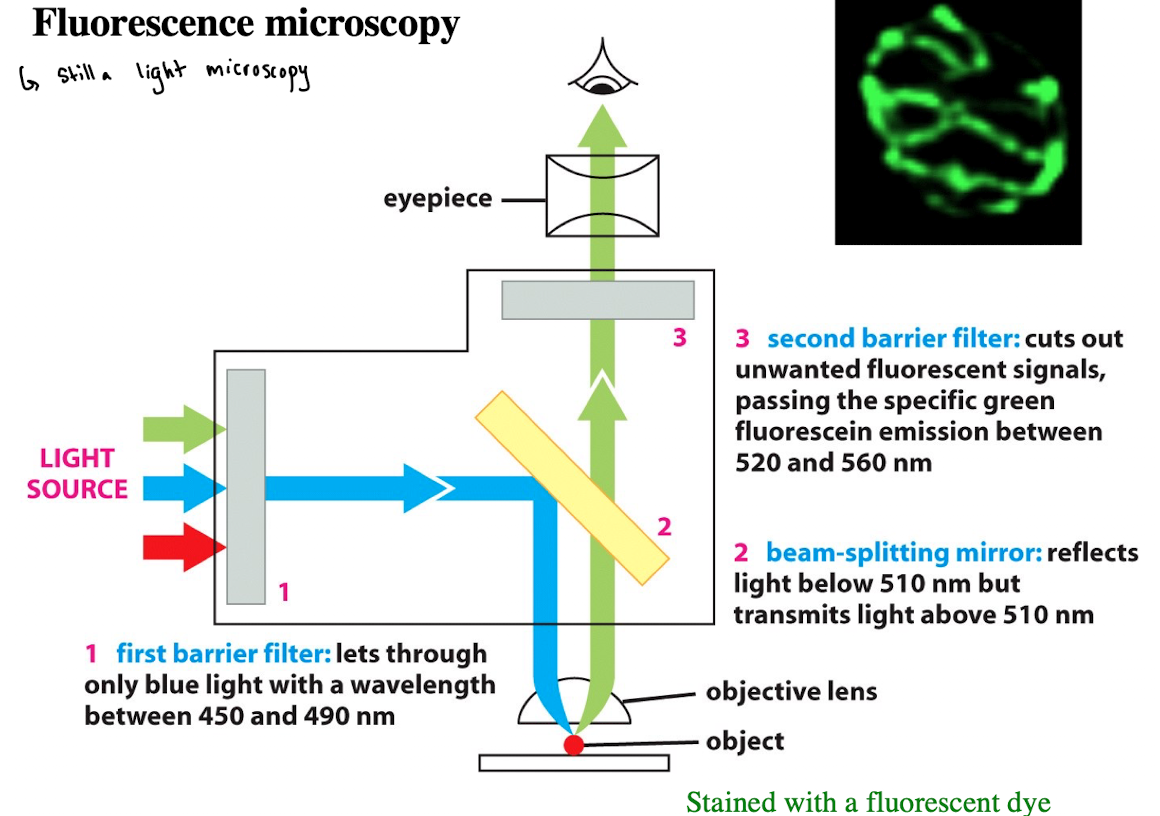
Fluorescence Microscopy: 2) Beam-Splitting Mirror
Reflects light below 510 nm but transmits light above 510nm
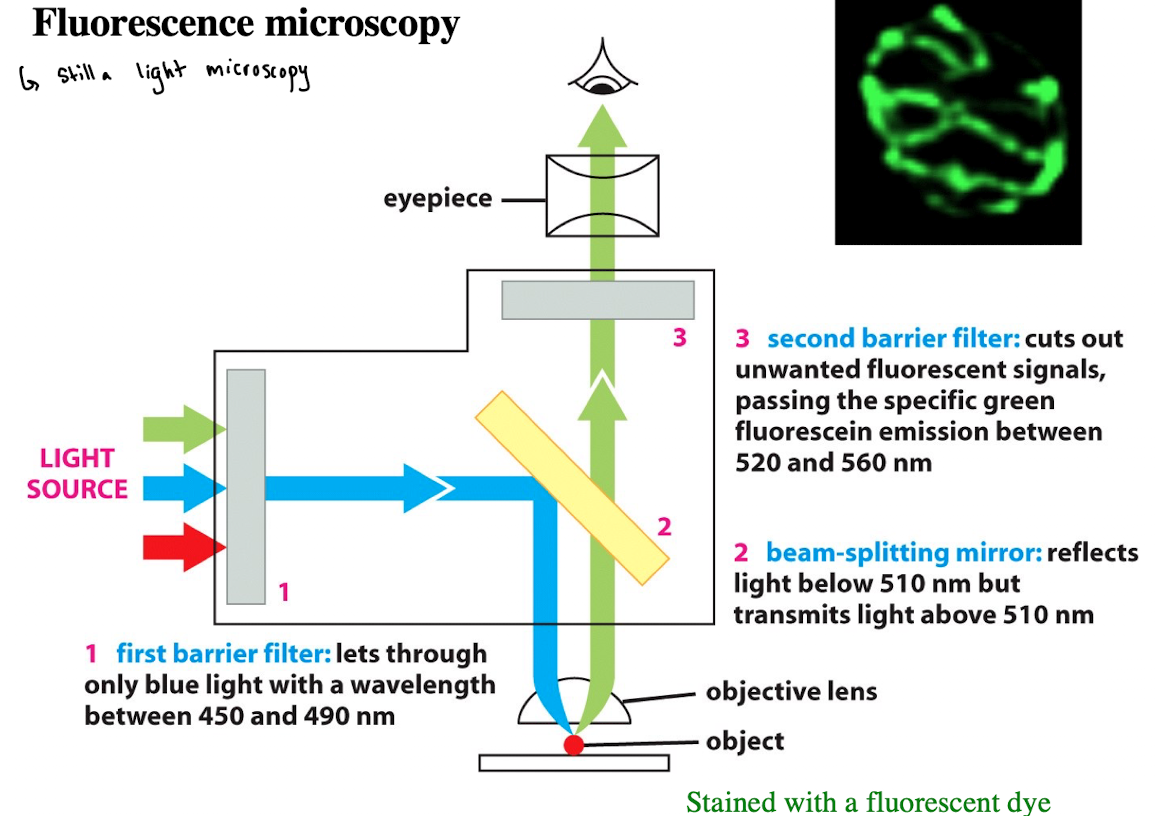
Fluorescence Microscopy: 3) Second Barrier Filter
Cuts out unwanted fluorescent signals, passing the specific green fluorescein emission between 520- 560 nm
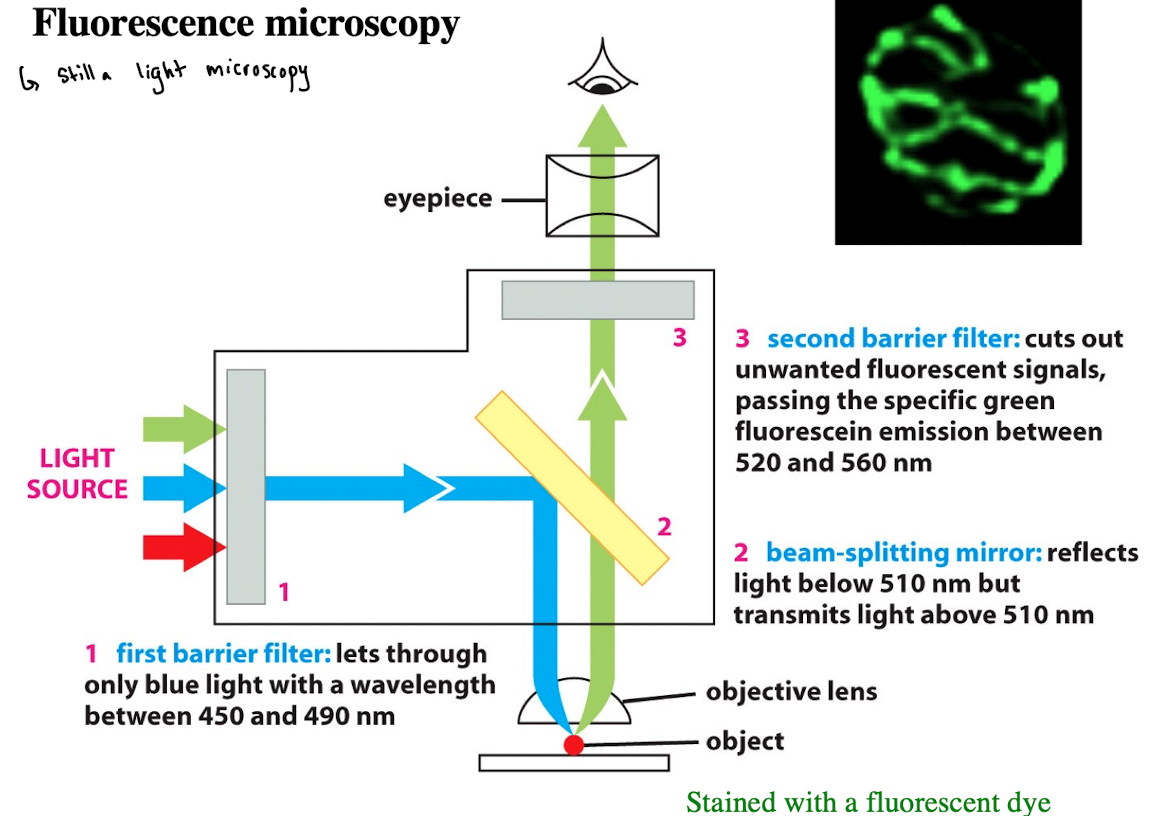
Fluorophores
Molecule that absorb photons of a specific wavelength and release a portion of the energy in longer wavelengths
Fluorescence Components
Fluorophores: Used to visualize otherwise transparent cells/structures
Fluorescein: excite w/blue light (450-490 nm) → emit green (520-580 nm)
Rhodamine: excite w/green light (535 nm)→ emit red (610 nm)
Fluorescence Microscopy: Common Dyes
DAPI: stains chromatin → nucleus appears blue
Mitotracker Red: stains mitochondria
Other dyes based on target and desired emission color
Immunofluorescence Microscopy
Used when specific dyes don’t exist for certain proteins
A technique that uses fluorescently labeled antibodies to visualize specific molecules. Relies on the principle that antibodies bind specifically to their target antigens
Fluorochrome-conjugated antibodies are used to locate specific cellular structures
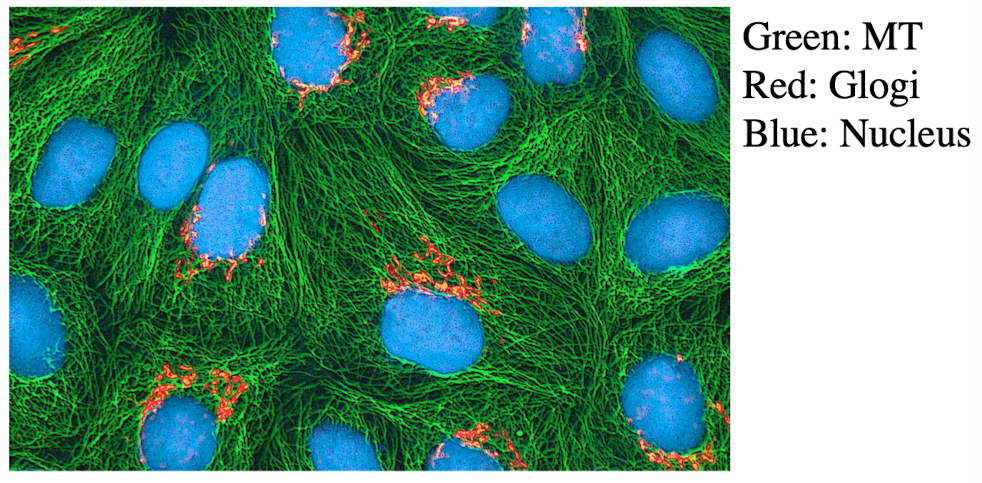
Direct Immunofluorescence
Antibody binds target protein
Antibody is directly conjugated to fluorophore
Simple but weaker signal
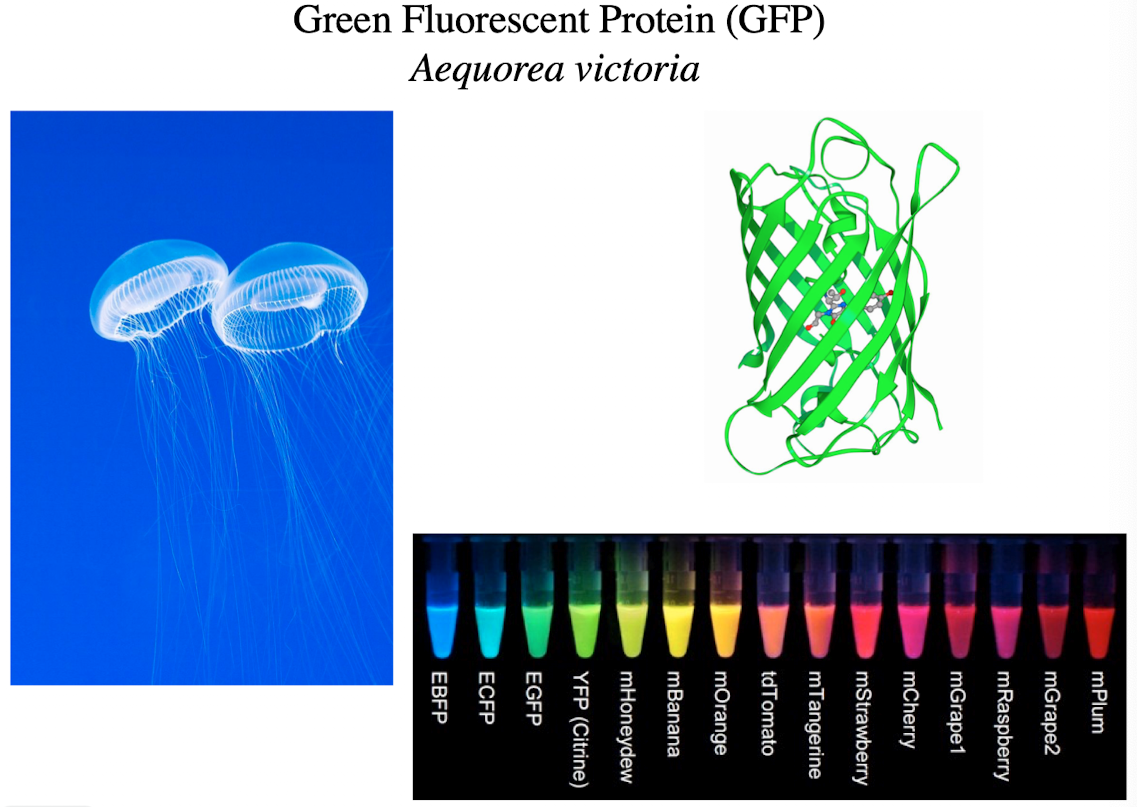
Green Fluorescent Protein (GFP)
Can be recombined with genes of interest in model organisms
Expressed with the host gene of interest
Used to follow a gene of interest
Indirect Immunofluorescence
Primary antibody binds proteins
Secondary antibody (w/fluorophore) binds primary antibody and carries fluorescent dye
Signal Amplification: multiple secondaries can bind one primary
Immunofluorescence Microscopy Primary/Secondary Antibody Example
Labeling Golgi with an antibody to a known Golgi protein
Use DAPi (nucleus), rhodamine (Golgi), fluorescein (tubulin) for multi-color labeling
Primary antibody has to be very specific to mitochondria
Lecture 5 04/09
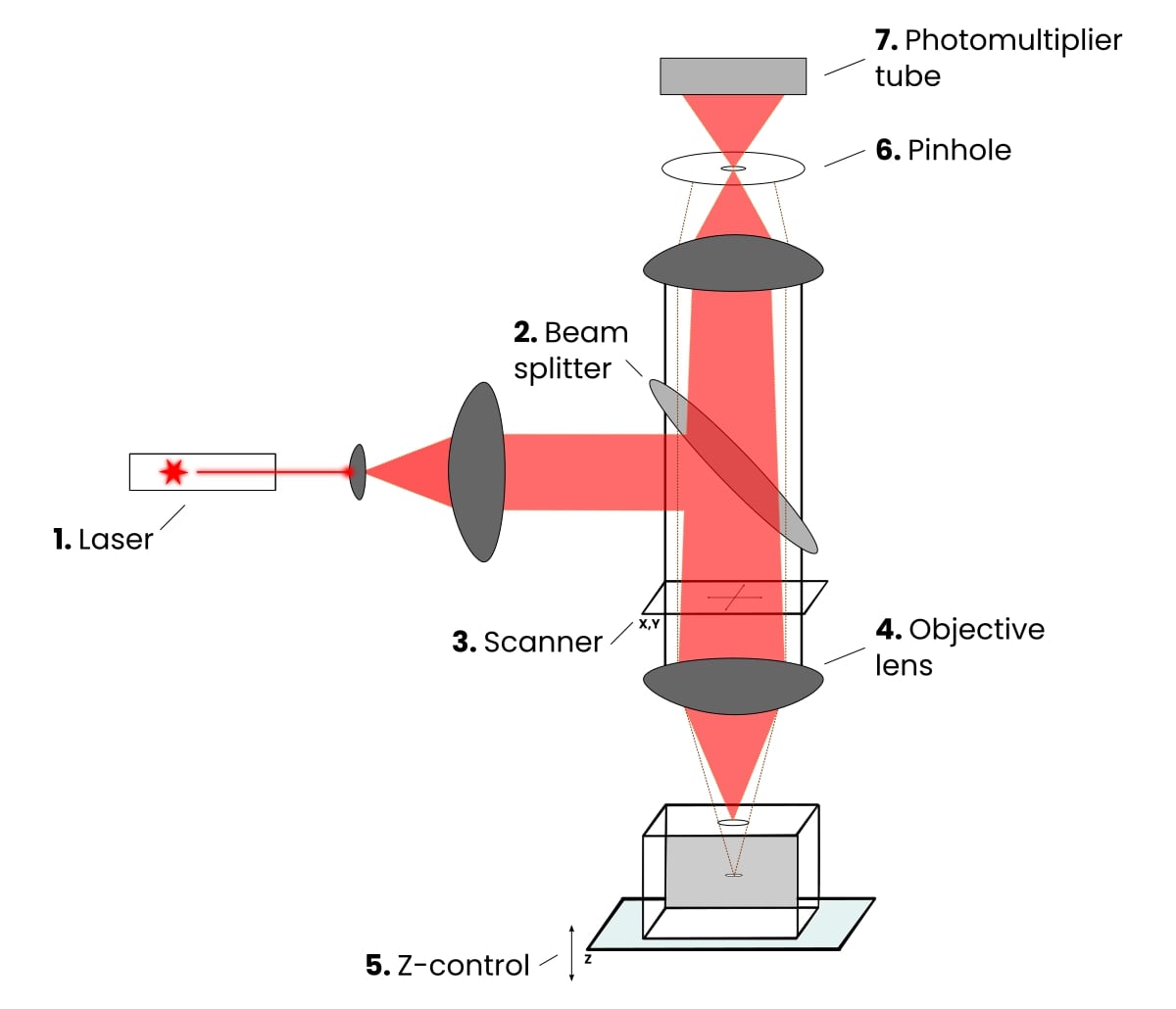
Laser Scanning Confocal Microscopy
Produces an image of a thin plane located within a much thicker specimen
Solves thickness/image clarity issue in fluorescence microscopy
a laser beam is used to examine planes at different depth in a specimen
computer can compile images for 3-D modeling
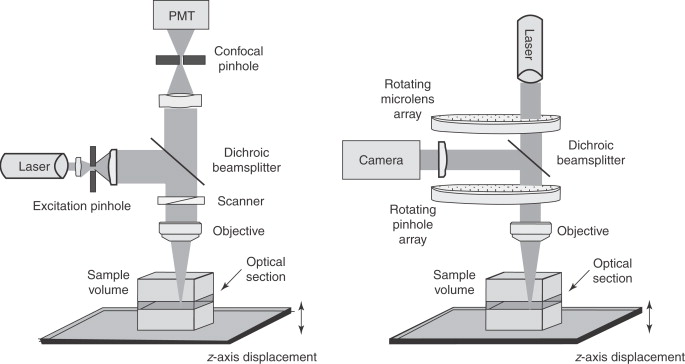
Key advantages and disadvantage of Confocal Laser Scanning Microscopy
Advantages:
pinpoint illumination using lasers
better control of light wavelength via illuminating aperture
pinhole instead of barrier filter to precisely collect light
allows optical sectioning (taking images at multiple focal planes
Uses photomultiplier to combine signals into crisp 3D images
Disadvantage:
Very expensive ($600k-1M), usually available through core facilities
Live Cell Imaging
A technique that allows researchers to observe living cells (cell activity) over time
Avoids traditional dyes (which kills cells)
Uses YFP, RFP, GFP
Proteins are fused genetically (gene A + GFP gene → fusion protein)
GFP Excitation and Emission Wavelengths in Live Cell Imaging
Excitation
375nm - 480nm
Emission
510 nm
Yellow Fluorescent Protein (YFP) Excitation and Emission Wavelengths in Live Cell Imaging
Excitation
513 nm
Emission
527 nm
Red Fluorescent Protein (RFP) Excitation and Emission Wavelengths in Live Cell Imaging
Excitation
558 nm
Emission
583 nm
Fluorescent Protein Spectra Problems and Solutions
Problem: Spectral overlap; too similar to be used together (GFP and YFP)
Solution: Use fluorophores with non-overlapping emission spectra (GFP and RFP)
GFP may affect…
localization
function of the protein
How would you confirm GFP fusion is accurately representing the original protein?
Knockout gene A, reintroduce A-GFP, check if function is restored (valid)
Change GFP position (N-terminus vs. C-terminus), if only one works then position matters
Biochemical methods to confirm location (subcellular fractionation)
Differential Centrifugation
Allows the isolation of particular organelles in bulk quantity
Isolated organelles can be used in cell-free systems to study cellular activities
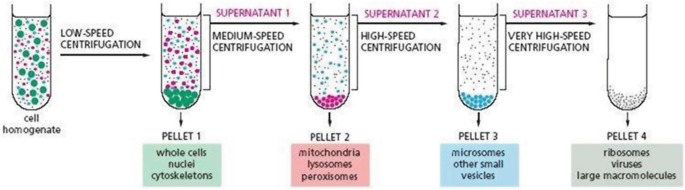
Pellet
Heavier components settle to the bottom depending on speed
Supernatant
Lighter materials or liquid above the pellet
Subcellular Fractionation (Differential Centrifugation) Spin Speed (pellet and supernatant)
600g
Pellets: Nucleus
Supernatant: Everything else
15,000g
Pellet: mitochondria, lysosomes, peroxisomes
Supernatant: ER, Golgi, Plasma Membrane, Cytosol
100,000g
Pellet: membranes (ER, Golgi, plasma membrane)
Supernatant: Cytosol
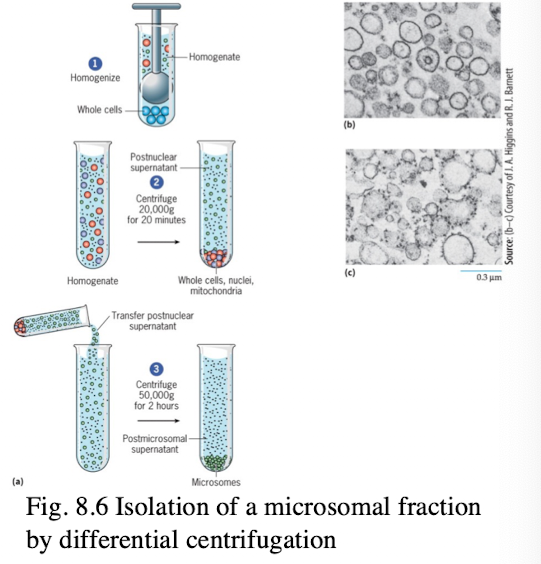
Subcellular Fractionation Process
cell homogenization fragments cytoplasmic membranes
Vesicles derived from the end-membrane system from similar sized vesicles (microsomes)
microsomal fraction can be fractionated into smooth and rough fractions
Once isolated, the biochemical composition of various lipid and protein fractions can be determined
Sucrose Density Centrifugation
Organelles separate into bands at their equilibrium density in sucrose gradient
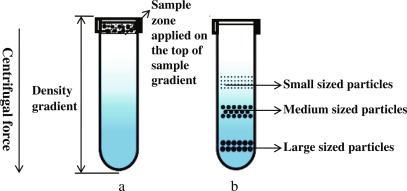
Sucrose Density Gradient Centrifugation Involves
solubilizing membranes using mild detergents
layering on a 0.5-5M sucrose gradient
Spinning at high speed (~65,000)
Sucrose Density Gradient Centrifugation Bands
ER - Upper Layers
Golgi - Middle
Plasma Membrane - lower
Mitochondria - Even lower
Lysosomes - Around same as mitochondria
How to detect proteins in Isolated Fractions
SDS-PAGE (sodium dodecyl sulfate, polyacrylamide gel electrophoresis)
SDS denatures and gives proteins uniform negative charge (unfolds into linear protein)
Proteins separated by size only
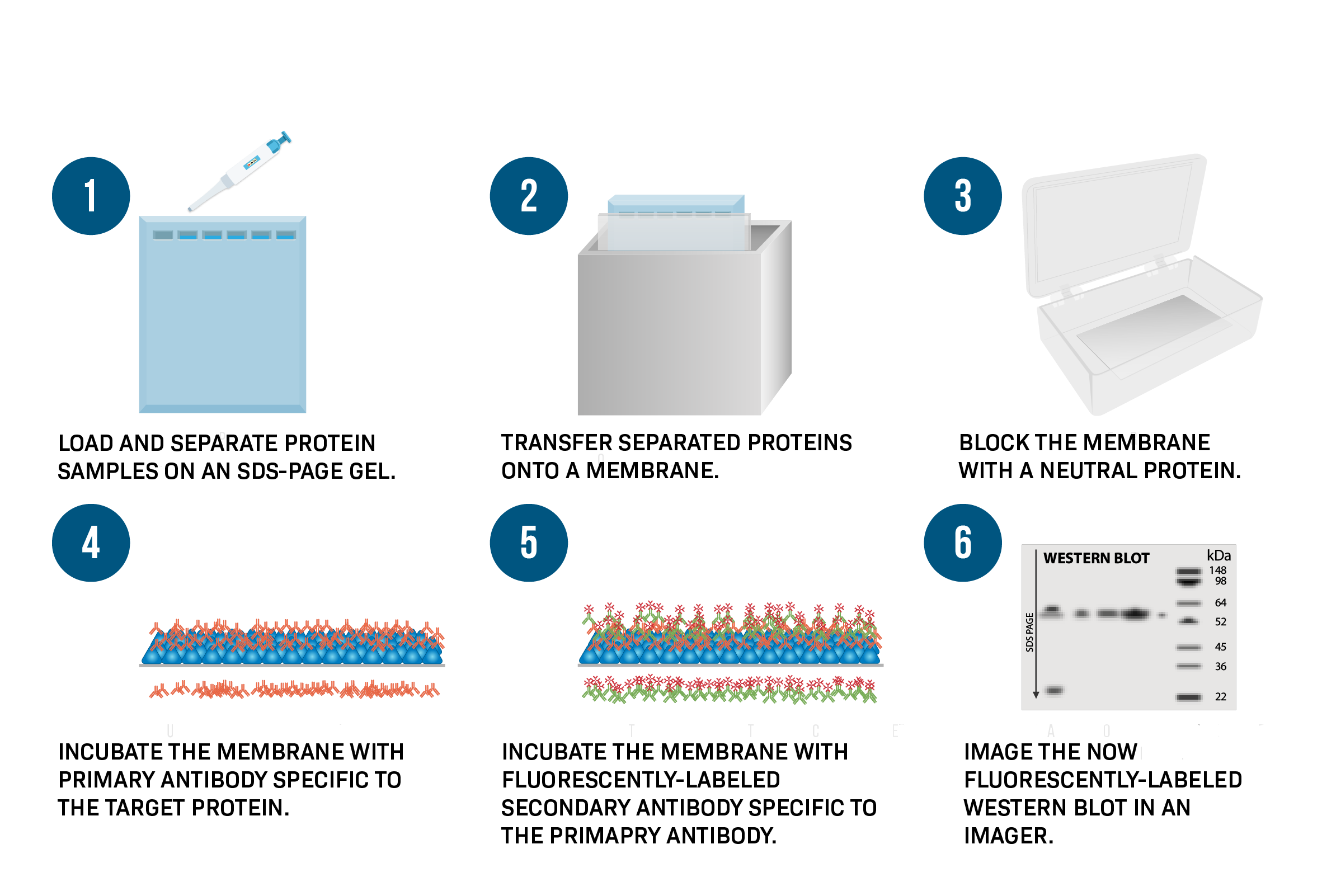
SDS-PAGE Steps ( Part of Western Blotting Process)
Run SDS-PAGE
transfer proteins to a membrane
Incubate with primary antibody (specific to target protein)
Detect with secondary antibody conjugated to HRP (enzyme)
Add substrate → produces color/light at protein band

Where in the cell does protein C reside?
A. Cytosol
B. Golgi
C. ER
D. Nucleus
E. Mitochondria
E. Mitochondria

Where in the cell does protein A reside?
A. Cytosol
B. Golgi
C. None of the above
D. Nucleus
E. Mitochondria
B. Golgi

Where in the cell does protein B reside?
A. Cytoplasm
B. Golgi
C. None of the above
D. Nucleus
E. Mitochondria
D. Nucleus
Positive and Negative Control for Protein localization
Positive Control:
Known nuclear protein detected to confirm successful nuclear isolation
Negative Control:
Known cytoplasmic protein should not be detected in nuclear fraction
If detected → contamination of nucleus w/cytoplasmic material
Lecture 7 04/14
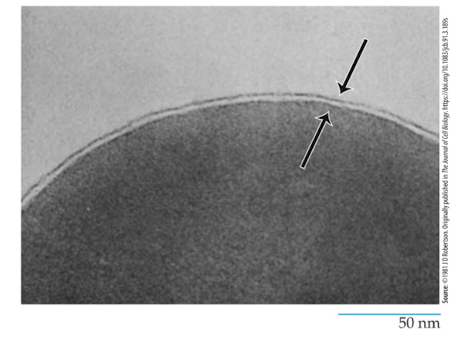
Plasma Membrane
the outer boundary of the cell that separates it from the world
is a thin, fragile structure about 5-10nm thick (which is why we wouldn’t use light microscopy-0.2um)
Need electron microscope to examine
All membranes examined closely from plants, animals or microorganisms have the same ultrastructure
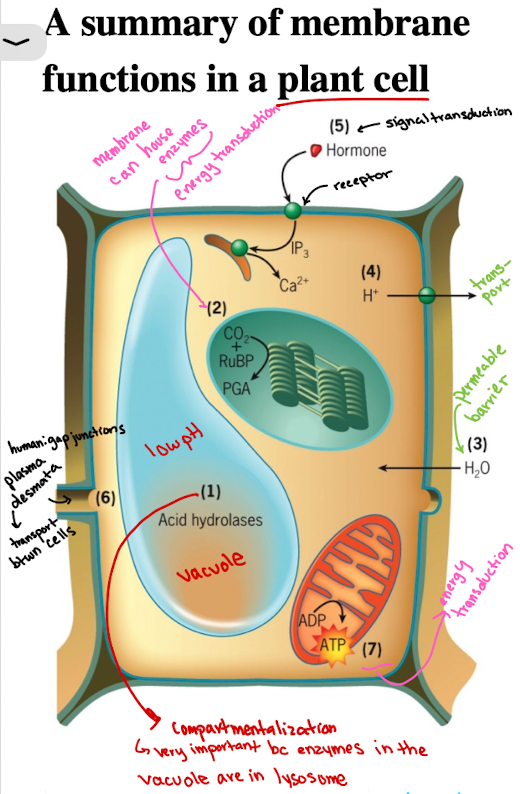
Functions of Cell Membranes in Plant Cell: Compartmentalization Example
Vacuoles/lysosomes contain acid hydrolases functioning at low pH (sequestered)
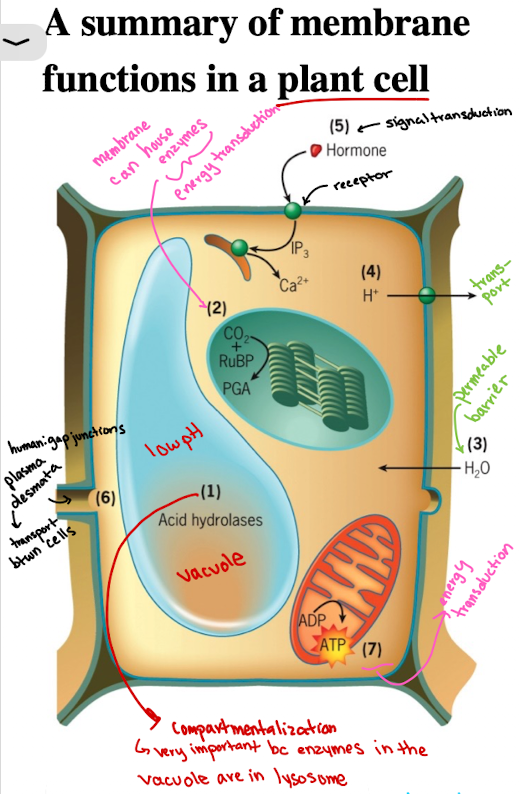
Functions of Cell Membranes in Plant Cell: Enzyme Housing (localization)
CO2 fixation catalyzed by enzyme associated with outer surface of thylakoid membranes of the chloroplasts (energy production)
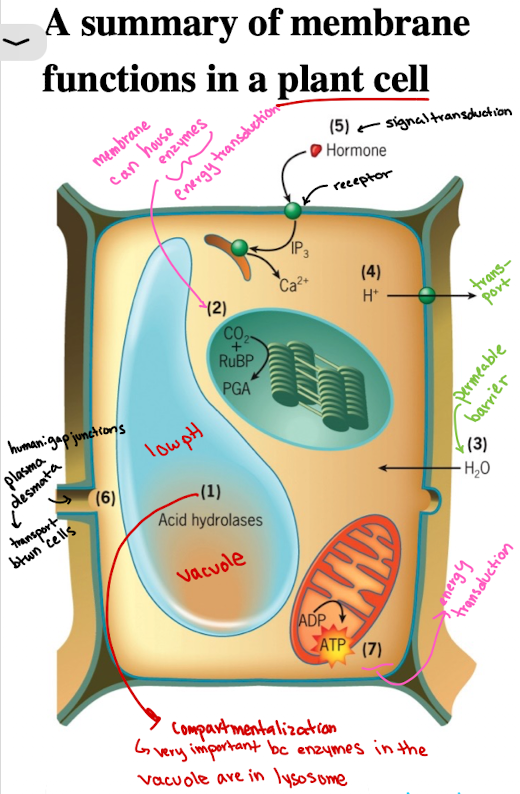
Functions of Cell Membranes in Plant Cell: Selective Permeability/Barrier
Water molecules are able to penetrate rapidly through plasma membrane, causing pressure against cell wall
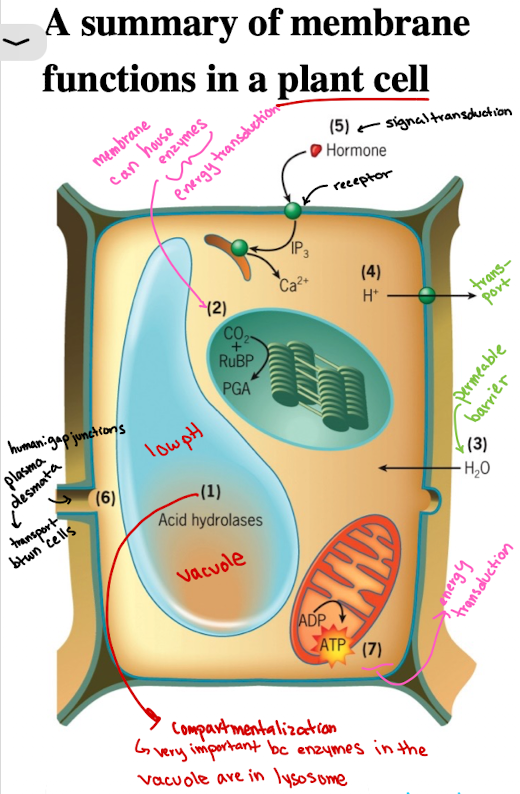
Functions of Cell Membranes in Plant Cell: Transport
Membrane transporters import/export ions, solutes (ex. H+) into extracellular space
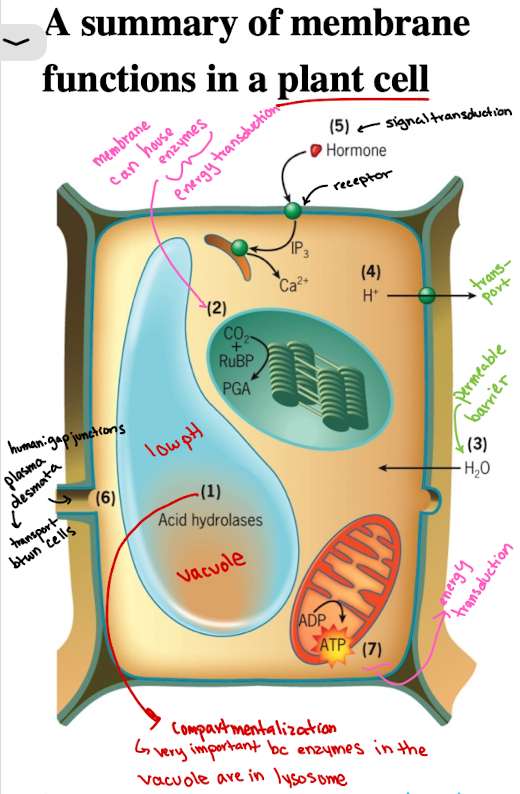
Functions of Cell Membranes in Plant Cells: Signal Transduction
Receptors (ex. growth hormones) on membranes initiate intracellular responses, as a result of response to external stimuli

Functions of Cell Membranes in Plant Cells: Cell-cell communication
Plants: Plasmodesmata
materials move directly from cytoplasm of one cell to its neighbors
Animals: gap junctions
Ernest Overton - 1895
Showed that membrane is semipermeable
More soluble the dye, the better it enters the cell
Tested on root hair cells where the more lipid-soluble a solute was, the more rapidly it entered
Membrane is made of of lipids
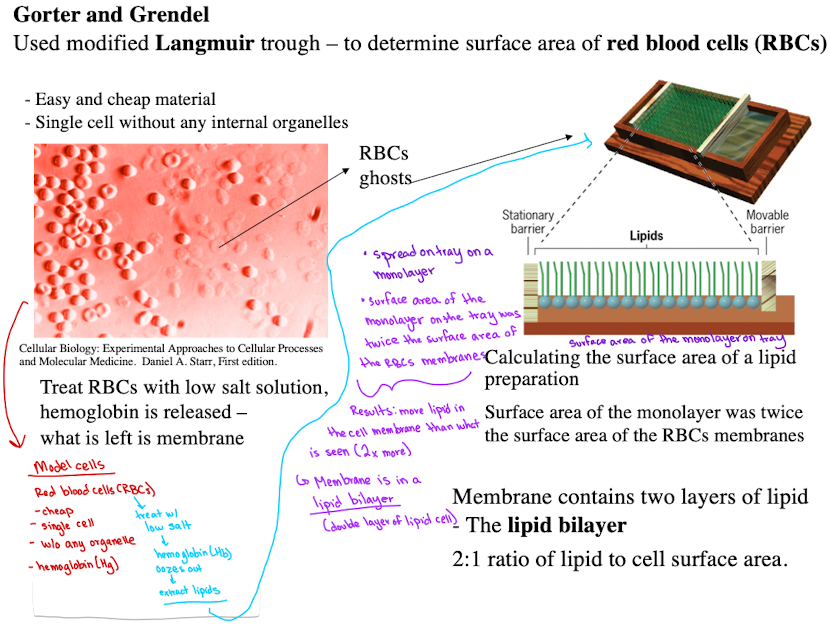
Gorter and Grendel - 1925
Used RBCs to extract lipids
Found lipid bilayer via monolayer spreading experiments through modified Langmuir trough.
Calculated surface area of RBC membranes, found 2:1 ratio of lipid to cell surface area
Membrane lipids are ____ which contain both hydrophilic and hydrophobic regions. Name three main lipid types.
amphipathic
Phospholipids (names based on polar head)
Sphingolipids
Cholesterol
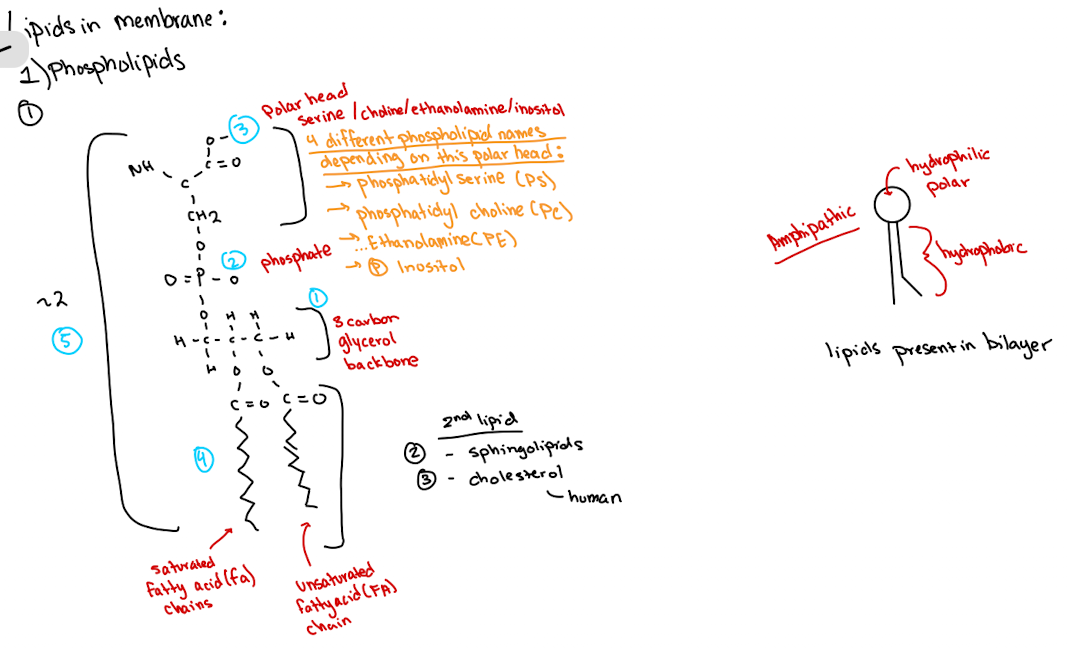
What is membrane fluidity dependent on?
Saturated vs. unsaturated fatty acids
The more unsaturated fatty acid you have, membrane will be more fluid
The less unsaturated fatty acid you have, membrane will be less fluid
Other factors include temperature, cholesterol
How would lipids move between bilayers
Flippase - energetically impossible without it
Cell membrane would likely be in liquid, crystalline state at warmer temps around 37ºC and rotate axis/move laterally through plane
Danielli A Davison 1935
discovered membranes also contain proteins
Experimented by observing oil droplets from fish eggs absorbing proteins
Frye and Edinin 1970
Discovered proteins are mobile within the bilayer
Experimented by fusing human and mouse cell
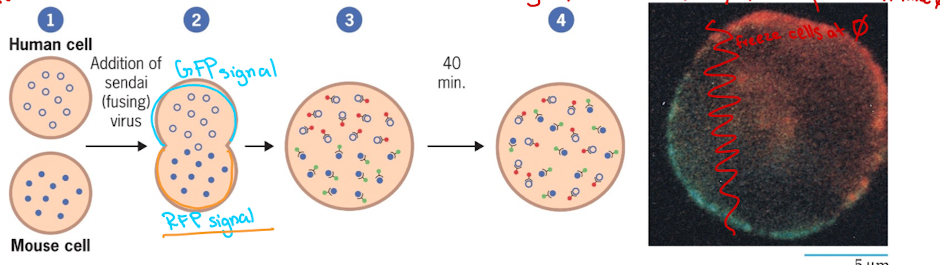
Lecture 8 04/16
Singer & Nicolas (1972)
Discovered Fluid Mosaic Model
Membrane is fluid: both lipids and proteins can move (mobile)
Membrane is a mosaic: contains different lipids and proteins interspersed
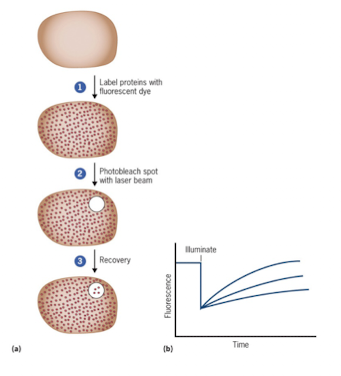
FRAP (Fluorescence Recovery After Photobleaching)
Used when you want to know whether protein is static or mobile on membrane
Study lateral movement of proteins within the membrane
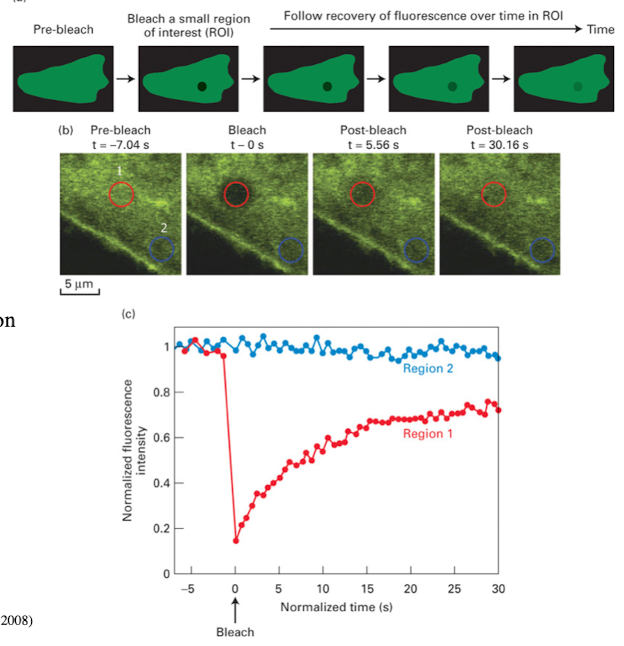
FRAP Steps
Fuse protein to GFP
Bleach a membrane region with laser (confocal microscopy)
If fluorescence recovers → protein is mobile
If it doesn’t recover → protein is static
List the Membrane Proteins
Multipass Membrane
Extracellular peripheral membrane protein
Singlepass Membrane
Intracellular peripheral membrane protein
Lipid-Anchored Proteins-
Acetyl
Prenylated
GPI
Beta Barrel Proteins
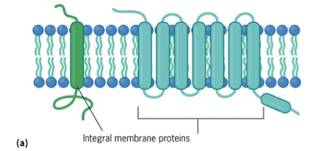
Integral Membrane Proteins (Transmembrane)
Typically contain one or more transmembrane helices
Function as receptors that bind ligands, channels, or transporters to move ions/solutes across the membrane
Amphipathic
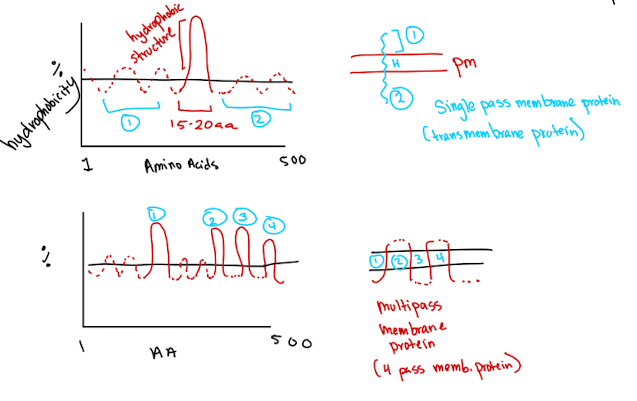
Single pass: One hydrophobic region → crosses membrane once
Multi-pass: Multiple hydrophobic regions → crosses multiple times (beta barrel proteins)
Hydrophobicity Plot
Used to identify hydrophobic alpa-helices (15-20 amino acids) in integral proteins
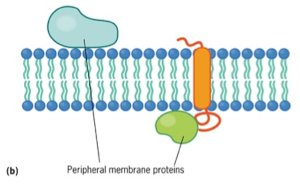
Peripheral Membrane Proteins
Noncovalently (loosely/weak electrostatic) bonded to polar head of lipid bilayer and/or integral membrane protein
Dynamic relationship with membrane, being recruited or released as needed
Lipid Anchored Proteins: What they do and lipid names
Covalently bonded to a lipid group that resides within the membrane
Myristoylation (or Acetylation)
Prenylation
GPI Anchored Protein
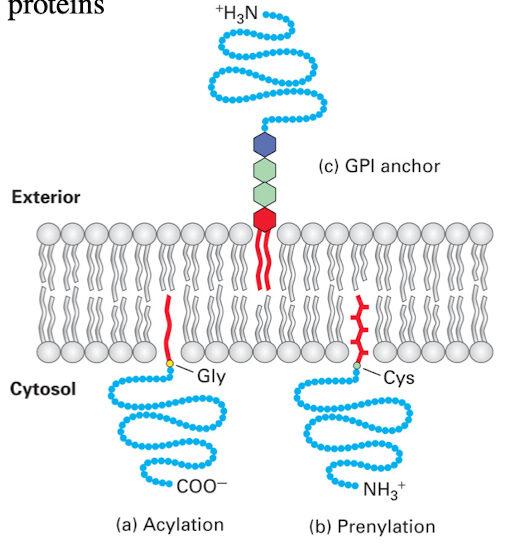
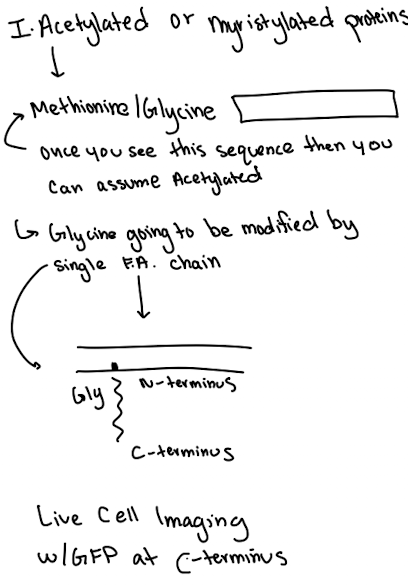
Lipid Anchored Protein: Myristoylation Example
Modification at glycine/methionine
Anchors protein to cytoplasmic side
GFP must be fused to C-terminus to avoid blocking anchoring
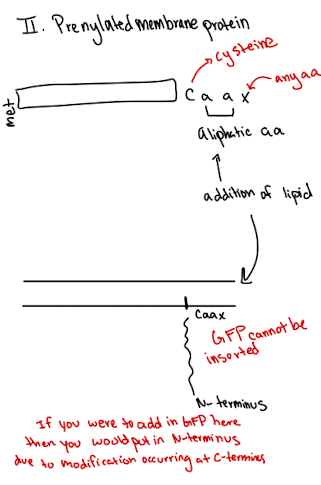
Lipid Anchored Protein: Prenylated Example
Modification at cysteine near the C-terminus (CAAX motif)
Anchors protein to cytoplasmic side
GFP must be fused to N-terminus
Lipid Anchored Protein: GPI Example
Added in Golgi
Anchors protein to Outer (extracellular) leaflet of membrane
No transmembrane domain required

Functional Importance of Membrane Proteins
Approx. 30% of human proteins are membrane-associated
Difficult to study due to solubility and extraction issues
Critical for signaling, transport, structure