Molecular techniques
1/23
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
24 Terms
Nucleic acid hybridisation
Radioactively labelled complementary ssDNA or ssRNAs form a duplex
biotin molecule is used to label non-radioactively
Detected via x-ray and assays
Northern blotting
Identify specific RNA sequences in a sample by size and sequence.
Separated by size in an agarose gel. Nucleic acids (-ve) move to negative pole.
Complementary target sequences stick to the radioactive probe
induces chaperon proteins by promoting protein refolding and assembly
however - not refined, needs lots of RNA
in situ hydridiation
Reveals the presence and location of specific nucleic acids in fixed tissues or cells.
Able to detect rare RNAS
However - less sensitive than PCR, and more complex
Gene expression changes
Dynamic - varies with development, environment, disease
Developmental - embryonic development
Environment - circadian rhythm
Disease conditions - can alter gene expression patterns, affecting cellular responses and behaviors.
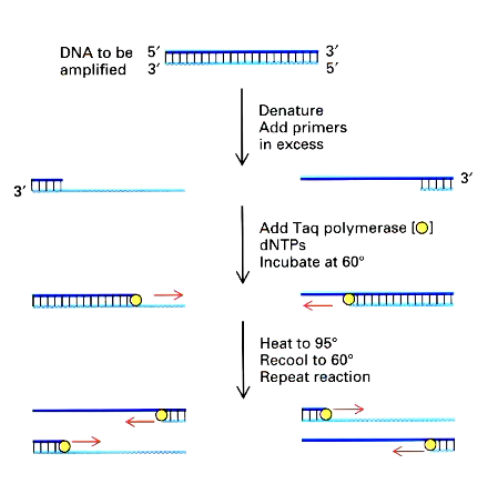
PCR and qPCR
Amplifies specific DNA sequences with primers
RNA is converted into complementary DNA (RT)
DNA amplified with specific primer to the target gene
Fluorescent dyes track amplification
highly sensitive and fast
Not quantitative
What is so special about PCR
It can detect tiny amounts of DNA and RNA
Allows researchers to determine gene expression with high precision
Laser capture microdissection
Adv - precise extraction of specific cells/tissues using a laser. Very sensitive with PCR
can see intestinal villi
Disadv - limited amount of sample can be obtained at once
Microarrays
Analyse large sets of labelled genes at once. via specific probes .
Gene chips use photolithography to synthesise specific DNA sequences onto a silicon chip. Measures t
Scan across the whole chip: more transcript = more cDNA = higher intensity of the fluorescence
Application of microarrays
Gene profiling - Patients with different genetic expression patterns respond differently to treatment.
Large-scale expression profiling helped identify distinct subtypes of lymphoma.
Limitations - noisy, and subjective due to variability in intensity
Sanger sequencing
chain terminators that block copying of DNA using dideoxynucleotides.
Gel electrophoresis determines sequence order by adding radioactivity to nucleotides
can read 10,000 bases a day.
Slow and labour intensive
High-throughput sequencing
Illumina sequencing - simultaneous sequencing of millions of DNA fragments,
Ion torrent & pyrosequencing -detects nucleotide incorporation based on pH
Nanopore - pass ssDNA through membranes. bases detected based on their affect on ionic current flow
What is a major application of single-cell RNA-Seq?
Identifying tumours, understanding developmental gene expression, and uncovering disease pathways
Summary of techniques
Technique | Use | Strengths | Limitations |
PCR | Detects specific DNA/RNA sequences | High sensitivity, simple | Not quantitative |
qPCR | Measures gene expression levels | Quantitative, real-time tracking | Limited to known targets |
Microarrays | Large-scale gene expression profiling | Simultaneous analysis of thousands of genes | Fluorescence variability, limited to known sequences |
RNA-Seq | Whole transcriptome sequencing | Digital quantification, novel transcript detection | Expensive, requires computational analysis |
Post-translational modifications
Alter protein activity, stability, and location
ex. cleavage, GPI linkages phosphorylation, ubiquitination, methylation, acetylation
Cannot be studied by DNA/RNA analysis bc they aren’t encoded in the genome
What must be studied to understand protein function?
Location, interactions, function, and quantity.
Protein analysis methods
Dyes to stain proteins
Antibodies to recognise target proteins (immunofluorescence and western blots)
Electron microscopy reveals protein organisation
biochemical purification (isolated via synaptosomes, endosomes, protein complexes
SDS-PAGE Gel electrophoresis
RNA separated by size in polyacrylamide gel
Large proteins move slower than small
SDS page denatured proteins and coats them with a negative charge to strandardise protein charge
2D gel electrophoresis
proteins placed in pH gradient and move to their isoelectric point (no net charge) and stop.
Proteins then separate by size in SDS-PAGE
Each spot in the gel represents a protein. Each PTM appears as a different spot for the same protein
Types of antibodies
Feature | Polyclonal Antibodies | Monoclonal Antibodies |
|---|---|---|
Source | Derived from multiple B-cell clones | Derived from a single B-cell clone fused with a cancer cell |
Target Recognition | Recognize multiple epitopes (parts) of the same protein | Recognize one specific epitope on the protein |
Production | Easier and faster to produce | More complex and time-consuming to produce |
Specificity | Less specific | Highly specific |
Batch Consistency | Varies between batches | High consistency across batches |
Applications | General detection, research use | Diagnostic tools, therapeutic drugs, research |
Western blotting
Proteins separated by SDS-PAGE
transferred to membrane for Ab incubation
primary Ab binds to target protein, secondary Ab binds to Fc region
Fluorescence/enzymes are used to detect
Specific Ab’s can detect phosphorylated proteins and ubiquitinated proteins
Protein-protein interactions
Use Abs to purify proteins and their partner
Ab binds to target proteins in mix
Ab is captured on beads, and complex is isolated
Western blot detects interacting proteins
Maps protein interaction networks
Mass Spectrometry
Identifies and quantifies proteins my measuring mass-to-charge ratio
digest proteins into peptides via trypsin
Analyse peptide masses via spectrometer
Compare to protein database to identify protein
used in proteomic analysis, tracking ptms, and studying protein interactions
What do chemical cross-linking and mass spectrometry reveal together
Which protein domains interact
How protein assemble into complexes
Used to build protein interaction network
Summary
SDS-PAGE → Separates proteins by size.
2D-PAGE → Separates proteins by charge + size.
Western Blot → Detects specific proteins using antibodies.
Co-IP → Identifies protein-protein interactions.
Mass Spectrometry → Identifies proteins and modifications without assumptions.
Immunofluorescence → Visualizes protein localization.
Cross-linking + Mass Spec → Reveals protein complex structure.