Section 2 Lectures 4-6
5.0(1)
Card Sorting
1/70
Earn XP
Description and Tags
Beyond Mendelian Genetics, DNA Discovery and Replication, Genetic Code
Last updated 4:44 PM on 4/12/23
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
71 Terms
1
New cards
Limitation’s of Mendel’s Ideas
* Did not explain continuous variation
* By law of independent assortment and meiosis genes must be on different chromosomes?
* Humans have 23 pairs of homologs, so do we only have 23 genes?
* By law of independent assortment and meiosis genes must be on different chromosomes?
* Humans have 23 pairs of homologs, so do we only have 23 genes?
2
New cards
Sex-Linked Traits
* Traits that run in families and predominantly affect males
* Both parents must be heterozygous or it would not be visible
* Both parents must be heterozygous or it would not be visible
3
New cards
Red Green Colour Blindness
* Appears in 1/4 children of unaffected parents (so recessive)
* Overwhelmingly appears in males
* Only males of unaffected parents are colour blind
* Colourblind females have affected parents
* Overwhelmingly appears in males
* Only males of unaffected parents are colour blind
* Colourblind females have affected parents
4
New cards
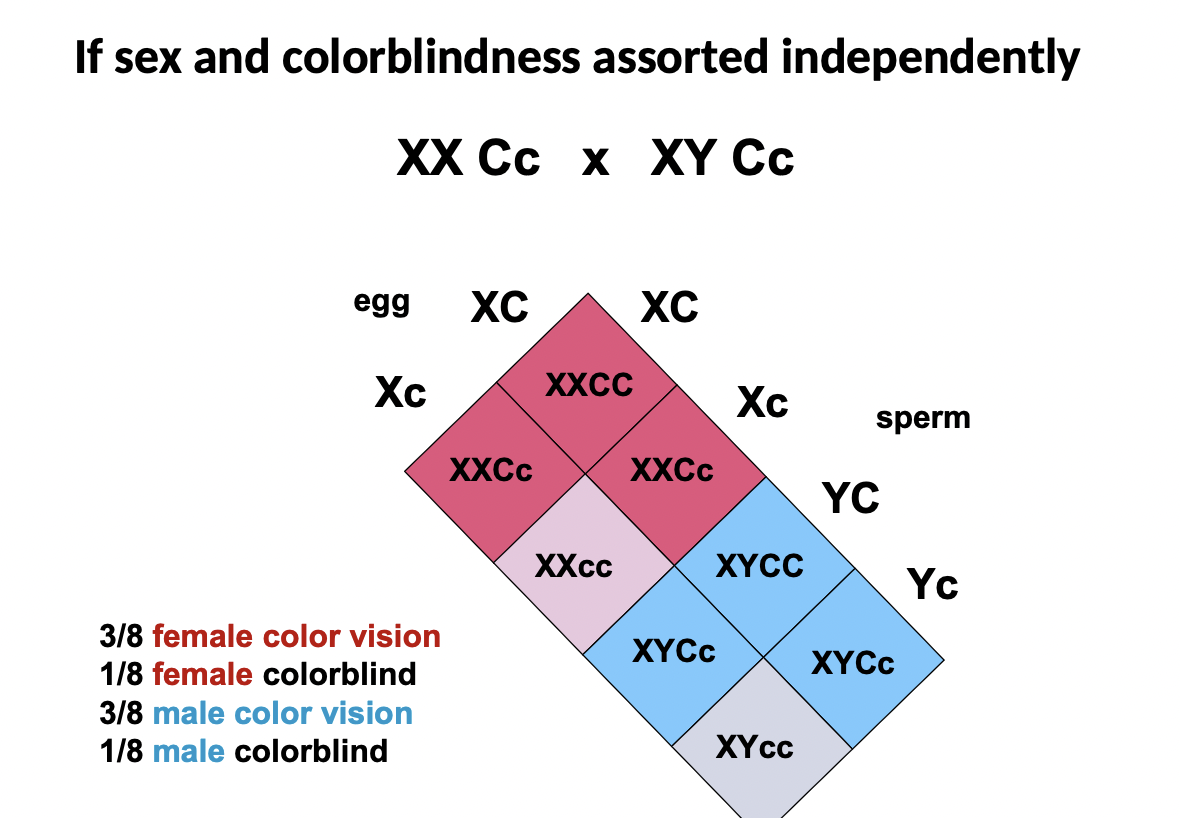
What would it look like if sex and colour blindness assorted independently?
* XX Cc crossed with XY Cc
5
New cards
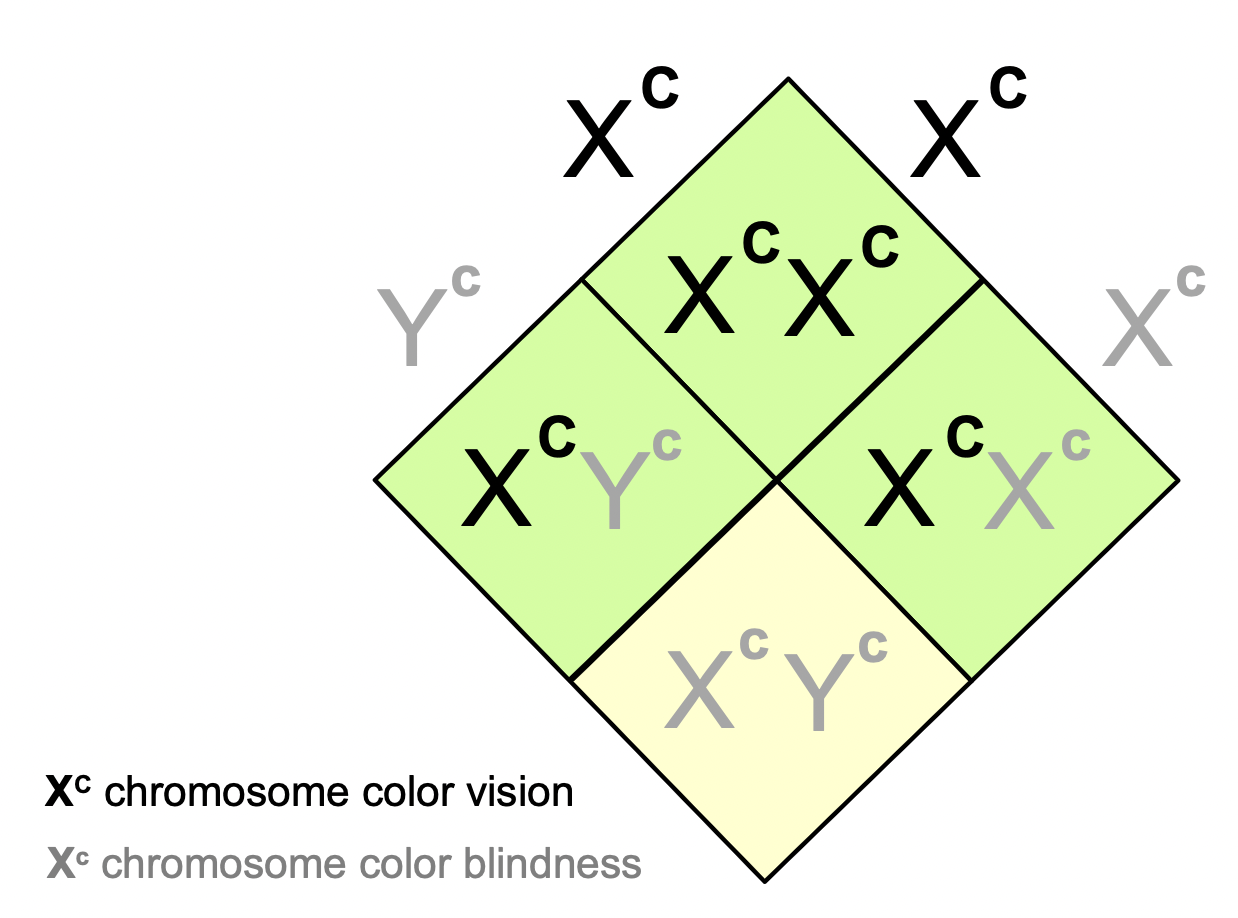
What is the actual ratio for colourblind progeny?
* The Y chromosome always has the recessive allele because it does not have a gene for colourblindness
6
New cards
Why does Y always have the recessive? (colourblindness)
* It is tiny compared to the X chromosome
* It does not have a gene for colourblindness
* The absence of a gene is also an allele
* It does not have a gene for colourblindness
* The absence of a gene is also an allele
7
New cards
What are the three types of alleles?
Wild Type: The predominant allele in a population (>99%)
Mutant: A change from the wild type allele, typically the result of a recent mutation. Can refer to alleles that cause disease.
Polymorphic: An allele that is present in
Mutant: A change from the wild type allele, typically the result of a recent mutation. Can refer to alleles that cause disease.
Polymorphic: An allele that is present in
8
New cards
What is the exception to the law of independent assortment?
* Only applies to genes on different chromosomes
9
New cards
What happens when someone has the colourblindness trait?
* Colourblindness gene make a protein (opsin) that detects green light. The colourblindness trait results from a gene on the X chromosome that is not functional and cannot make the protein properly. It is mutated.
10
New cards
Hemizygous
* When a gene is missing from one of the chromosomes
11
New cards
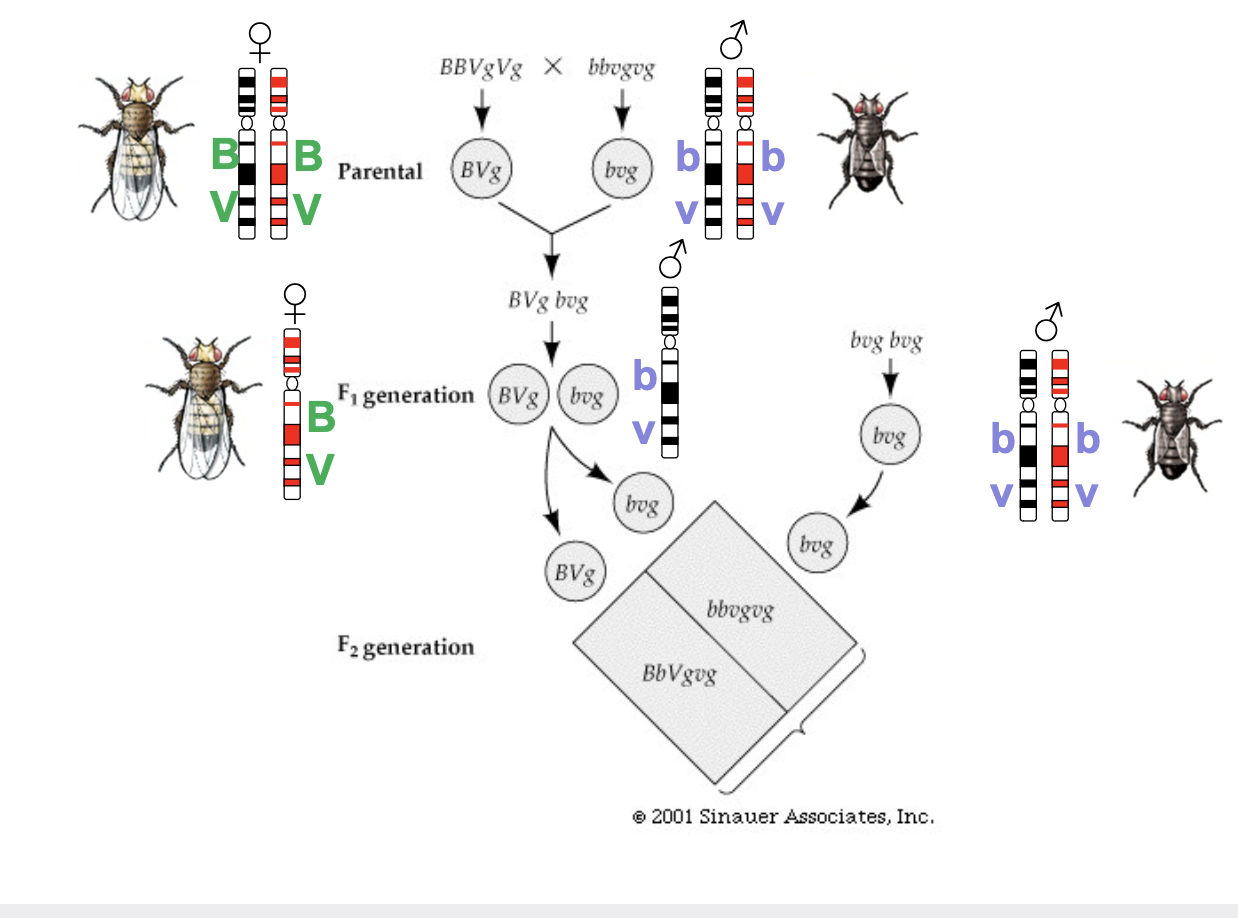
Thomas Hunt Morgan and Fly Test Cross
* Discovered linkage and surmised it was the result of 2 genes being on the same chromosome
* After crossing fruit flys, and examining their colour (black, grey) and wing type (vestigial, normal) there was anomalies in the result, so the genes do not independently assort but are linked on the same chromosome
* Anomalies because the chromatids exchange arms in prophase of meiosis 1, so if the breaking and rejoining occurs between the B and V in a BV chromatid that is exchanging with a bv chromatid, you get end up with Bv on one and bV on the other
* After crossing fruit flys, and examining their colour (black, grey) and wing type (vestigial, normal) there was anomalies in the result, so the genes do not independently assort but are linked on the same chromosome
* Anomalies because the chromatids exchange arms in prophase of meiosis 1, so if the breaking and rejoining occurs between the B and V in a BV chromatid that is exchanging with a bv chromatid, you get end up with Bv on one and bV on the other
12
New cards
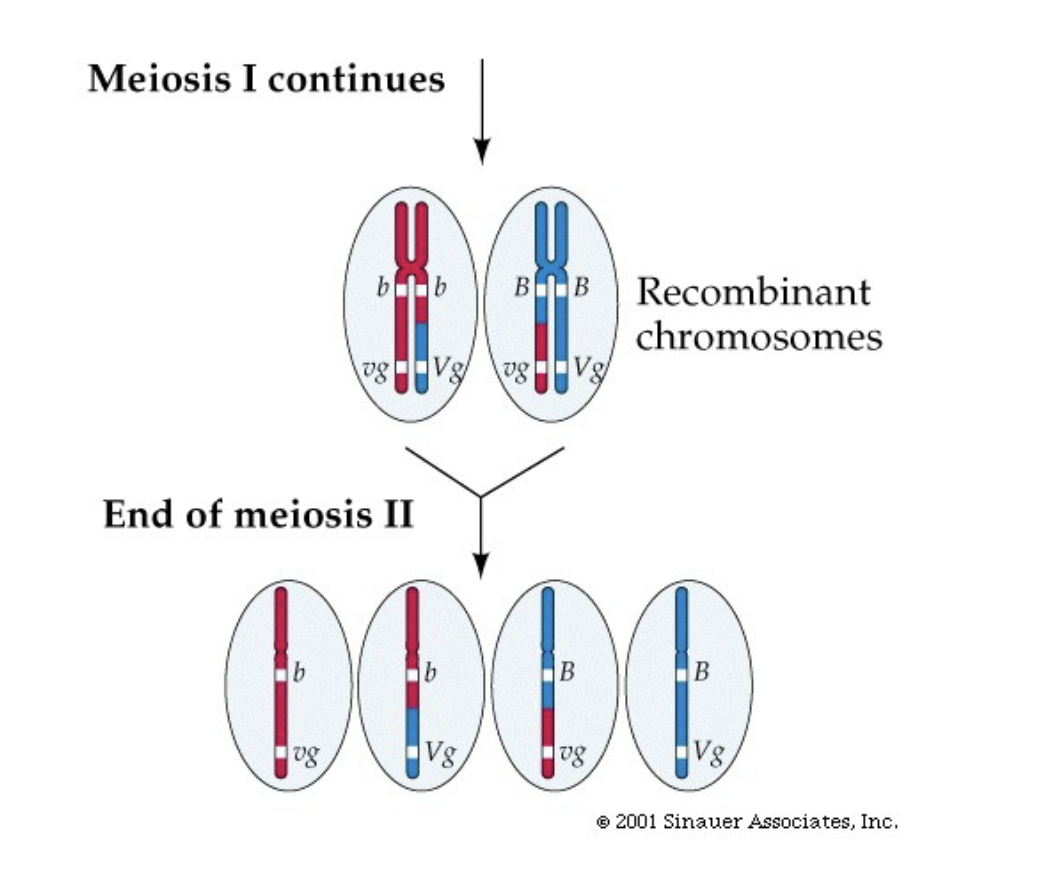
Recombination/Recombinant Phenotypes
* Chromatids exchange arms in prophase of meiosis 1, so if the breaking and rejoining occurs between the 2 genes you get an unexpected mix (like in fruit fly test)
13
New cards
Genes Linked in Trans
* Alleles on different homologous chromosomes
14
New cards
Genes Linked in Cis
* Alleles on same homologous chromosome
15
New cards
Recombination Rate
* Recombination occurs at random points
* A measure of physical distance on the chromosome
* The probability/rate of recombination occurring between 2 genes depends on how far apart the two genes are on the chromosome
* A measure of physical distance on the chromosome
* The probability/rate of recombination occurring between 2 genes depends on how far apart the two genes are on the chromosome
16
New cards
Locus
* Specific position on a chromosome where a particular gene or genetic marker is located
17
New cards
Formula for Distance Measured in centiMorgans
100( # of recombinants/ # total progeny)
18
New cards
Linkage Group
* Set of genes shown to be linked together on the same chromosome
19
New cards
Linear Order of Genes
* Genes have a linear order on the chromosome that is invariant
* Every individual of a species has the same genes linked together on the same chromosomes in the same linear order
* Every individual of a species has the same genes linked together on the same chromosomes in the same linear order
20
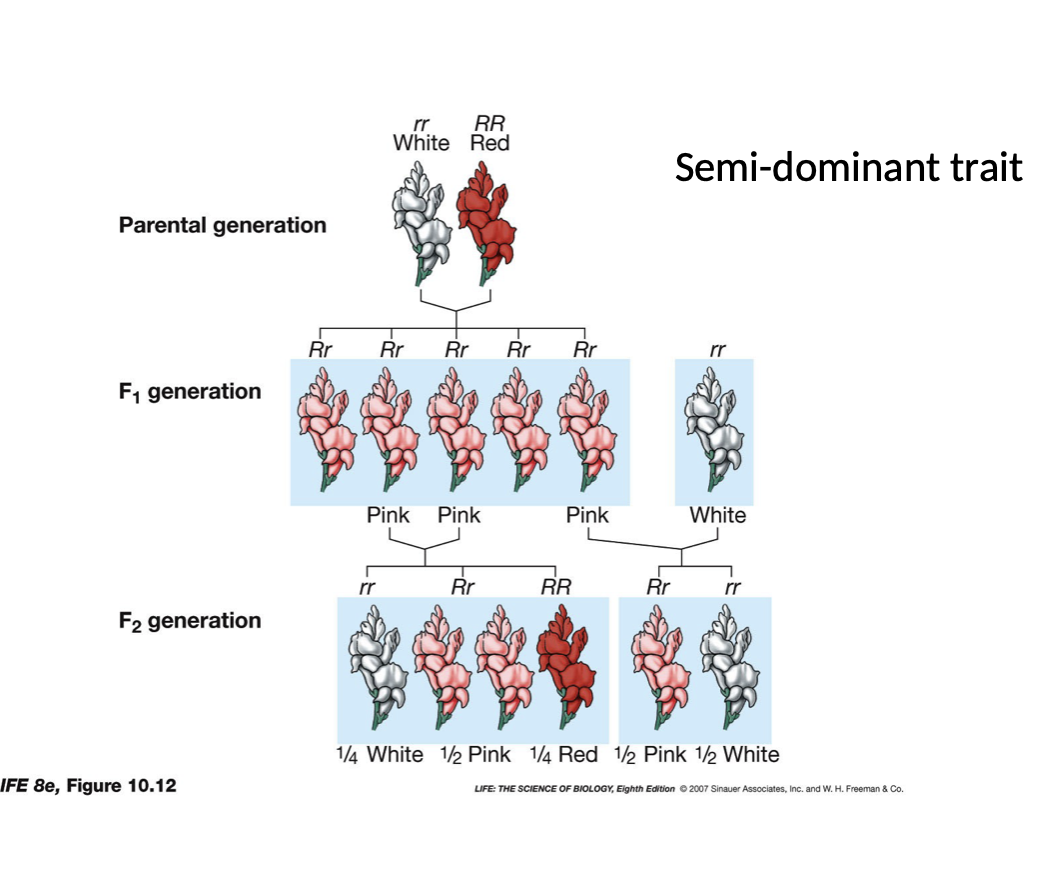
New cards
Incompletely Dominant or Semi-dominant
* Contribution to continuous variation
* Ex. Snap Dragon flower with rr white and RR red
* Homozygous progeny are white or red
* But heterozygous progeny (Rr) turn pink
* Due to the non functional genes from the recessive allele
Summary: Three phenotypes for one gene, two alleles
* Ex. Snap Dragon flower with rr white and RR red
* Homozygous progeny are white or red
* But heterozygous progeny (Rr) turn pink
* Due to the non functional genes from the recessive allele
Summary: Three phenotypes for one gene, two alleles
21
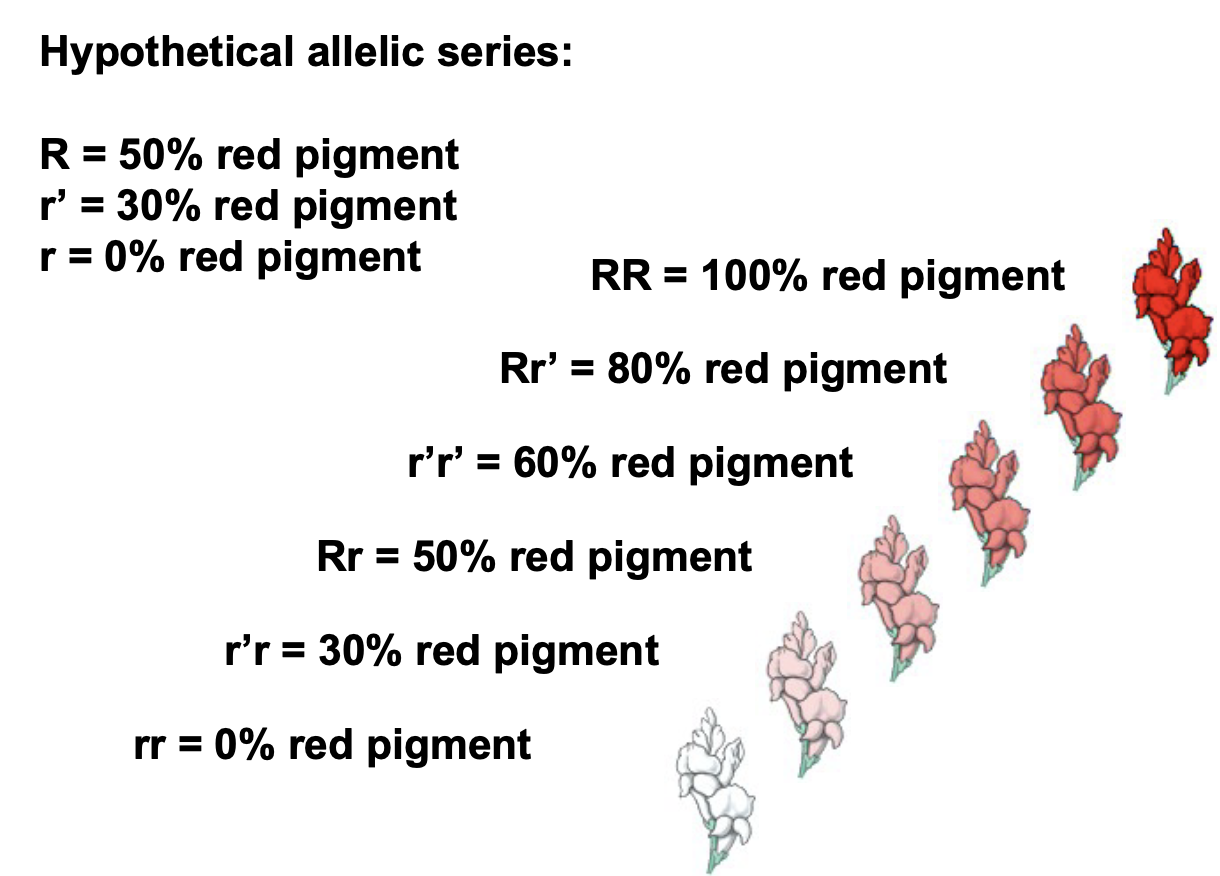
New cards
Allelic Series
* Contribution to continuous variation
* More than 2 alleles per gene (in a population)
* Alleles encode gene products with varying degrees of partial function who’s effects are additive
* Ex. Alzheimer’s can be related to an allelic series
* More than 2 alleles per gene (in a population)
* Alleles encode gene products with varying degrees of partial function who’s effects are additive
* Ex. Alzheimer’s can be related to an allelic series
22
New cards
Environmental Contribution to Phenotype
* Contribution to continuous variation
* Penetrance, Expressivity, Polygenic/Multigenic, and Epistasis all tie into this
* Penetrance, Expressivity, Polygenic/Multigenic, and Epistasis all tie into this
23
New cards
Penetrance
The % of individuals of a genotype that show the phenotype at all, sometimes you can have the same genotype and still not display the phenotype
24
New cards
Expressivity
Degree to which a phenotype is expressed. As individuals with the same genotype vary in their degree of expression.
Ex. shades of pink in a flower
Ex. shades of pink in a flower
25
New cards
Polygenic/Multigenic
* Multiple genes that may contribute to a train
* Ex. at least 6 genes that influence heart attack risk
* Ex. at least 6 genes that influence heart attack risk
26
New cards
How to Purify Something
* Separate components of a complex mixture
* Assay each component to determine which has the desired property
* Assay each component to determine which has the desired property
27
New cards
Assay
* Way of measuring something
* Can measure a substance, like an assay for starch
* Can measure an abstract phenomenon, like an assay for memory (ex. mice in a maze)
* Can measure a substance, like an assay for starch
* Can measure an abstract phenomenon, like an assay for memory (ex. mice in a maze)
28
New cards
How to Purify DNA
* grind up organism
* extract lipids and proteins with organic solvent (phenol)
* precipitate with ethanol
* extract lipids and proteins with organic solvent (phenol)
* precipitate with ethanol
29
New cards
Friedrich Miescher isolated nuclei from pus cells…
found that the main constituent of the nucleus was a compound he called nuclein (turned out to be DNA)
30
New cards
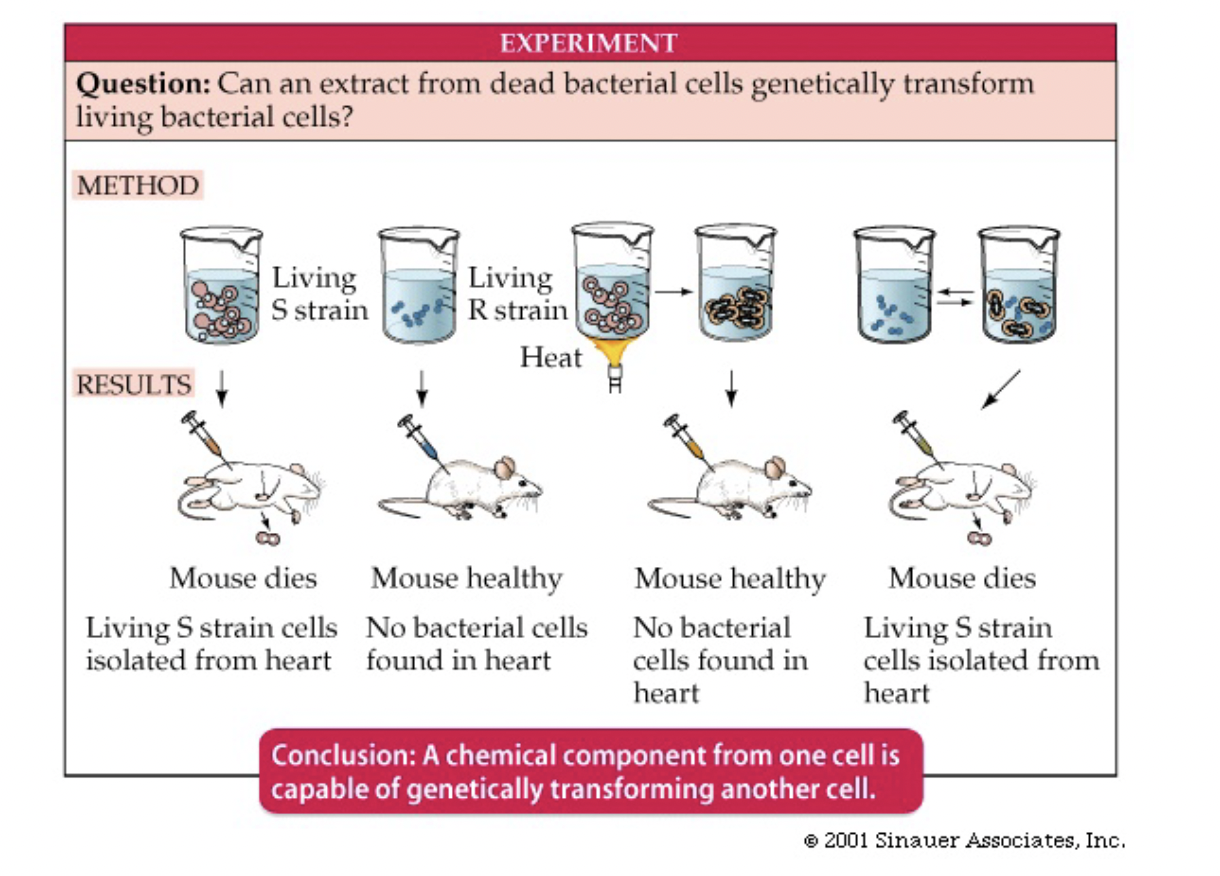
Fredrick Griffith Mice Experiment
* interested in developing a vaccine for pneumonia with mice
* injected mice with live or heat killed pneumonia bugs
* either smooth (S) strain or rough (R) strain
* S would kill injected mice, R would not
* mice injected with dead S and live R would die because R transformed into S
* injected mice with live or heat killed pneumonia bugs
* either smooth (S) strain or rough (R) strain
* S would kill injected mice, R would not
* mice injected with dead S and live R would die because R transformed into S
31
New cards
Transforming Principle
* Substance that transformed R to S
* So S had genetic material that was heritable to R
* Can conclude transforming principle = genetic material
Aside: ability to transform R to S is an assay for genetic materialness
* So S had genetic material that was heritable to R
* Can conclude transforming principle = genetic material
Aside: ability to transform R to S is an assay for genetic materialness
32
New cards
Oswald Avery
* identified that transforming principle was DNA
33
New cards
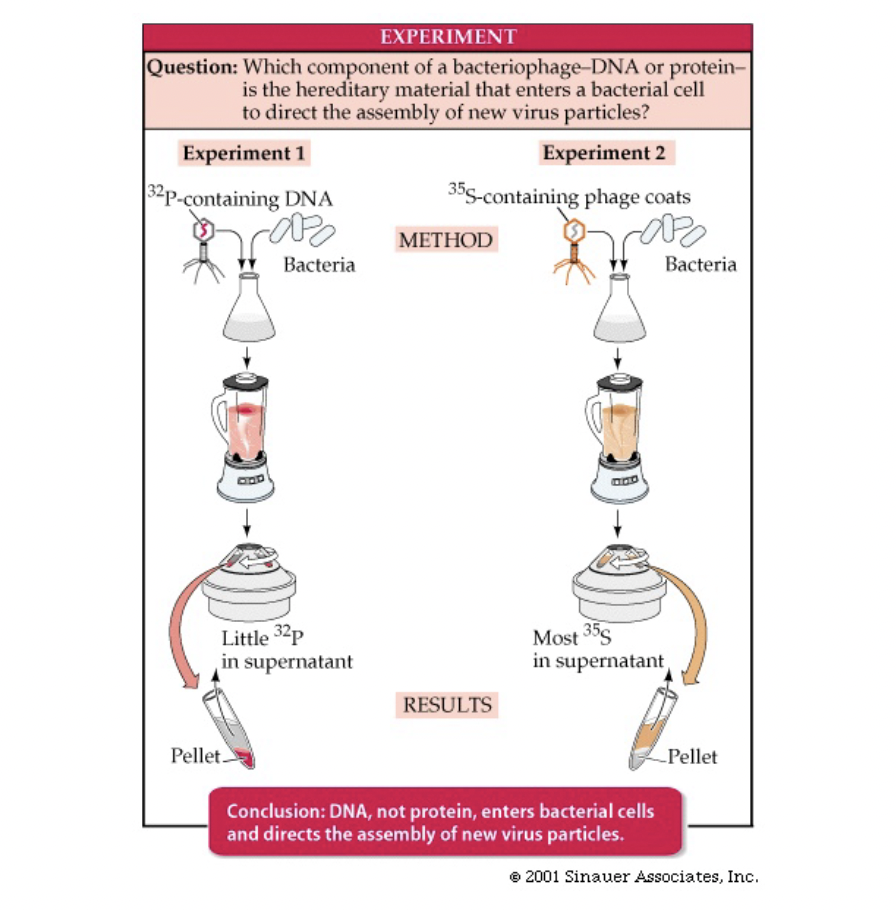
Hershey Chase Experiment
* only 2 things in a virus (DNA and protein)
* viruses attach to bacterial and insert material to get bacteria to create more virus
* wondered if material inserted was DNA or protein
* labeled protein and DNA with two different radioactive elements
* infected bacteria with virus and threw it in a blender
* caused protein coat to be sheared off of the bacteria
* put into super natant, idea was that whatever stayed back in the pellet with the bacteria was the genetic material
* now the bacteria produced more virus containing DNA marker, thus DNA must be the viral genetic material
* viruses attach to bacterial and insert material to get bacteria to create more virus
* wondered if material inserted was DNA or protein
* labeled protein and DNA with two different radioactive elements
* infected bacteria with virus and threw it in a blender
* caused protein coat to be sheared off of the bacteria
* put into super natant, idea was that whatever stayed back in the pellet with the bacteria was the genetic material
* now the bacteria produced more virus containing DNA marker, thus DNA must be the viral genetic material
34
New cards
Watson & Crick
* Tried to figure out how DNA replicates and determines phenotype, and 3D structure
* used Rosalind Franklin and Maurice Wilkin’s unpublished data
* used Rosalind Franklin and Maurice Wilkin’s unpublished data
35
New cards
Clues to Mystery of DNA Structure
1) chemical structure of monomers
* DNA is a polymer made of 4 subunits (nucleotides AGCT)
* Polarity defined by which end of the sugar backbone was sticking out: 3’ OH or 5’ phosphate
2) Chargaff’s Rule
* showed that the amount of A = T & G = C but A+T/G+C can vary between organisms
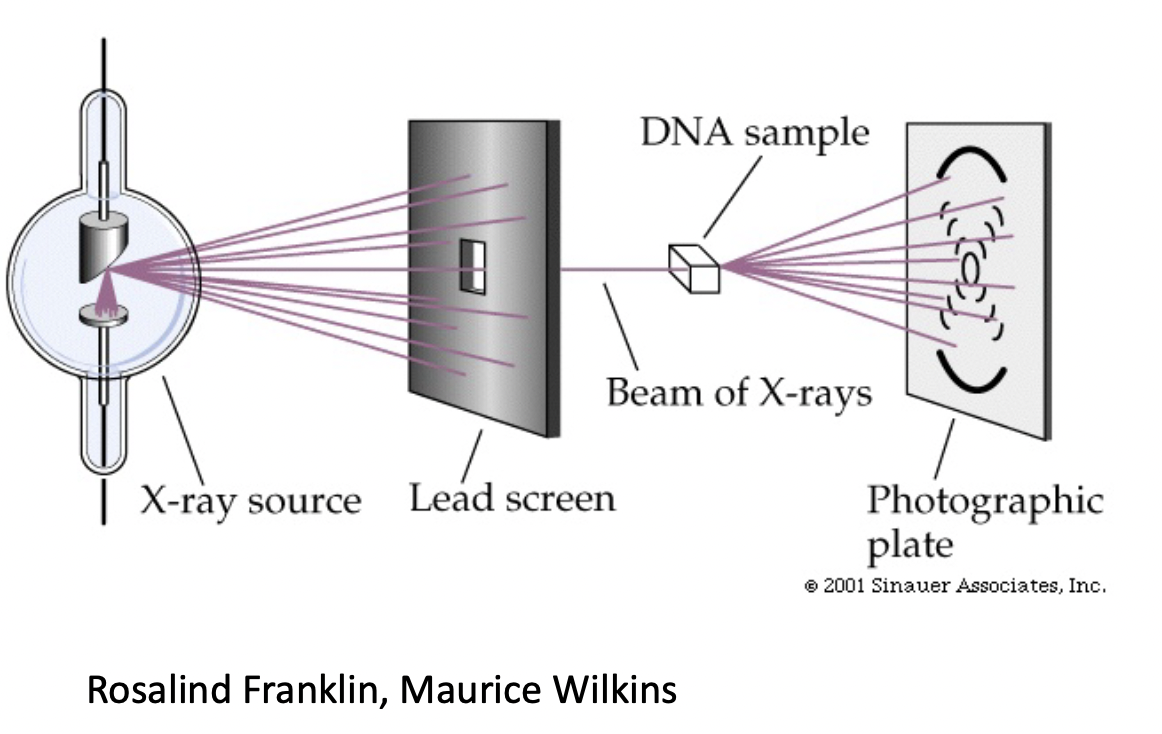
3) Crytalography - Franklin & Wilkins
* used x-ray diffraction patterns on a photographic plate
* found DNA consists of 2 strands
* strands twist around each other in a double helix
* phosphates are likely on the outside (they are(
* strands run antiparallel to eachother
* DNA is a polymer made of 4 subunits (nucleotides AGCT)
* Polarity defined by which end of the sugar backbone was sticking out: 3’ OH or 5’ phosphate
2) Chargaff’s Rule
* showed that the amount of A = T & G = C but A+T/G+C can vary between organisms
3) Crytalography - Franklin & Wilkins
* used x-ray diffraction patterns on a photographic plate
* found DNA consists of 2 strands
* strands twist around each other in a double helix
* phosphates are likely on the outside (they are(
* strands run antiparallel to eachother
36
New cards
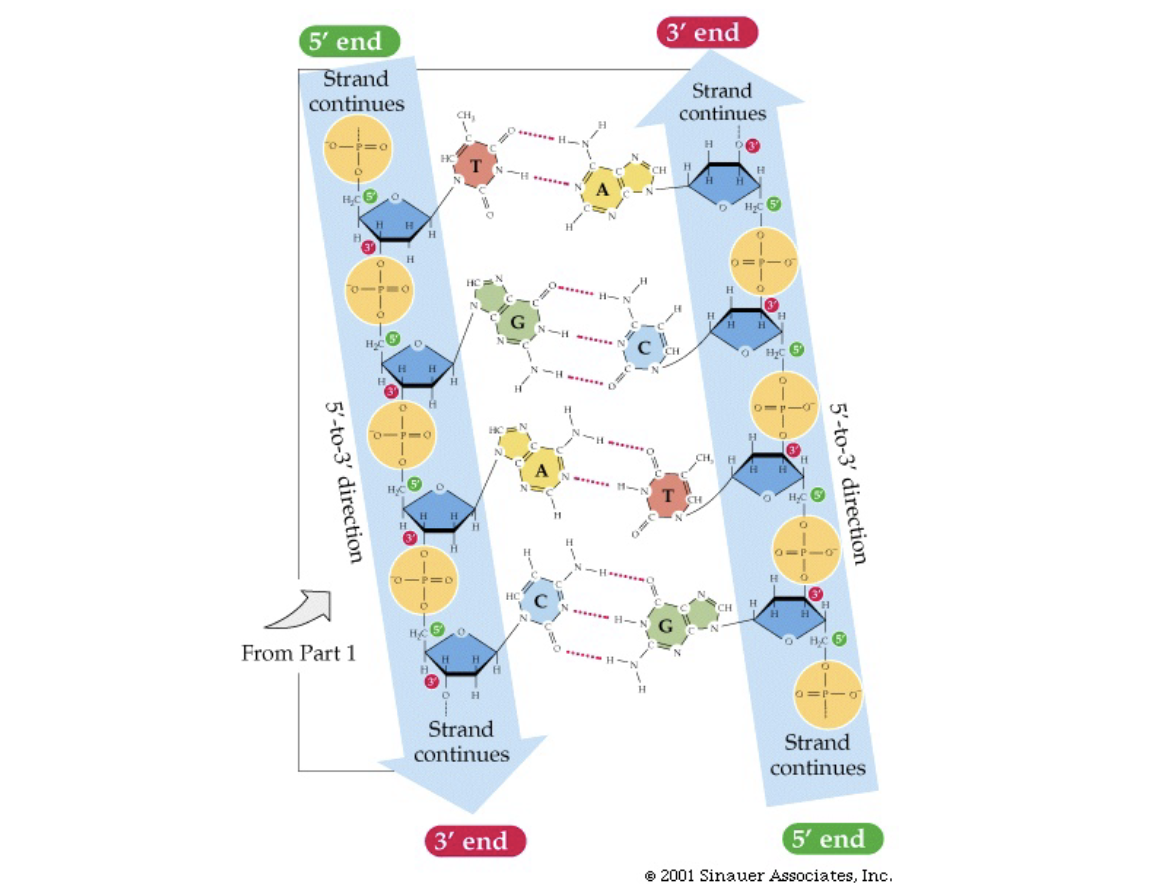
How were the structure clues used?
* came up with model where nitrogenous bases are in the middle and the phosphate backbone is on the outside
* charged phosphate backbone exposed to water
* bases are hydrophobic planar molecules, stacked on top of each other in the centre
* A always with T, G always with C
* this makes H bonds most stable
* always purine-pyrimidine pairs, keeps distance between strands constant
* charged phosphate backbone exposed to water
* bases are hydrophobic planar molecules, stacked on top of each other in the centre
* A always with T, G always with C
* this makes H bonds most stable
* always purine-pyrimidine pairs, keeps distance between strands constant
37
New cards
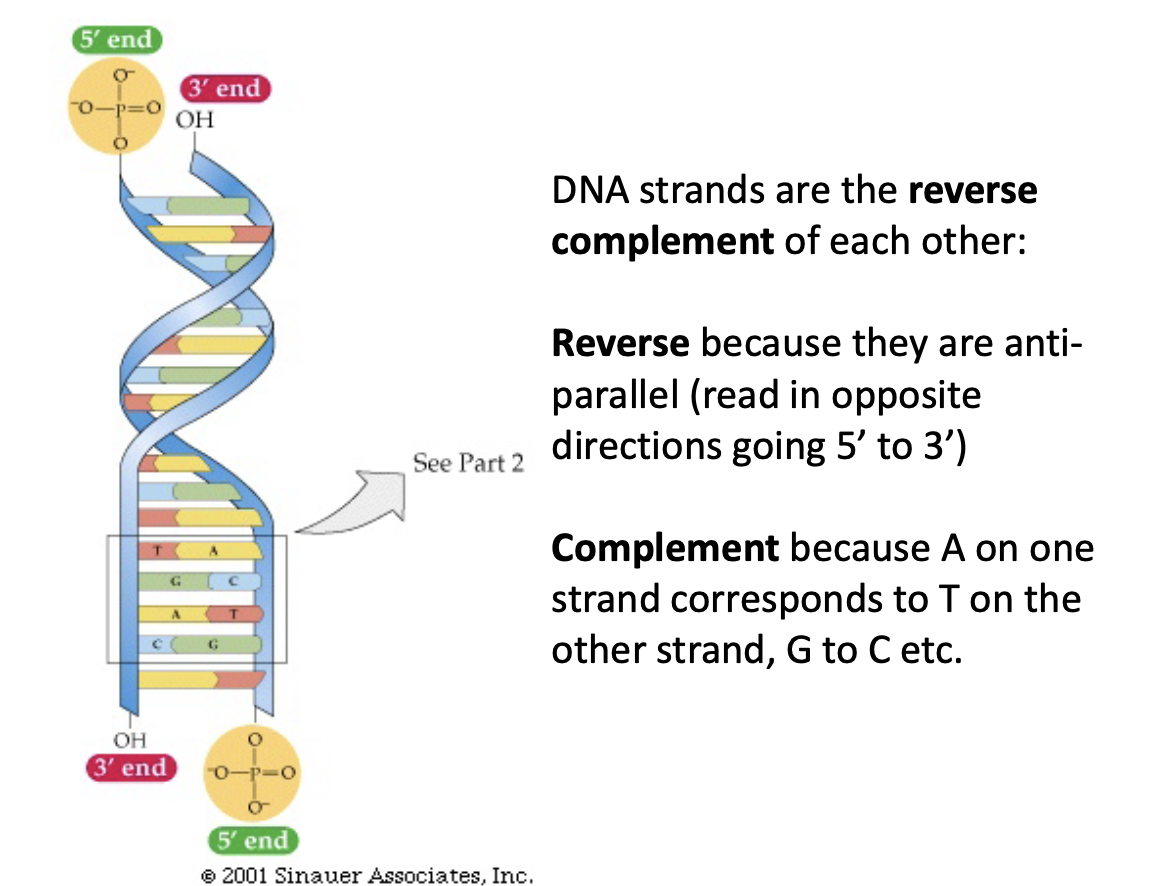
Reverse Complement and DNA
DNA are reverse complements of eachother.
Reverse: they are anti parallel (read in opposite directions)
Complement: A on one strand corresponds to T on the other, etc.
Reverse: they are anti parallel (read in opposite directions)
Complement: A on one strand corresponds to T on the other, etc.
38
New cards
What does the structure of DNA suggest?
1) nucleotide sequence doesn't affect the overall structure, information is encoded arbitrarily by the sequence of base pairs
2) 2 strands encode the same information in a complementary form suggesting a method of replication
2) 2 strands encode the same information in a complementary form suggesting a method of replication
39
New cards
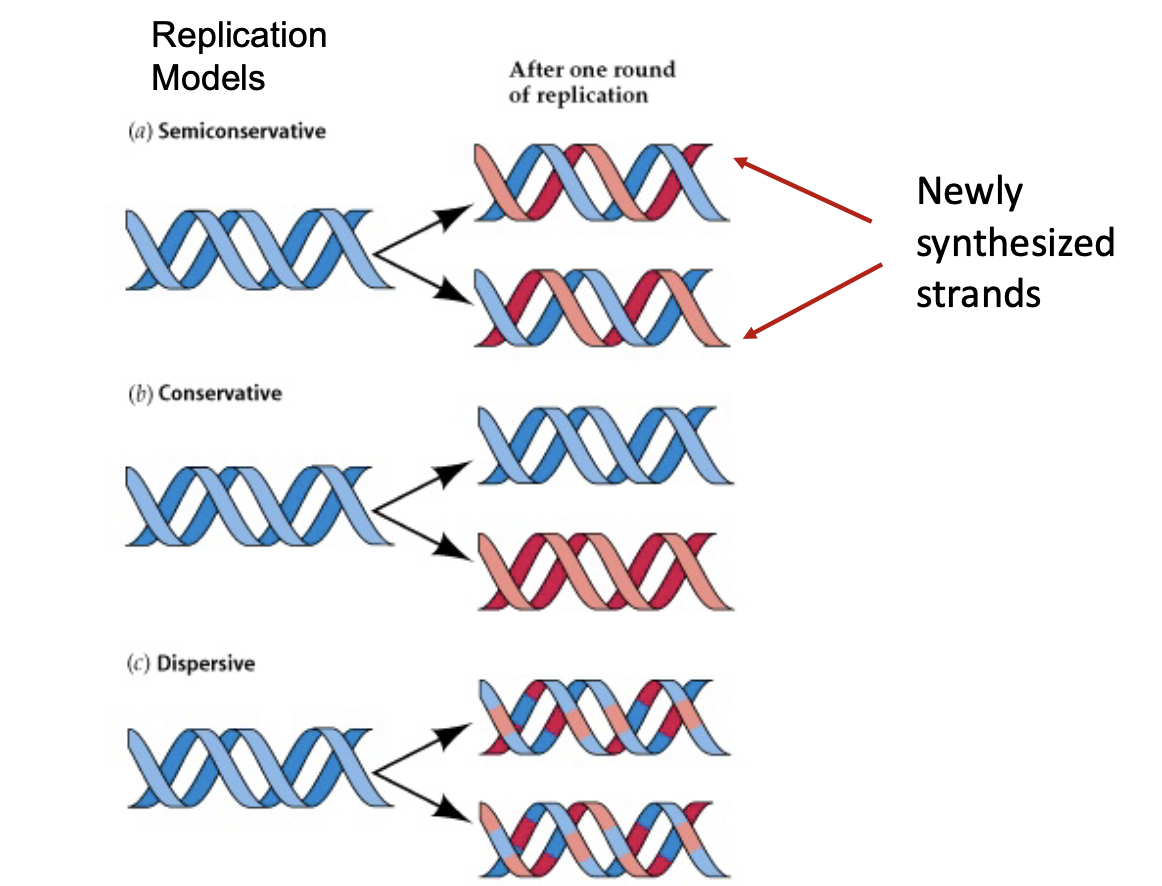
Propositions for DNA Replication Mechanisms (3)
1) Conservative Replication - 2 Strands unbounded, made a new strand and then re-annealed, and the 2 new strands re-annealed. Gives 1 old helix, 1 new helix.
2) Semiconservative Replication - Each old strand made a new strand and remained annealed to it (one that actually happens)
3) Dispersive Replication - DNA would break apart and rejoin to produce 4 strands, each with a mixture of old and new DNA
2) Semiconservative Replication - Each old strand made a new strand and remained annealed to it (one that actually happens)
3) Dispersive Replication - DNA would break apart and rejoin to produce 4 strands, each with a mixture of old and new DNA
40
New cards
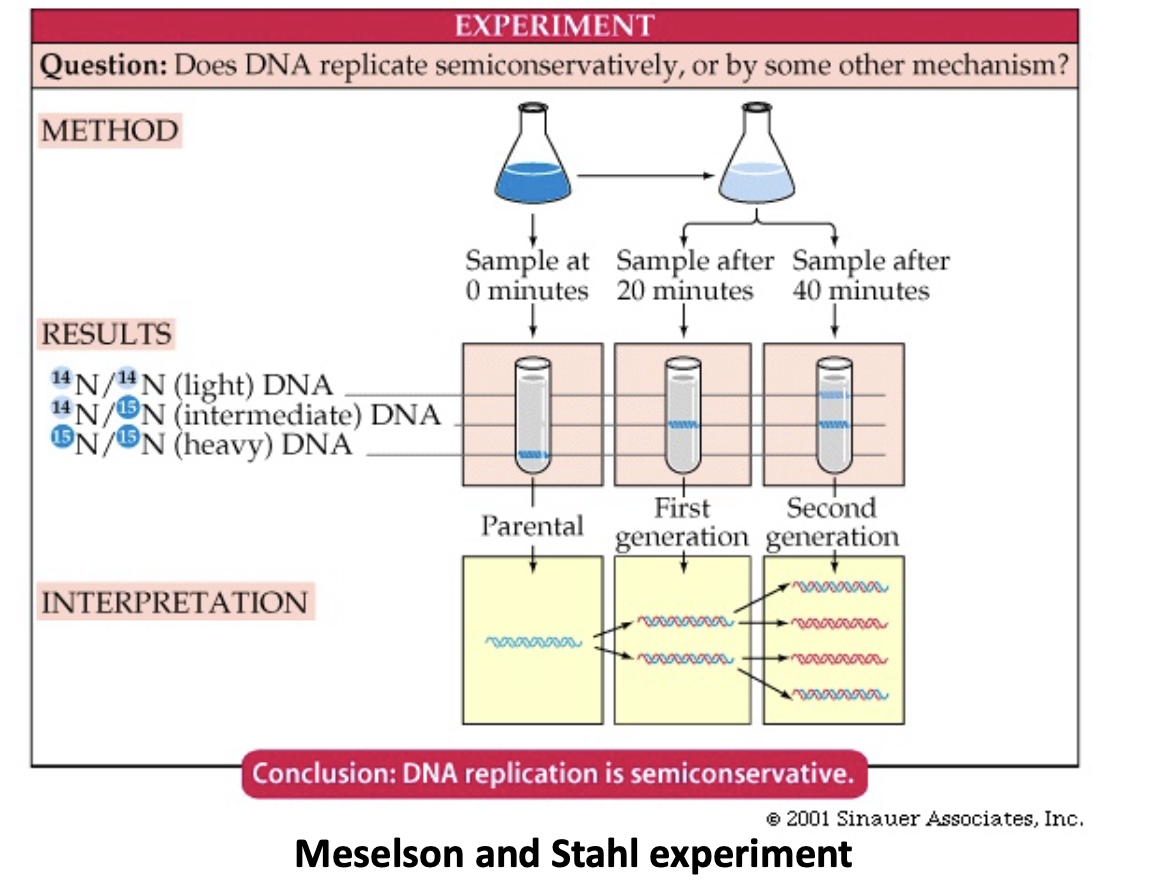
Meselson and Stahl Expiriement
* proved it was semiconservative
* labeled DNA by growing bacteria with a heaver isotope of Nitrogen
* 15N made the DNA heavy allowing it to be distinguished when centrifuged in cesium chloride
* after the 1st duplication, half as heavy, since half was normal weight
* after second duplication same thing occurred which could only occur if it was semiconservative replication
* labeled DNA by growing bacteria with a heaver isotope of Nitrogen
* 15N made the DNA heavy allowing it to be distinguished when centrifuged in cesium chloride
* after the 1st duplication, half as heavy, since half was normal weight
* after second duplication same thing occurred which could only occur if it was semiconservative replication
41
New cards
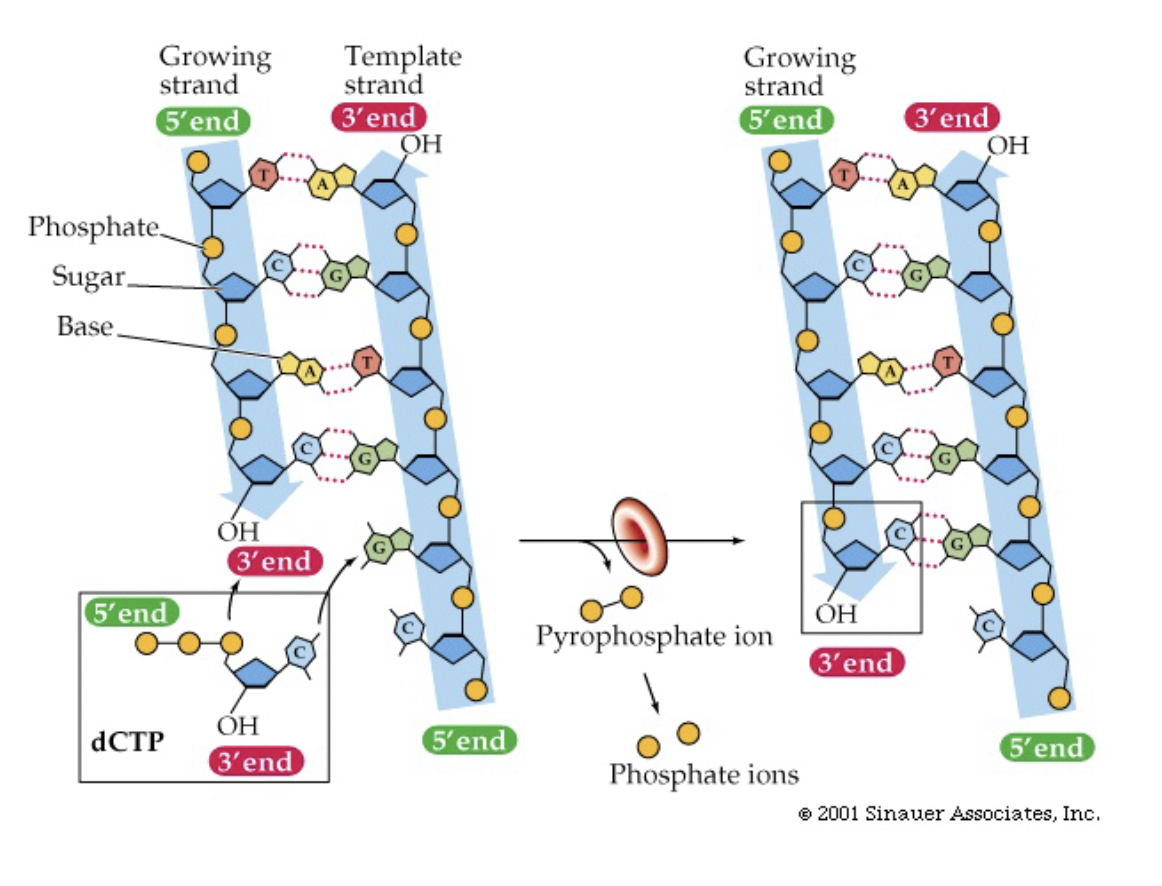
Making DNA in a Test Tube
* four nucleotides (in triphosphate nucleotide form)
* DNA polymerase - enzyme that does the polymerization reaction
* Template DNA with ragged ends so that DNA polymerase can attach the complementary nucleotide next in line
* The bond between the alpha phosphate (attached to sugar) and the beta phosphate of the nucleotide is broke
* then the alpha phosphate is attached to the 3’ hydroxyl group of the last nucleotide on the strand being extended
(DNA Synthesis 5’ → 3’)
\
Note: in the test tube the DNA usually comes from an organism already purified so it has ragged ends from the damage of this process
* DNA polymerase - enzyme that does the polymerization reaction
* Template DNA with ragged ends so that DNA polymerase can attach the complementary nucleotide next in line
* The bond between the alpha phosphate (attached to sugar) and the beta phosphate of the nucleotide is broke
* then the alpha phosphate is attached to the 3’ hydroxyl group of the last nucleotide on the strand being extended
(DNA Synthesis 5’ → 3’)
\
Note: in the test tube the DNA usually comes from an organism already purified so it has ragged ends from the damage of this process
42
New cards
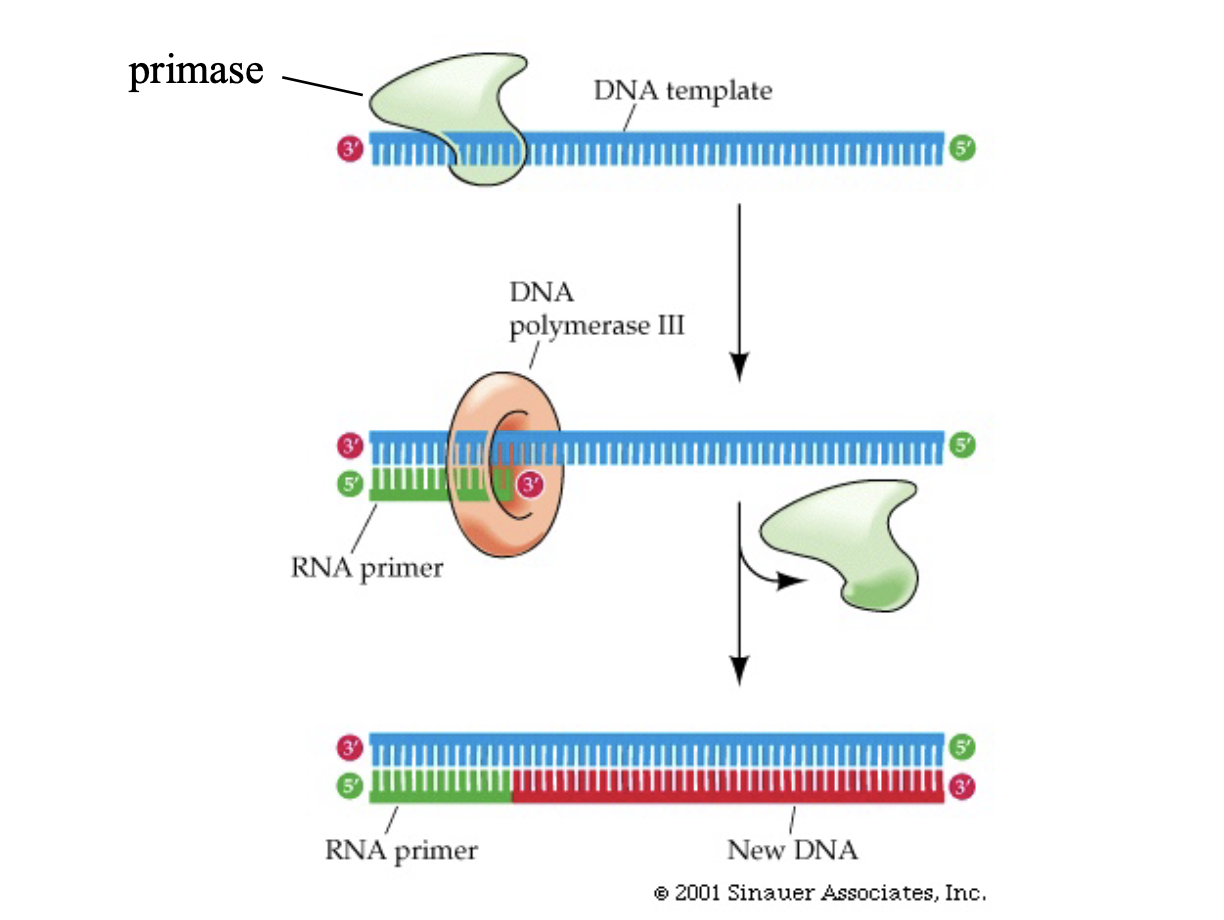
How to pry strands apart and how to make ragged ends?
enzyme helicase: unwinds the DNA strands by expanding energy
enzyme primase: makes short complementary RNA primers that act as a ragged end so DNA polymerase can go to work
\
Note: primase only acts on the lagging end
enzyme primase: makes short complementary RNA primers that act as a ragged end so DNA polymerase can go to work
\
Note: primase only acts on the lagging end
43
New cards
What are origins of replication?
* since helicase and primase do not randomly initiate replication, but at specific spots
* spots are called origins of replication
* bacteria have one, eukaryotes have many
* spots are called origins of replication
* bacteria have one, eukaryotes have many
44
New cards
Replication Forks
* helicases unwind the DNA in each direction from the origin of replication
* bubble forms with replication forks on either end where the old double stranded DNA is being split to act as the template for the formation of 2 new strands
* bubble forms with replication forks on either end where the old double stranded DNA is being split to act as the template for the formation of 2 new strands
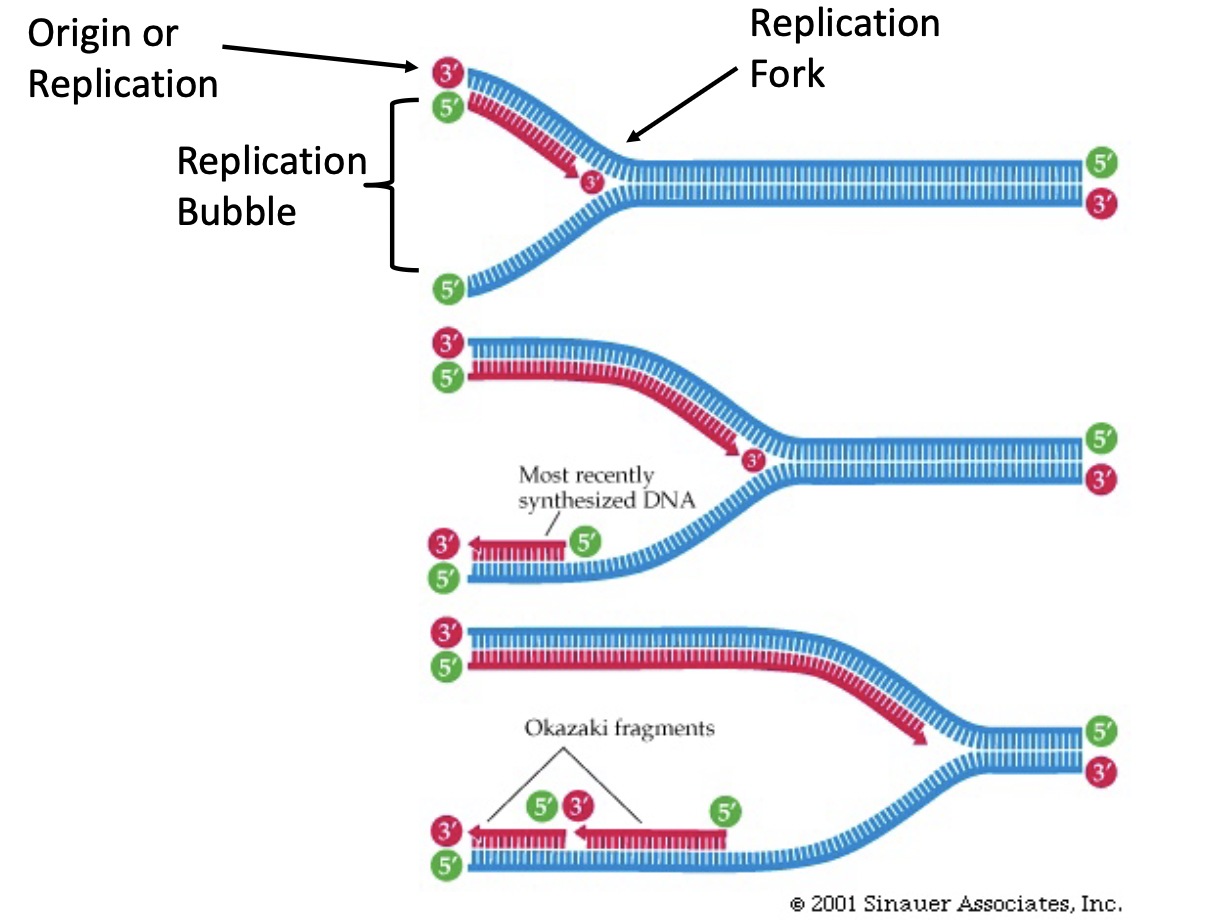
45
New cards
Lagging Strand
* primase puts down short RNA stretches (primers) every few hundred bases in 3’→ 5’ direction
* DNA polymerase 3 joins these fragments (Okazaki fragments) from 5’ → 3’ direction
* DNA polymerase 1 chews up the RNA primers and uses the newly synthesized DNA as a primer to fill in the gaps
* DNA ligase joins the ends of the newly synthesized strand and replication finishes
* (joins by making phosphate bond between strands)
* DNA polymerase 3 joins these fragments (Okazaki fragments) from 5’ → 3’ direction
* DNA polymerase 1 chews up the RNA primers and uses the newly synthesized DNA as a primer to fill in the gaps
* DNA ligase joins the ends of the newly synthesized strand and replication finishes
* (joins by making phosphate bond between strands)
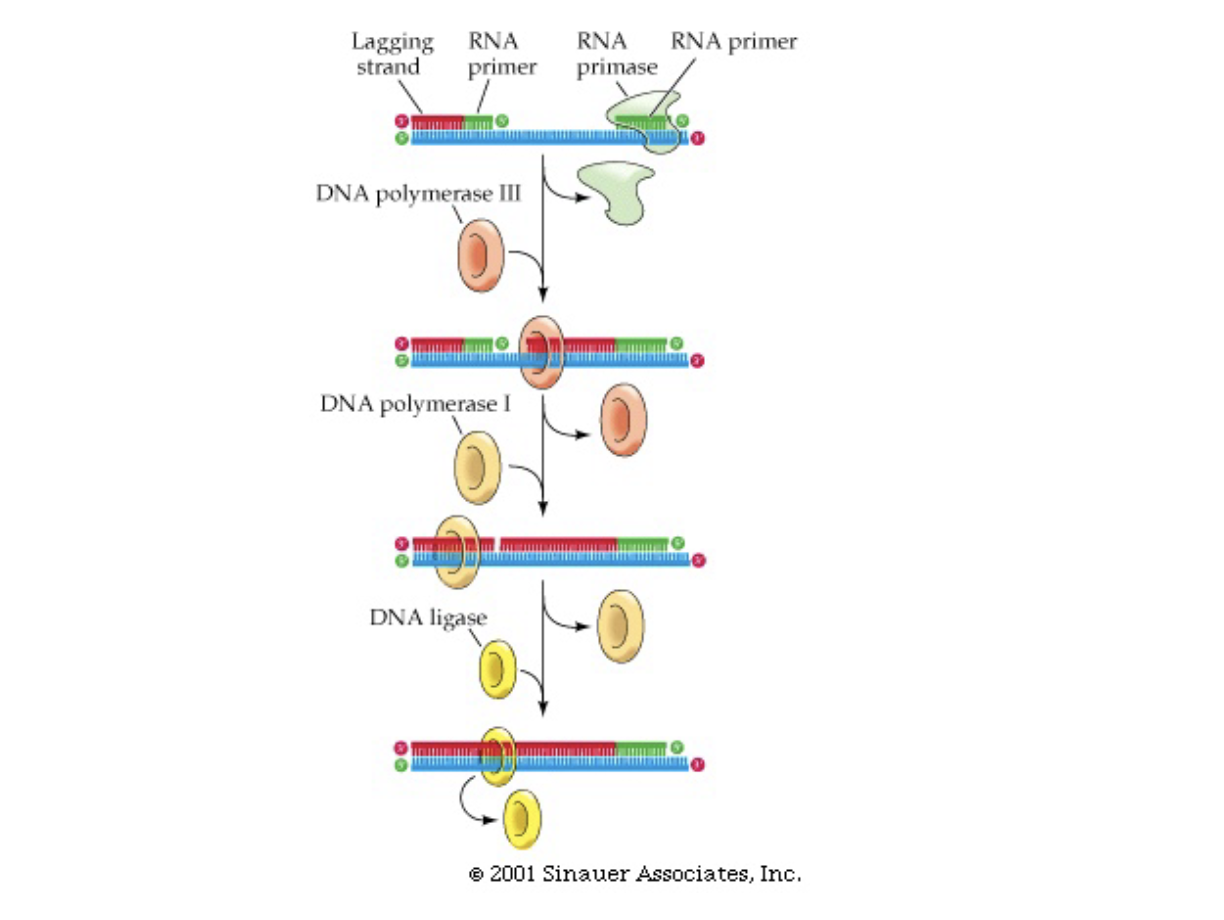
46
New cards
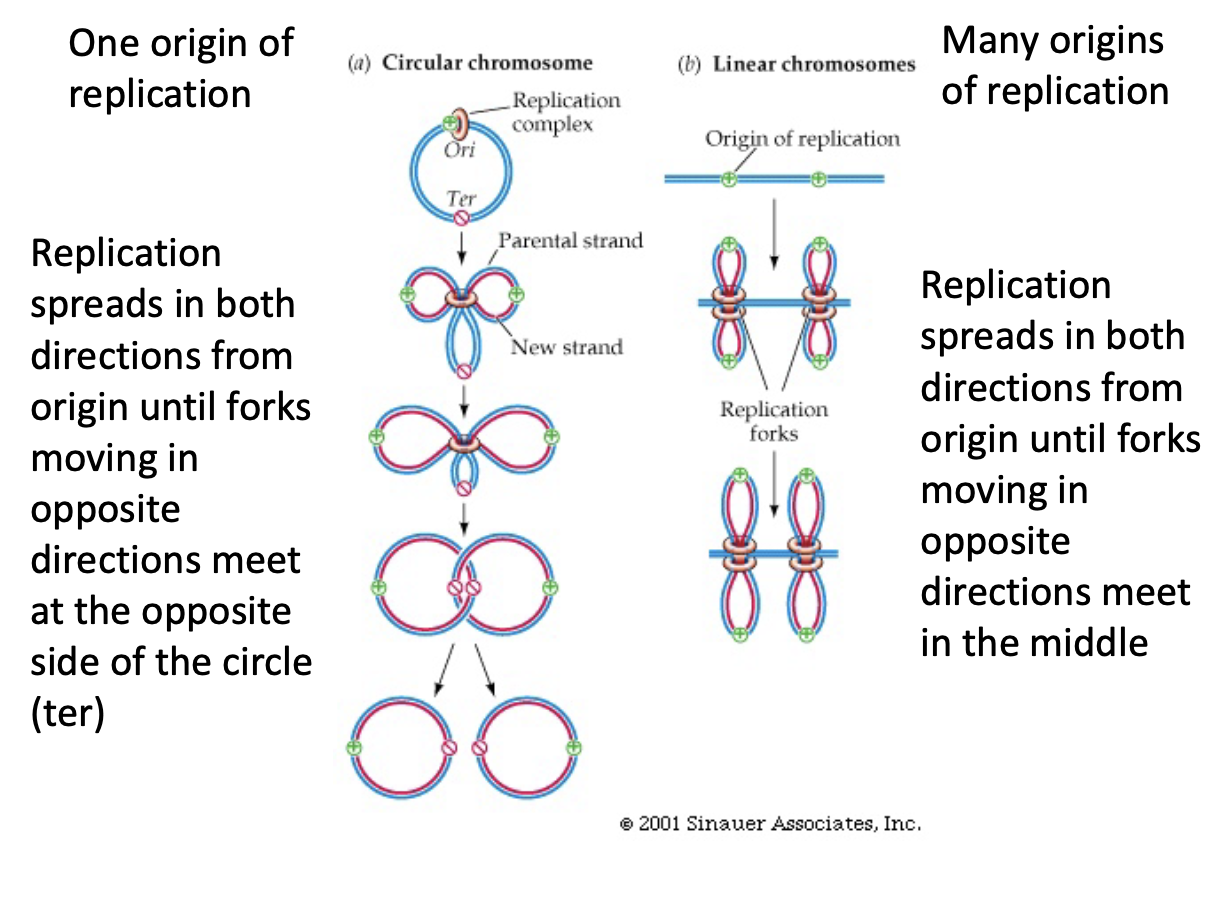
Circular Chromosome (DNA) Replication
* replication spreads in both directions from origin until forks moving in opposite directions meet at the opposite side of the circle
* this creates 2 linked circles that are broken and unlinked by and enzyme
* this creates 2 linked circles that are broken and unlinked by and enzyme
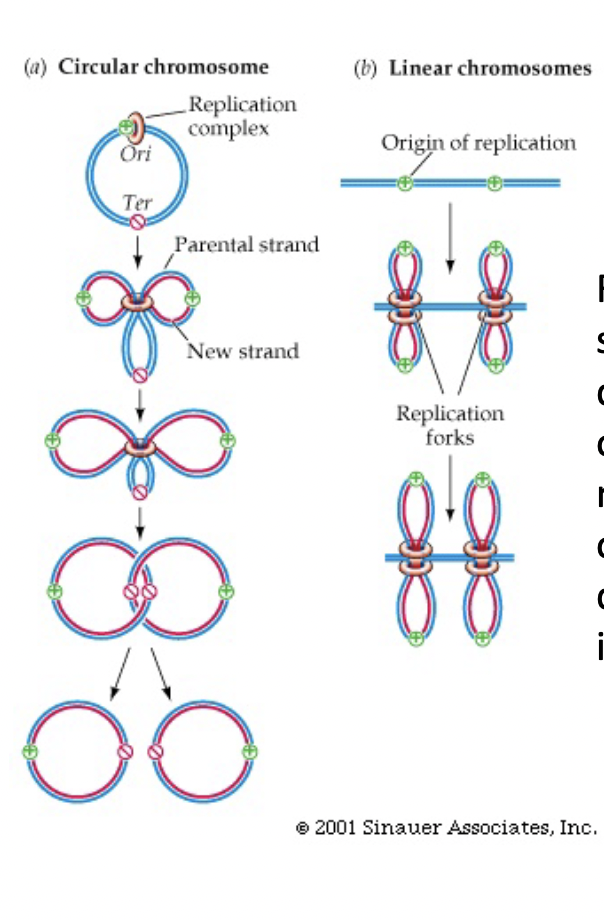
47
New cards
Linear Chromosome Replication
* replication spreads in both directions from origin(s) until forks moving in opposite directions meet in the middle
* DNA is replicated simultaneously in parallel, at multiple origins of replication
* DNA is replicated simultaneously in parallel, at multiple origins of replication
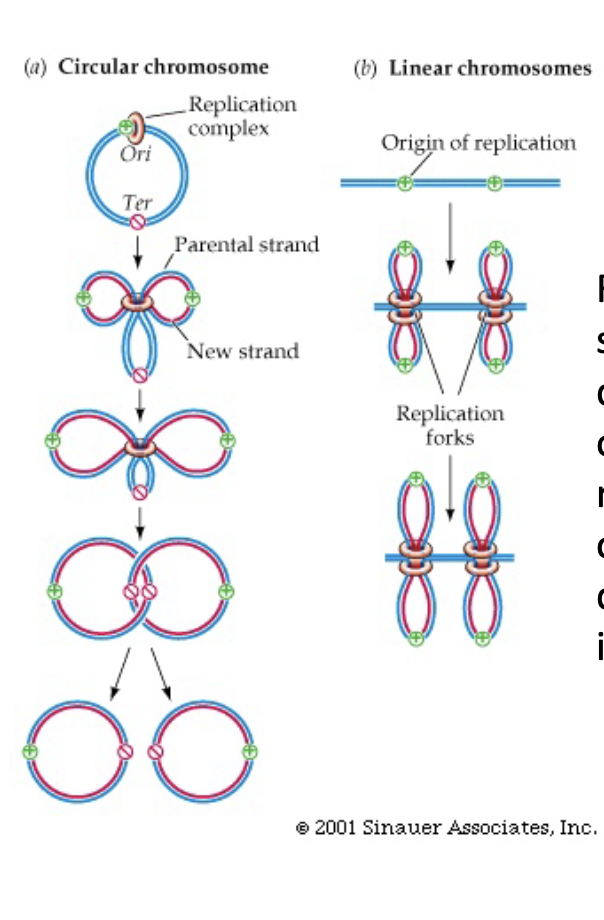
48
New cards
Fixing Replication Mistakes
1) Polymerase has proofreading mechanism that immediately removes bases that are not complementary to the template strand
2) mechanism at work during recombination called mismatch repair
3) mechanism at work the rest of the time called excision repair
\
Note: excision repair also works on thymine dimers formed by exposure to UV light
2) mechanism at work during recombination called mismatch repair
3) mechanism at work the rest of the time called excision repair
\
Note: excision repair also works on thymine dimers formed by exposure to UV light
49
New cards
Language of Genes
* must be encoded in the sequence of nucleotides
* code tells the cell what proteins to make
* code tells the cell what proteins to make
50
New cards
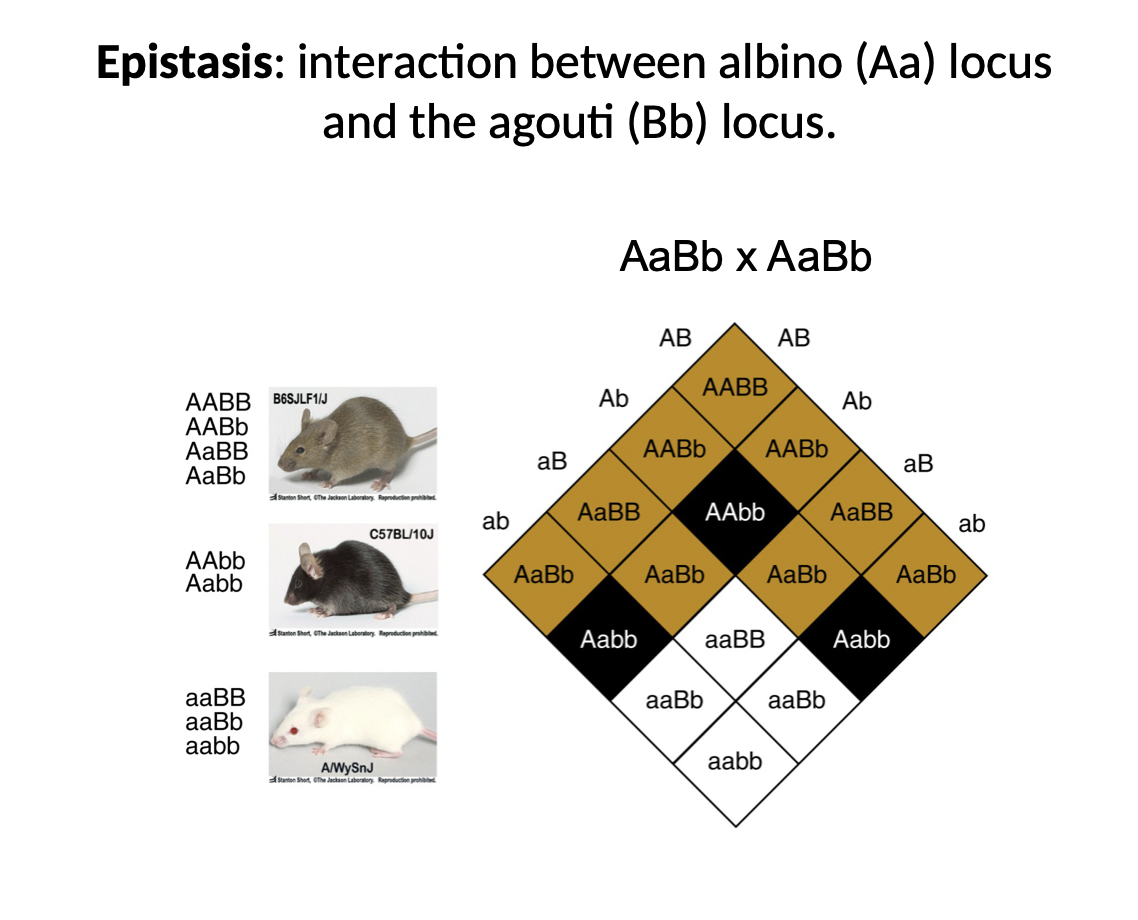
Epistasis
* kind of inheritance
* albino and agouti genes in mice have an epistatic relationship (not additive)
* so if you are homozygous recessive for the albino gene (aa) it doesn’t matter what you are for the agouti gene (always albino)
* albino gene has “super dominance”
* albino and agouti genes in mice have an epistatic relationship (not additive)
* so if you are homozygous recessive for the albino gene (aa) it doesn’t matter what you are for the agouti gene (always albino)
* albino gene has “super dominance”
51
New cards
Why do albino and agouti genes in mice have an epistatic relationship ?
* albino gene is necessary to make a colourless chemical and make it into a coloured pigment (that ends up in fur)
* if you don’t have the albino gene you can’t make the pigment and your fur never gets coloured
* the agouti gene takes the pigment (made by albino) and distributes it in the fur
* normally it distributes it in a way that gives brown fur, but a recessive allele (bb) gives black
* the two genes interact in a way where if you are missing the ability to make pigment (recessive albino) then the agouti gene is irrelevant because there is no pigment to distribute and you will always have white fur
* if you don’t have the albino gene you can’t make the pigment and your fur never gets coloured
* the agouti gene takes the pigment (made by albino) and distributes it in the fur
* normally it distributes it in a way that gives brown fur, but a recessive allele (bb) gives black
* the two genes interact in a way where if you are missing the ability to make pigment (recessive albino) then the agouti gene is irrelevant because there is no pigment to distribute and you will always have white fur
52
New cards
Alkaptonurea (Garrod)
* disease where urine is black
* if neither parent was affected the 1/4 of children was (not dominant)
* in one case an affected father has 4/8 affected children
* Garrod showed that it was a recessive hereditary trait
* deduced that it results from the absence of a specific enzyme in the metabolism
\
CONCLUDED THAT GENES MAKE ENZYMES
* if neither parent was affected the 1/4 of children was (not dominant)
* in one case an affected father has 4/8 affected children
* Garrod showed that it was a recessive hereditary trait
* deduced that it results from the absence of a specific enzyme in the metabolism
\
CONCLUDED THAT GENES MAKE ENZYMES
53
New cards
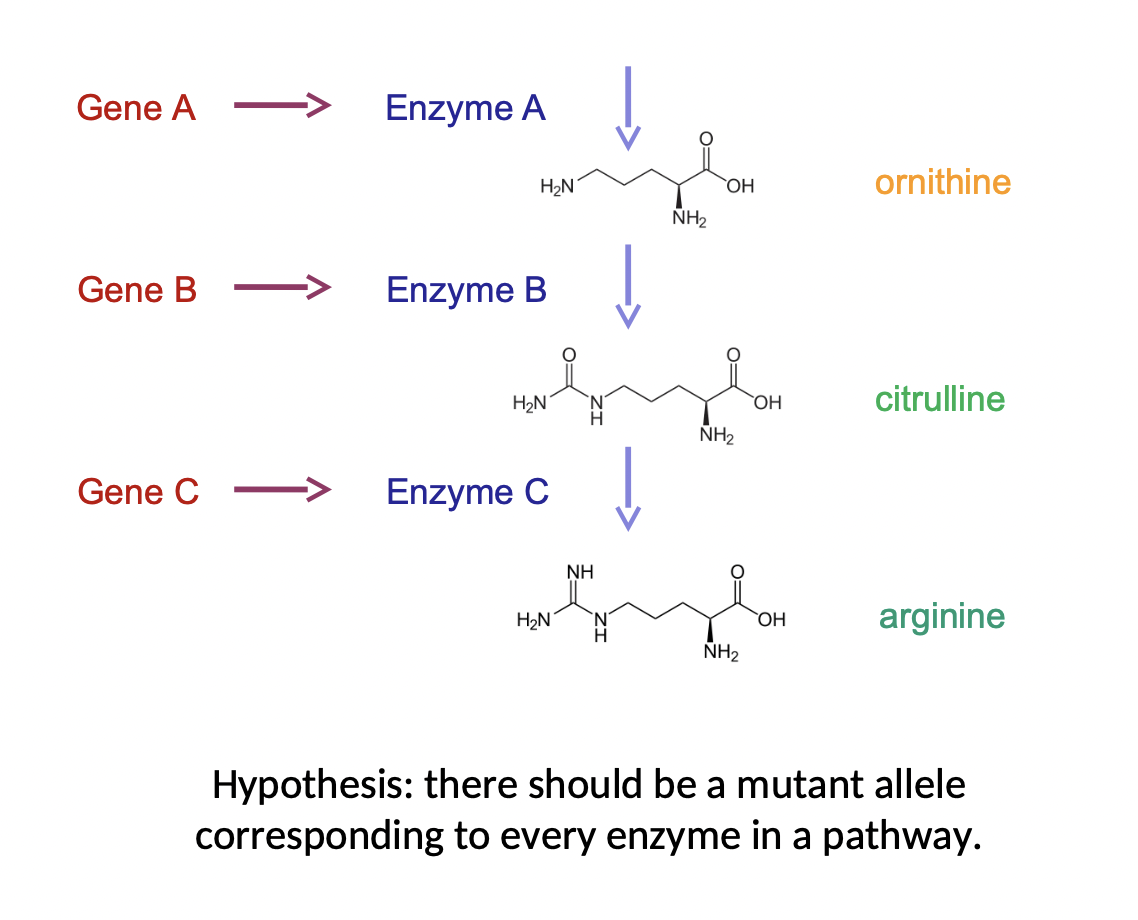
Alkaptonurea (Beadle & Tatum)
* expanded on Garrod’s idea
* if genes make proteins (enzymes) then we should be able to find a mutant corresponding to every enzyme in a pathway
\
One-gene, one-enzyme hypothesis → one-gene, one-protein
* if genes make proteins (enzymes) then we should be able to find a mutant corresponding to every enzyme in a pathway
\
One-gene, one-enzyme hypothesis → one-gene, one-protein
54
New cards
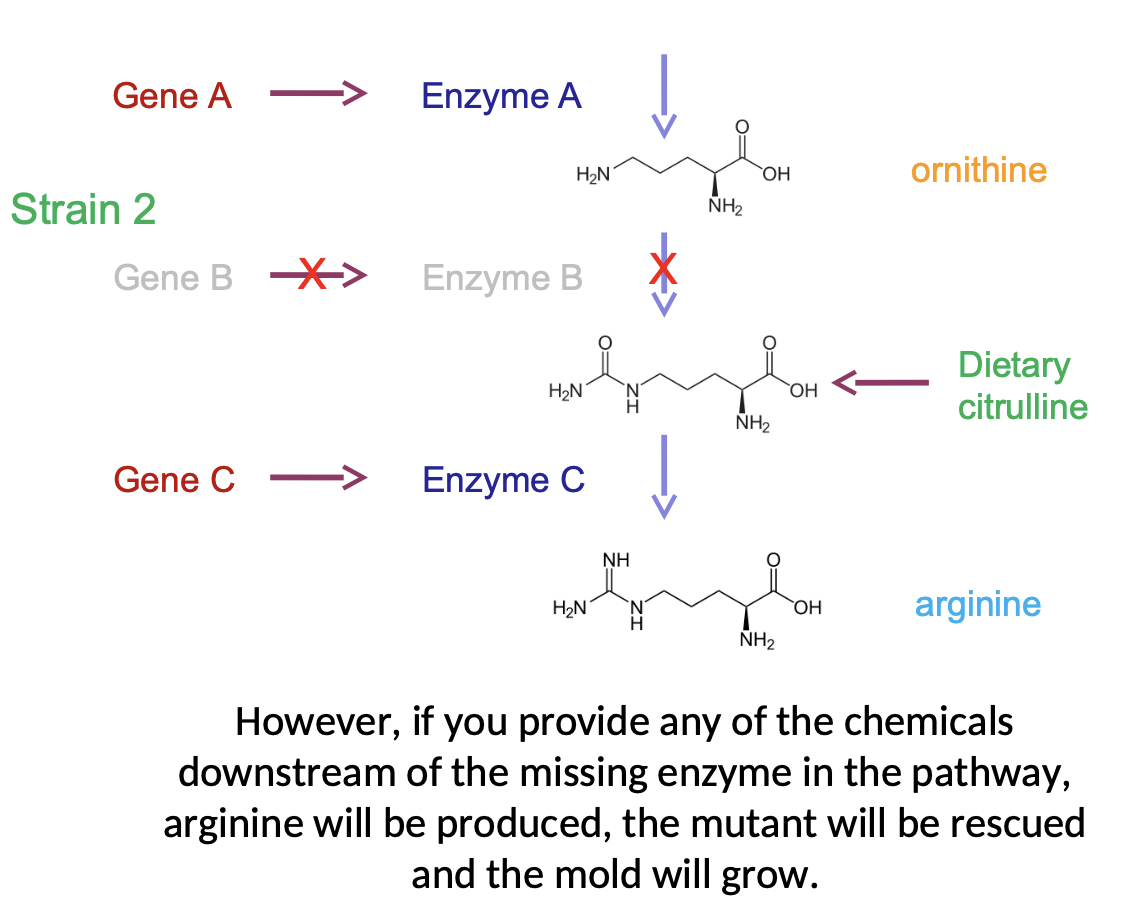
Beadle & Tatum Experiment
* used a mold, reasoned that mutants lacking enzymes necessary for a pathway would get stuck and not be able to produce the components downstream
* would prevent mold from growing, but could be rescued by providing the missing compounds in the growth media
* they isolated mutants that required certain metabolic precursors in order to grow (auxotrophs) as opposed to the wild type prototrophs
\
* mold needed Argenine, but not orthinine or citrilline, other than to make argenine
* ex. strain 2 could not make enzyme B (lacked gene b) so it could not turn orthinine to citrilline, but if given citrilline in its diet it could convert it with enzyme C into argenine
* would prevent mold from growing, but could be rescued by providing the missing compounds in the growth media
* they isolated mutants that required certain metabolic precursors in order to grow (auxotrophs) as opposed to the wild type prototrophs
\
* mold needed Argenine, but not orthinine or citrilline, other than to make argenine
* ex. strain 2 could not make enzyme B (lacked gene b) so it could not turn orthinine to citrilline, but if given citrilline in its diet it could convert it with enzyme C into argenine
55
New cards
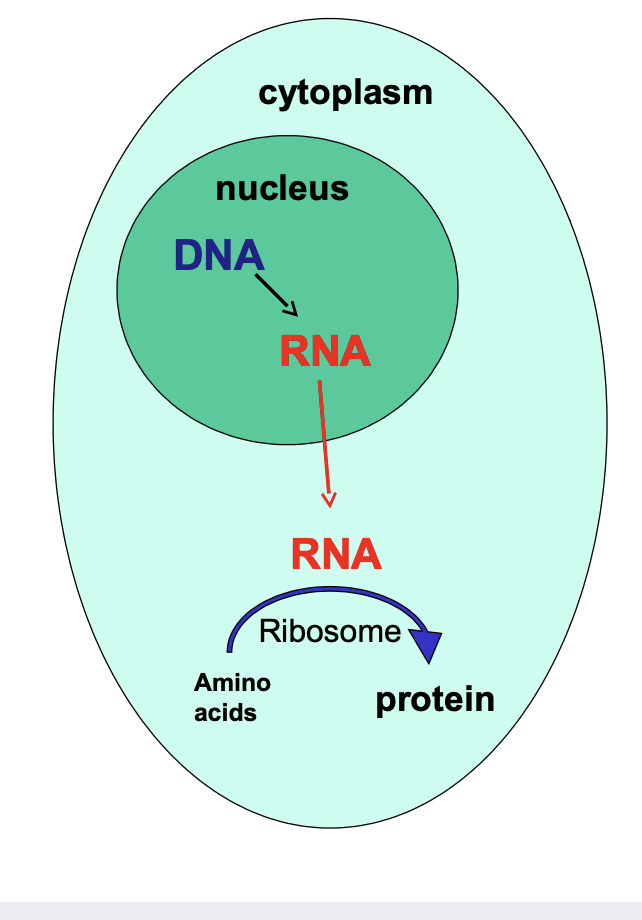
Central Dogma Overview
* information flow from gene to protein
* DNA is in the nucleus but proteins are manufactured in the cytoplasm by ribosomes
* RNA transfers the genetic information from the nucleus to the cytoplasm (where proteins are made)
* DNA is in the nucleus but proteins are manufactured in the cytoplasm by ribosomes
* RNA transfers the genetic information from the nucleus to the cytoplasm (where proteins are made)
56
New cards
Crick, Brenner, Jacob (Central Dogma)
* led to formulation of the central dogma
* idea that genetic information from from DNA → RNA → Protein
* proposed that the flow is unidirectional, it never goes in reverse
* idea that genetic information from from DNA → RNA → Protein
* proposed that the flow is unidirectional, it never goes in reverse
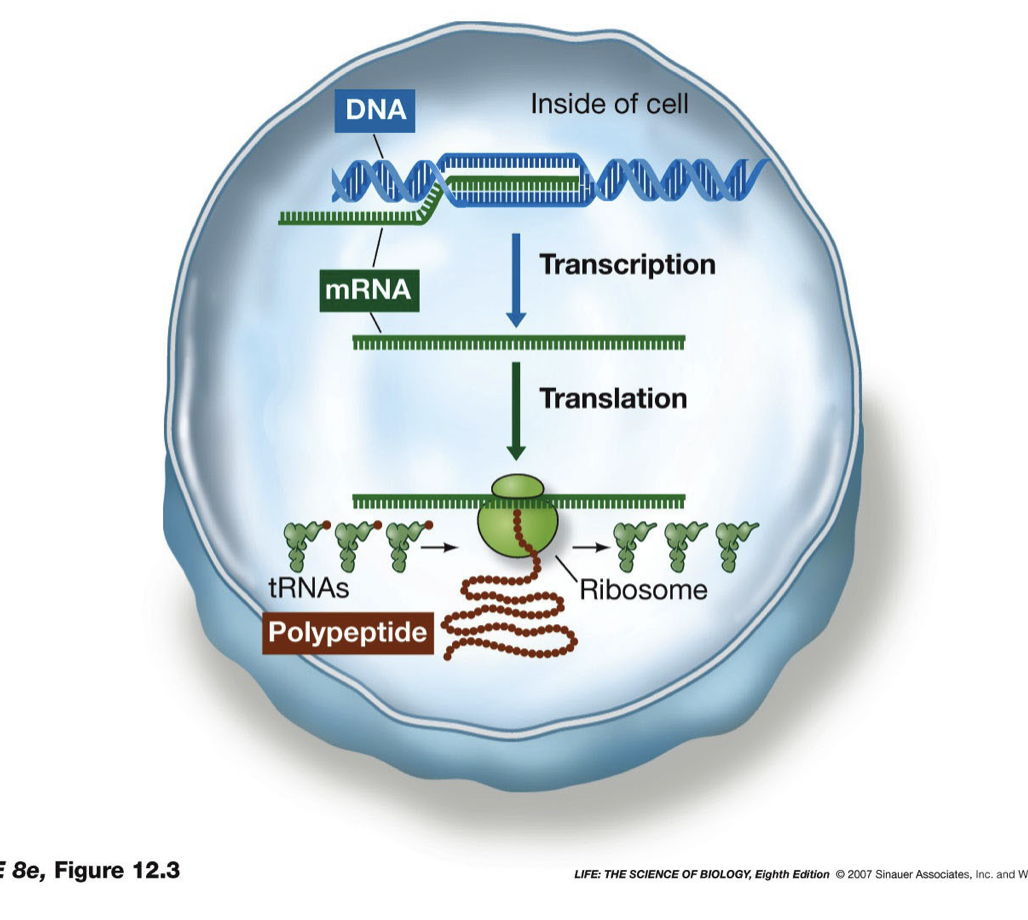
57
New cards
First Step of Central Dogma
* first step from genotype to phenotype is the formation of the messenger RNA
* RNA very similar to DNA (U instead of T), RNA has 2’ hydroxyl but that does not have an effect
* Similarity allows for information to be easily transferred
* RNA polymerase (enzyme) sits down on DNA at the beginning of a gene and unwinds
* Starts creating a complementary RNA strand, until it reaches the end of the gene
* RNA very similar to DNA (U instead of T), RNA has 2’ hydroxyl but that does not have an effect
* Similarity allows for information to be easily transferred
* RNA polymerase (enzyme) sits down on DNA at the beginning of a gene and unwinds
* Starts creating a complementary RNA strand, until it reaches the end of the gene
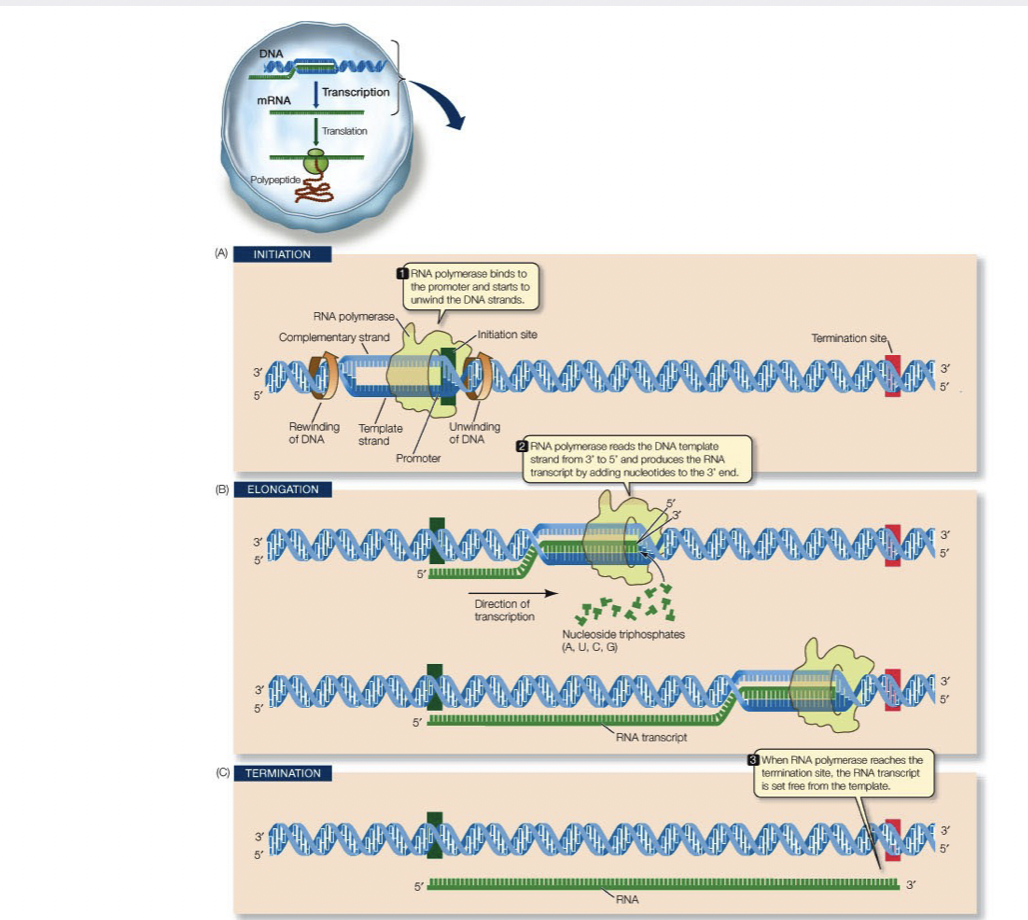
58
New cards
RNA Polymerase
* makes RNA like DNA polymerase but it doesn’t need a 3’ hydroxyl to get started and it incorporates triphosphate ribonucleotides
* occurs from 5’ → 3’
* new mRNA has all the info on the DNA for the gene
* mRNA leaves the nucleus and enters the cytoplasm
* ribosomes use it to make proteins
* occurs from 5’ → 3’
* new mRNA has all the info on the DNA for the gene
* mRNA leaves the nucleus and enters the cytoplasm
* ribosomes use it to make proteins
59
New cards
Coding Problem
Making of Amino Acids
* cannot be a 1 nucleotide per amino acid correspondence, would only make 4 amino acids (4^1)
* cannot be a 2 nucleotide per amino acid correspondence, would only make 16 amino acids (4^2)
* has to be a 3 nucleotide per amino acid correspondence, to make 64 amino acids (4^3)
* cannot be a 1 nucleotide per amino acid correspondence, would only make 4 amino acids (4^1)
* cannot be a 2 nucleotide per amino acid correspondence, would only make 16 amino acids (4^2)
* has to be a 3 nucleotide per amino acid correspondence, to make 64 amino acids (4^3)

60
New cards
Transcription
* process of making mRNA
* process where information from a DNA strand is transferred to an mRNA
* process where information from a DNA strand is transferred to an mRNA
61
New cards
First Protein that had its amino acid sequence determined
insulin
62
New cards
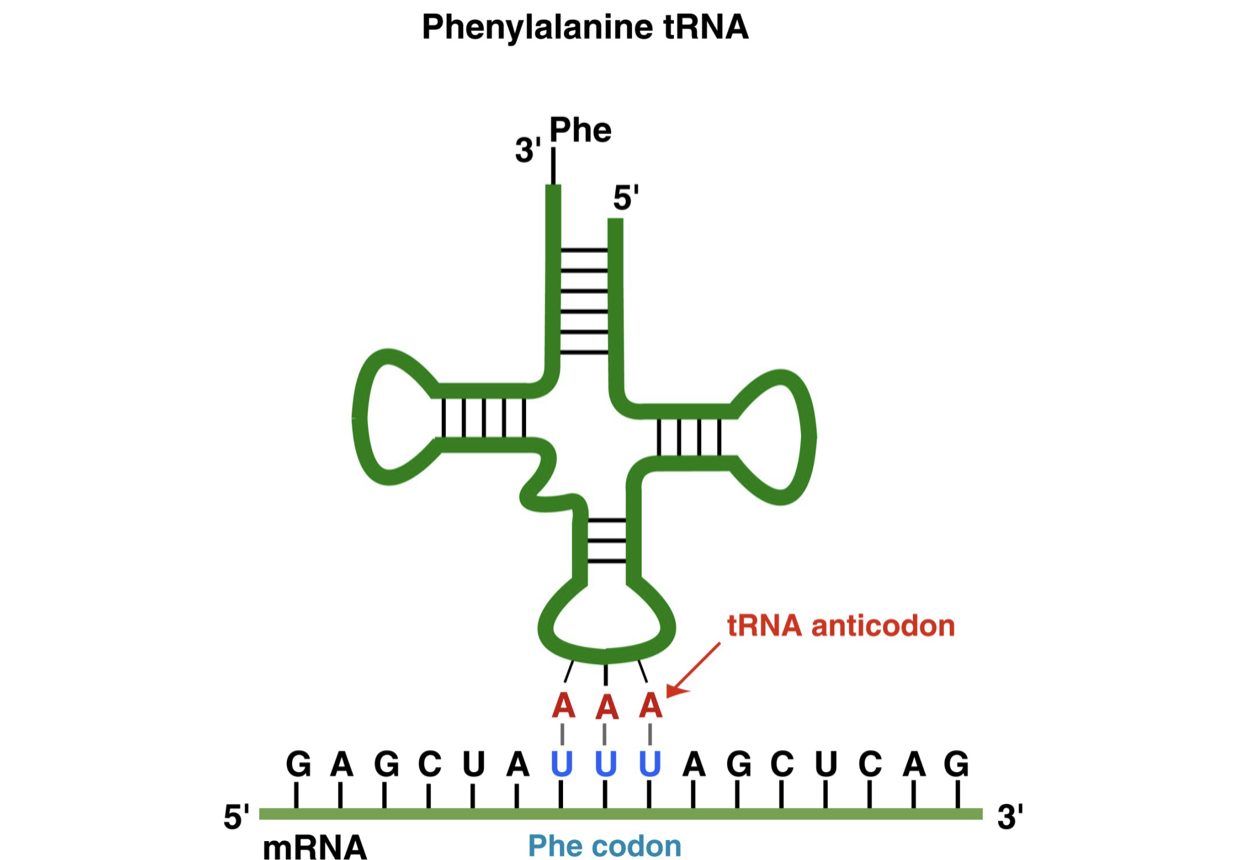
Transfer RNA Role and Structure
Transfer RNA (tRNA)
* acts as an adapter between nucleic acid and protein (float around in cell)
* at least one tRNA for each amino acid (often 1+)
* tRNA for an amino acid contains an anticodon sequence that is the reverse complement of the codon for that amino acid
* each tRNA has unique sequences that causes it to have a unique overall shape
* they all have an amino acid attachment site on the 3’ end
* acts as an adapter between nucleic acid and protein (float around in cell)
* at least one tRNA for each amino acid (often 1+)
* tRNA for an amino acid contains an anticodon sequence that is the reverse complement of the codon for that amino acid
* each tRNA has unique sequences that causes it to have a unique overall shape
* they all have an amino acid attachment site on the 3’ end
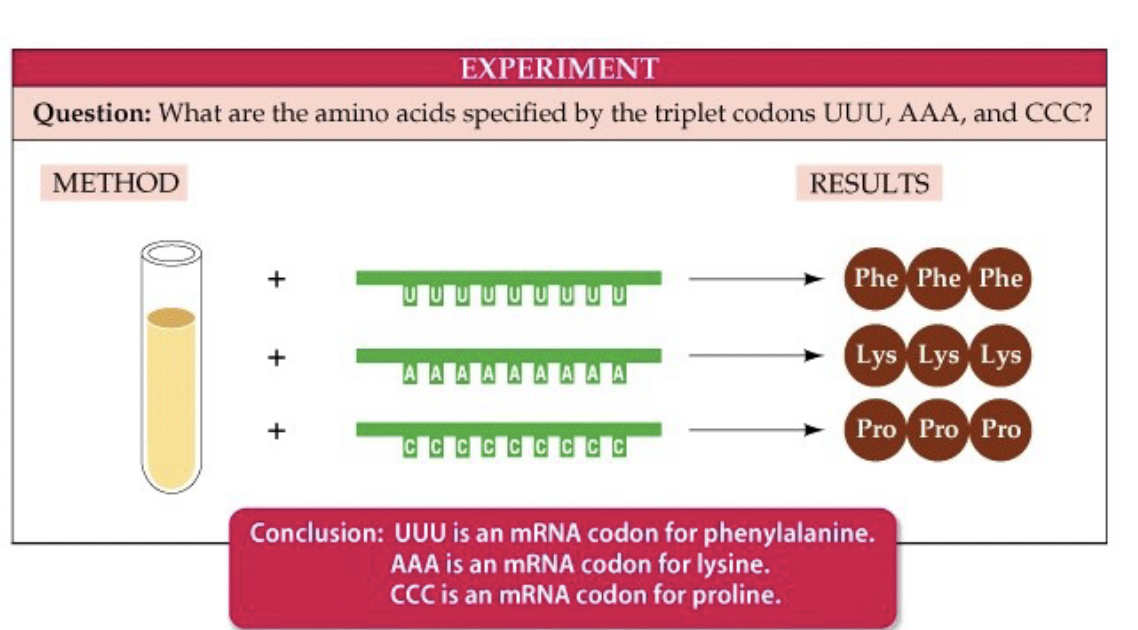
63
New cards
Experiment to figure out genetic code
* test tube has things from a cell that make proteins (ex. ribosomes)
* add a strand of RNA that you made with a repeating sequence ex. AAA, AGA
* analyze the results, which is a protein chain composed of amino acids corresponding to the RNA sequence
* ex. UUUUUUUUUUUUUU → PhePhePhe…
* add a strand of RNA that you made with a repeating sequence ex. AAA, AGA
* analyze the results, which is a protein chain composed of amino acids corresponding to the RNA sequence
* ex. UUUUUUUUUUUUUU → PhePhePhe…
64
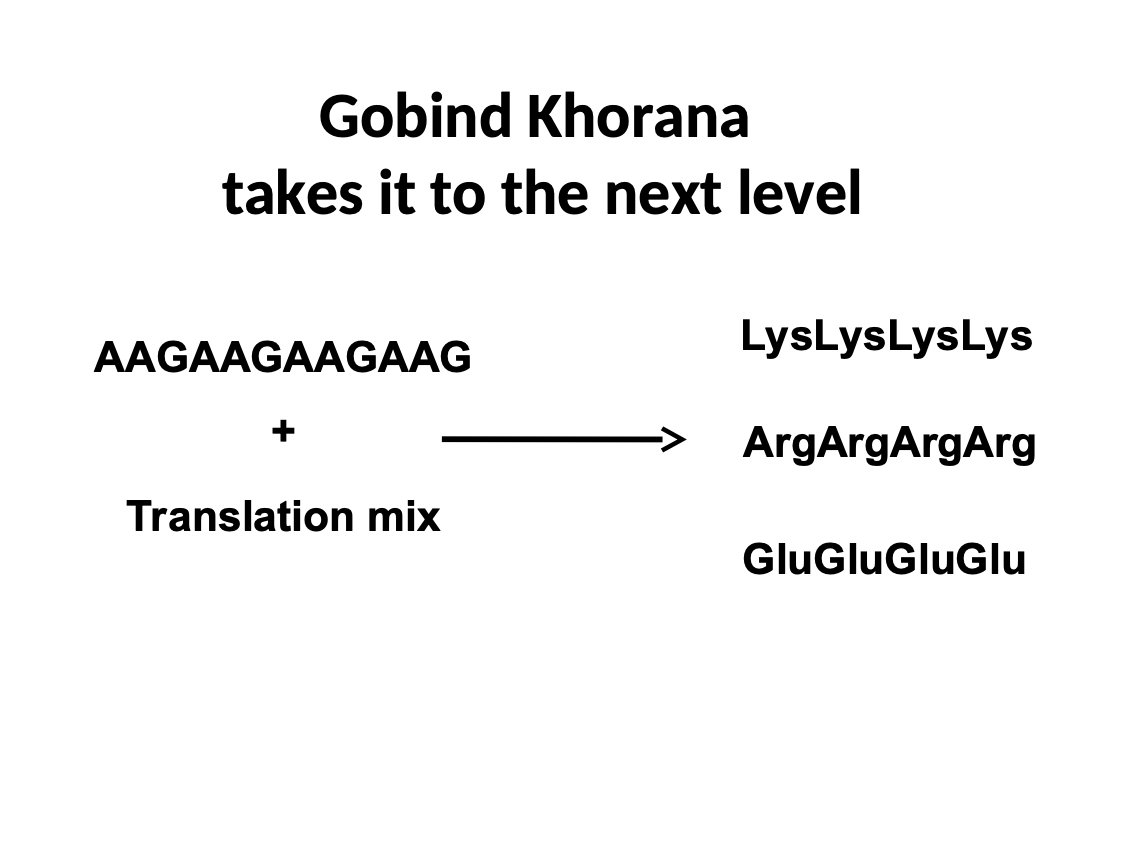
New cards
Reading Frames
* Gobind Khorana realized that a sequence can yield 3 different sequences
* can be read as AAG, AGA, or GAA
* now we know there is a frame for reading the code that can be shifted
* can be read as AAG, AGA, or GAA
* now we know there is a frame for reading the code that can be shifted
65
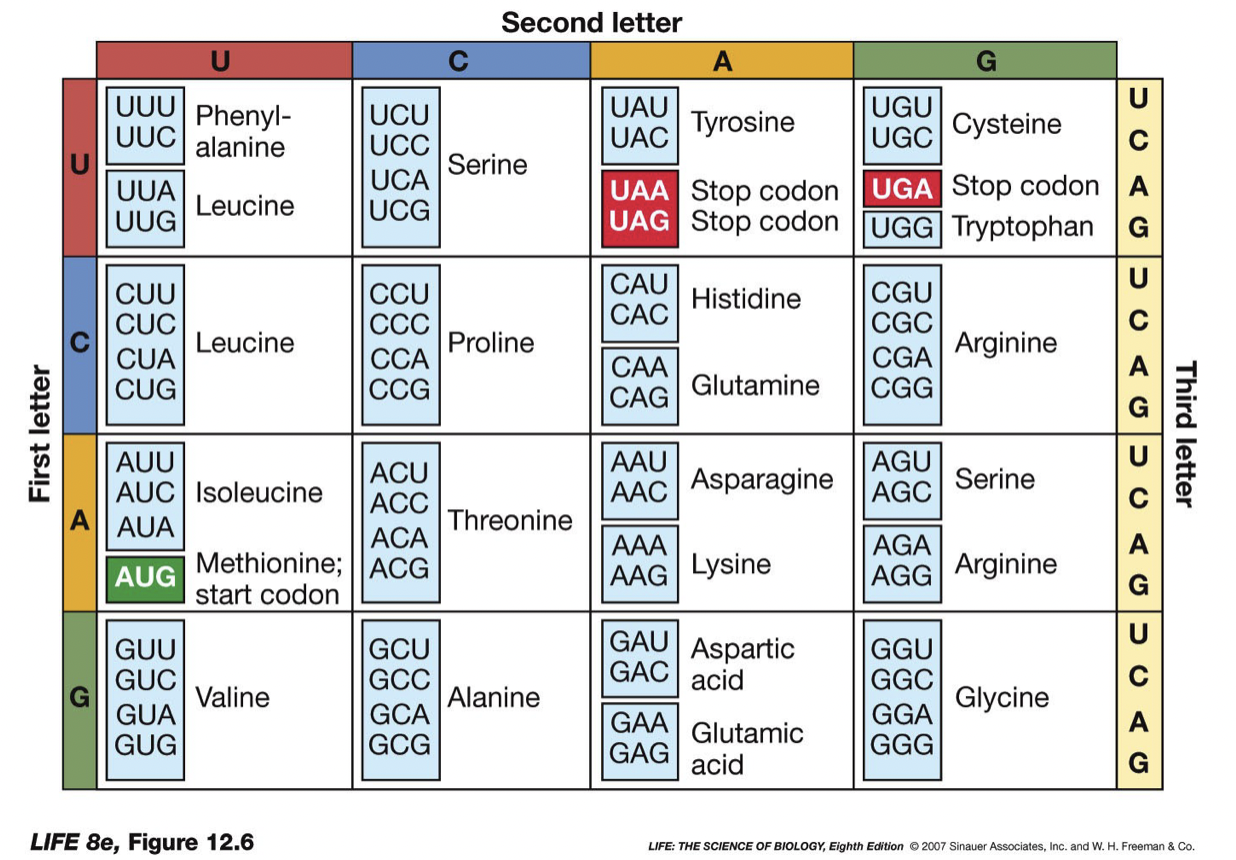
New cards
Codon
* three DNA letter combination
* Stop codon: UAA, UGA, UAG, tell the ribosome the protein is done
* several different codons for the same amino acid, thus the code is said to be degenerate
* Stop codon: UAA, UGA, UAG, tell the ribosome the protein is done
* several different codons for the same amino acid, thus the code is said to be degenerate
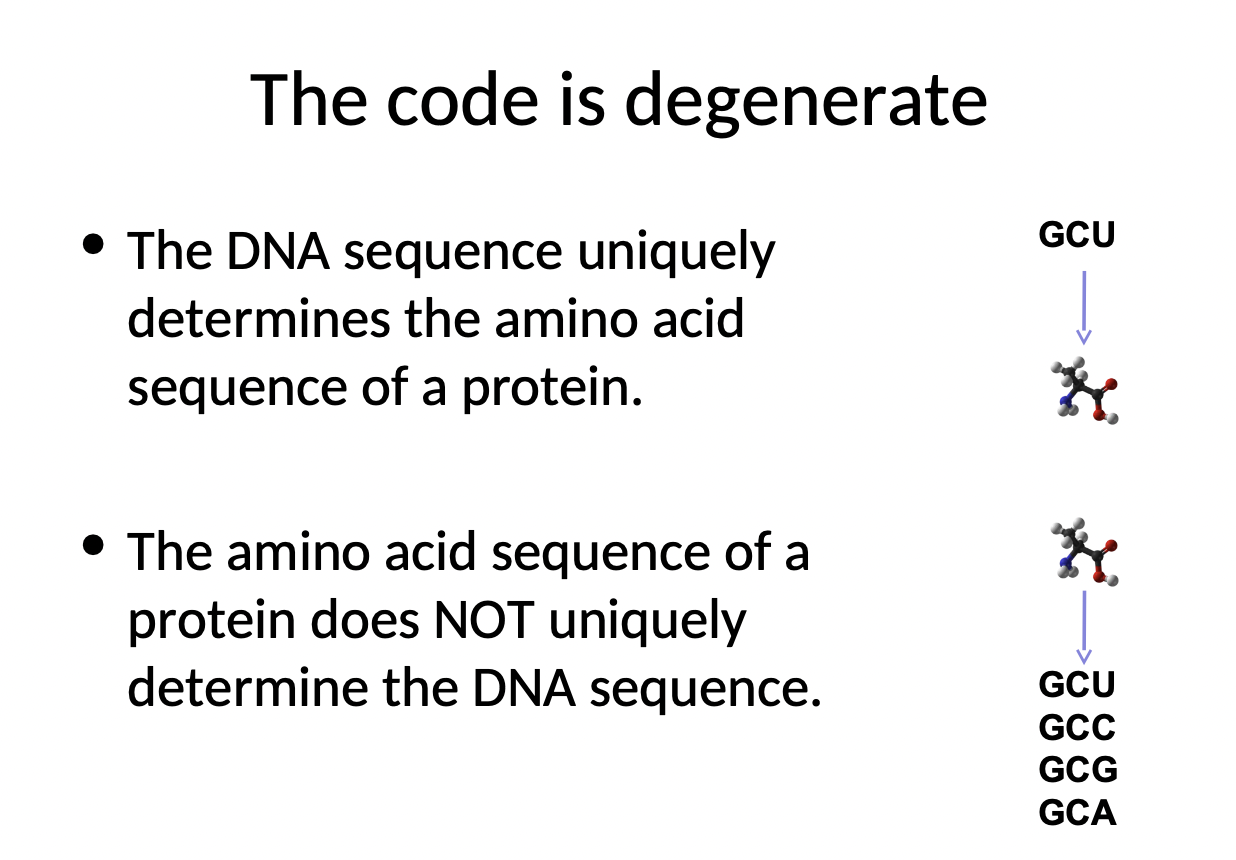
66
New cards
Why is the code degenerate?
* DNA sequence uniquely determines the amino acid sequence of a protein
* amino acid sequence of a protein does not uniquely determine the DNA sequence
* amino acid sequence of a protein does not uniquely determine the DNA sequence
67
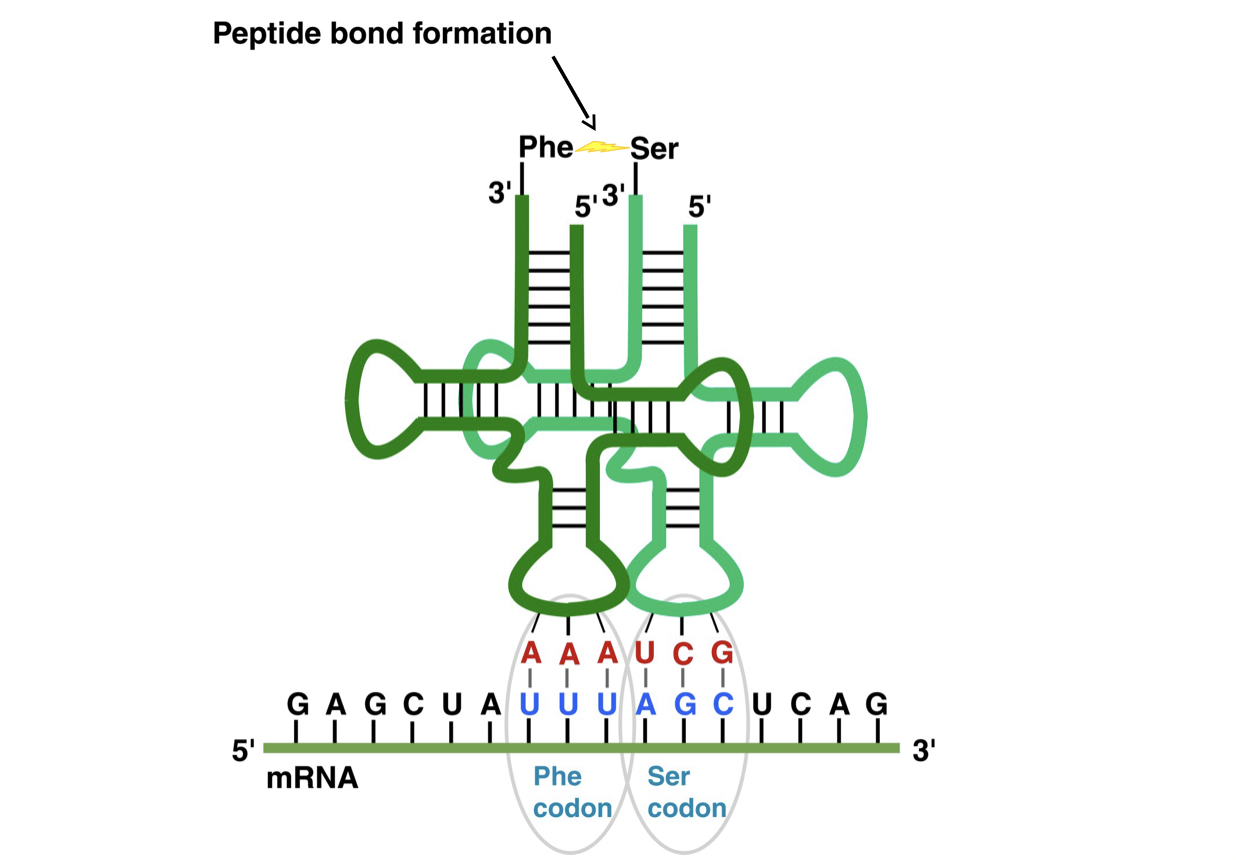
New cards
mRNA and tRNA interactions
* ribosome causes tRNA to sequentially bind to mRNA codons
* the catalyzes the polymerization of the amino acids on the other end of the tRNAs
* creates a string of amino acids, whose sequence is determined by the mRNA sequence
* the catalyzes the polymerization of the amino acids on the other end of the tRNAs
* creates a string of amino acids, whose sequence is determined by the mRNA sequence
68
New cards
Translation
* process of converting the mRNA sequence to the protein sequence
* when the ribosome converts the mRNA sequence with tRNAs into a protein polypeptide
* nucleotide sequence → amino acid sequence
* when the ribosome converts the mRNA sequence with tRNAs into a protein polypeptide
* nucleotide sequence → amino acid sequence
69
New cards
How do amino acids get attached to tRNA?
* class of enzymes, one for each amino, catalyzes the attachment of a specific amino acid to a tRNA
* enzymes = aminoacyl tRNA sunthases
* this is what the code is truly determined by
* enzymes = aminoacyl tRNA sunthases
* this is what the code is truly determined by
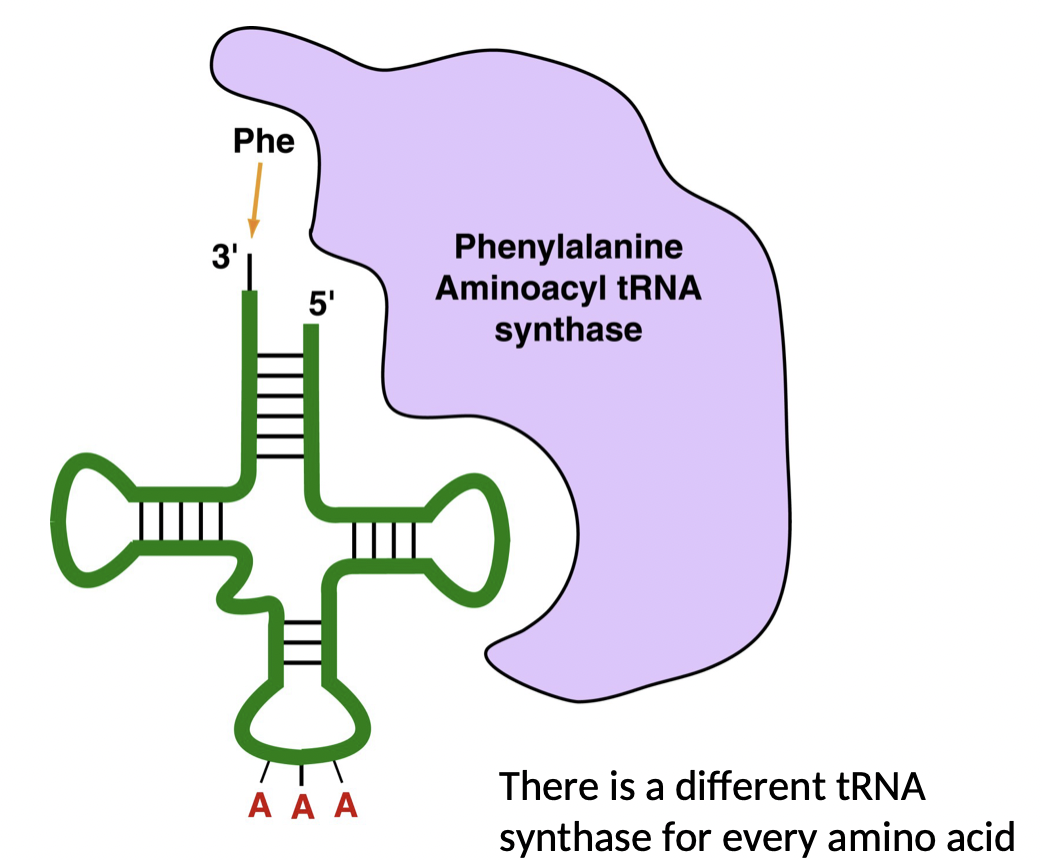
70
New cards
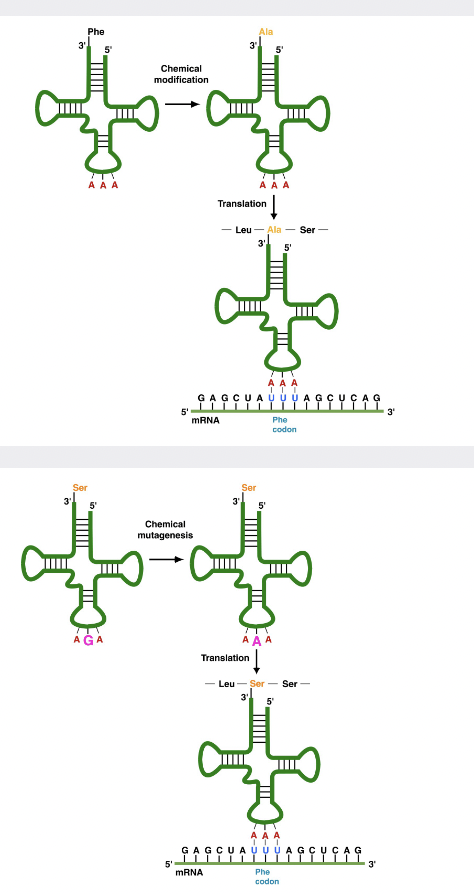
What is the code truly determined by, and how do we know?
* enzymes = aminoacyl tRNA sunthases
* take a tRNA with phenylalanine and chemically convert to alanine
* adding modified tRNAs to a reaction mix with ribosomes, and a known mRNA, the ribosome puts an alanine everywhere there is supposed to be a Phe
\
* also, can mutate the anticodon of tRNA
* ex. codon for serine to codon for Phe
* synthase will still charge the mutant tRNA with serine, but the serine will be incorporated into the protein where Phe should be
* take a tRNA with phenylalanine and chemically convert to alanine
* adding modified tRNAs to a reaction mix with ribosomes, and a known mRNA, the ribosome puts an alanine everywhere there is supposed to be a Phe
\
* also, can mutate the anticodon of tRNA
* ex. codon for serine to codon for Phe
* synthase will still charge the mutant tRNA with serine, but the serine will be incorporated into the protein where Phe should be
71
New cards
Conclusion on Origins of the Genetic Code
* genetic code is arbitrary
* codon used for each amino acid was decided by chance many eons ago and became fixed
* fixed because changing the code in any organism would generate nonfunctional proteins
* code is almost universal, so all organisms must have a common ancestor billions of years ago with the same genetic code that we use today
* codon used for each amino acid was decided by chance many eons ago and became fixed
* fixed because changing the code in any organism would generate nonfunctional proteins
* code is almost universal, so all organisms must have a common ancestor billions of years ago with the same genetic code that we use today