Genetics: Bacterial Gene Control
1/117
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
118 Terms
constitutive gene expression
genes that are always active, their product is in constant demand
regulated gene expression
- their products are rarely needed
- only expressed when needed
genes are hardly ever active
operons
genes that coordinate the regulation of gene expression
polycistronic and operons
several genes transcribed from same promoter and regulated by the same conditions
make products that function in similar ways
structural genes
- genes that are to be transcribed
- may be enzymes or structural proteins
- these genes are occasionally needed
Promoter (P)
- site that is a fixed # nts upstream
- -35, TATA, etc boxes located here
- place RNA polymerase recognizes
- remember: all genes have a promoter-- in operon or not
Operator Site (O)
- site that is fixed in place, near or adjacent to the initiation site of operon structural genes
- site which determines if transcription will occur
- may be more than 1 present
what a protein regulator will bind to
Regulator (R)
- protein that acts at the operator site to regulate transcription
- gene encoding regulator does NOT have to be near operon genes
- regulators may be represented with other letters/ info
Operon organization
1. inducible
2. repressible
both of these can have positive and neg regulation
for repressible mostly neg
positive regulator recruits RNA polymerase by:
1. direct interaction
2. making promoter accessible
Negative regulation blocks RNA polymerase by:
- positive and negative regulation describe effect of regulator on transcription
- inducible system can have both types of regulation
inducer molecules cause
transcription in both regulation types
positive regulation on gene x without inducer
activator cannot bind DNA --> no transcription
positive regulation on gene x with inducer
activator and inducer bind and are able to bind to DNA which allows for transcription to happen --> mRNA
negative regulation on gene Z without inducer molecule
the repressor remains on gene Z and no transcription occurs
negative regulation on gene Z with inducer molecule
inducer molecule binds the repressor so it cannot bind to DNA
Gene Z continues on with transcription and mRNA is made
inducible systems: keyed to substrates (Gene X)
Gene X - often encodes catabolic enzyme
Substrate serves as inducer of gene X expression
Repressible systems: keyed to end products (Gene Z)
Gene Z - often encodes anabolic enzyme
End product of enzyme serves as a repressor of gene Z expression
lac operon
- first operon to be discovered
- inducible
- contains 3 structural genes > each encode for an enzyme that allows the cell to use lactose as an energy source
lac operon expression
genes are expressed unless repressor is bound at the operator
if E. Coli are grown in the presence of glucose
then the glucose is suddenly replaced with lactose
E.coli in which glucose is replaced with lactose
- cells will seem unable to metabolize lactose for short time
- lag in growth, cells appear unable to survive
- during this time the cells are turning on the lac operon
3 critical genes of lac operon
Z- ß-galactosidase
Y- permease
A- transacetylase
Z- ß-galactosidase
breaks lactose into 2 monosaccharides (glucose and galactose)
Y- permease
transports lactose into cells
A- Transacetylase
transfers an acetyl group from acetyl-CoA to galactosides; prevents buildup of toxic product of ß-galactosidase
LacI+ gene
encodes the operon regulator protein
Protein made from lacI+
bound to operator unless inducer molecule is present (lactose)
what kind of regulation is lac operon under
positive and negative
negative regulation of lac operon
implies something is turning off gene transcription by binding at a control element
what is the repressor of lac operon
protein encoded by regulatory site lacI
in the absence of lactose the Lac operon is
mostly off
repressor is bound at the operator when lactose is not present
lac operon expression on vs off
on: lactose present and glucose is not
off: glucose is present
homotetramer repressor
- can bind 2 of 3 operator sequences at once (O1, O2, O3)
- if bound to O1 and O3, intervening DNA forms a repressor loop
maximum repression of lac operon occurs when
all 3 O sites are bound
- lacO2 is at +412
which gene is expressed at low levels constitutively in lac operon
repressor gene
- has weak promoter
- a few repressors always bound
- repressor molecules bind and unbind
- even without inducer present, RNA pol. could initiate transcription before another repressor binds
what must happen to repressors for operator to turn an operon on
they must be prevented from binding operator
lac operon in presence of lactose
allolactose binds the repressor
allolactose acts as what
the inducer molecule
allolactose binds the repressors:
- some lactose is converted by ß-galactosidase into allolactose
- binds repressor at operator, changes its shape
- also binds free repressors
what is more efficient for the cell to use? lactose or glucose?
Glucose
Energy for biochemical reactions comes from
Glucose
operon is at basal level unless
something intervenes to turn it on
- neg control is like a brake
- removing brake is not enough to activate expression
How does E. Coli keep lac operon at basal level if glucose is present
- influence of a breakdown product, a catabolite of glucose metabolism
- called catabolite repression or glucose effect
what is the best positive controller of the lac operon
a substance that senses the lack of glucose and activates the operon
lac operon: catabolite repression with cAMP
- cAMP formed from cytosolic ATP by adenyl cyclase
- cAMP has numerous roles in signal transduction pathways
- Glucose inhibits adenyl cyclase
- cAMP has a crucial role in catabolite repression
-↑ [glucose] results in ↓ [cAMP]= basal lac operon transcription
what happens if cAMP is added to bacteria
it can overcome catabolite repression of the lac operon (even in the presence of glucose)
cAMP also has a role in positive control of
gal and ara
process of positive control is complex, what are the two parts?
1. cAMP
2. protein factor
CAP
catabolite activator protein
also called cAMP receptor protein
gene name of CAP
crp
Mechanism of CAP and cAMP action
- When present in high amounts, CAP and cAMP associate
- This complex binds just 5 ́ of operon promoter
- Now RNA pol can strongly recognize promoter and considerable transcription takes place
The CAP binding sites in lac, gal, and ara operon promoters all contain what sequence
TGTGA
What sequence is important for the CAP-cAMP complex binding
TGTGA
Operons activated by CAP and cAMP have remarkably weak promoters
- their -35 boxes are not very similar to consensus sequences
- almost not recognizable
Why does weak promoter at lac operon make sense
> constitutively, basal expression level
- glucose levels drop, cAMP rises
> cAMP associates with CAP, complex
- makes stable RNA polymerase promoter complex
- increased production of lactose digesting enzymes
what does it mean that the lac operon is still under operator control
if repressor binds, basal level occurs
FINAL regulation of lac operon depends on what
presence of inducer:
appropriate inducer specifically selects which operon will work
lac operon: cAMP present and lactose present
CAP and cAMP are bound together, lactose makes the repressor inactive
>> binding of RNA pol is facilitated by CAP and transcription occurs
lac operon: cAMP present and lactose absent
CAP is bound to cAMP but without lactose the repressor is active and binds >> transcription is blocked by represor
lac operon: cAMP is absent and lactose is present
CAP is inactive and repressor is inactive >> transcription inhibited by lack of CAP
lac operon: cAMP is absent and lactose is absent
CAP is inactive and the repressor is active >> transcription inhibited by lack of CAP and the presence of repressor
lac operon mRNA translation
- structural genes in lac operon close together
- once ribosome is loaded onto transcript, it does not release until all 3 genes are translated
- once ribosome reaches UGA of one gene, it stays on long enough to reach AUG of next gene
nonsense mutation in structural gene
- ribosome will not reach next AUG
- genes following nonsense mut. will not be translated
making stop codon mutation where one shouldn't be
polar mutations (polar effects)
always only affect downstream
missense mutations
point mutation, changes one amino acid into another amino acid
Conjugation: the F-plasmid
Physical contact between 2 cells is essential*
• Contact is the initial step
- established by structure called F sex pilus
- F+ cells have cell extensions covering external surface = F-pili
- Once a pilus is in contact with F- cell, it elongates and becomes an F sex pilus
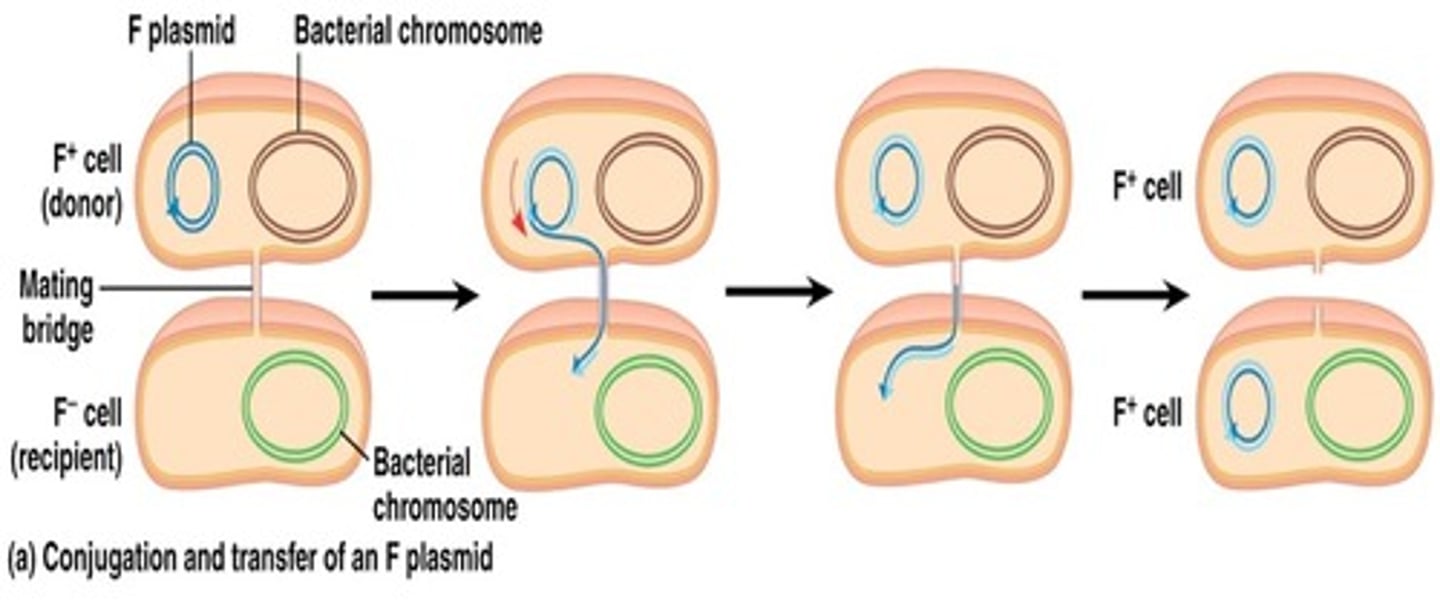
Conjugation of the F plasmid
a copy of the F factor is almost always transferred from F+ to F- cell, converting it to F+ state
F' Factor (lac)
plasmid formed of the F factor + bacterial gene that loop out of the chromosome
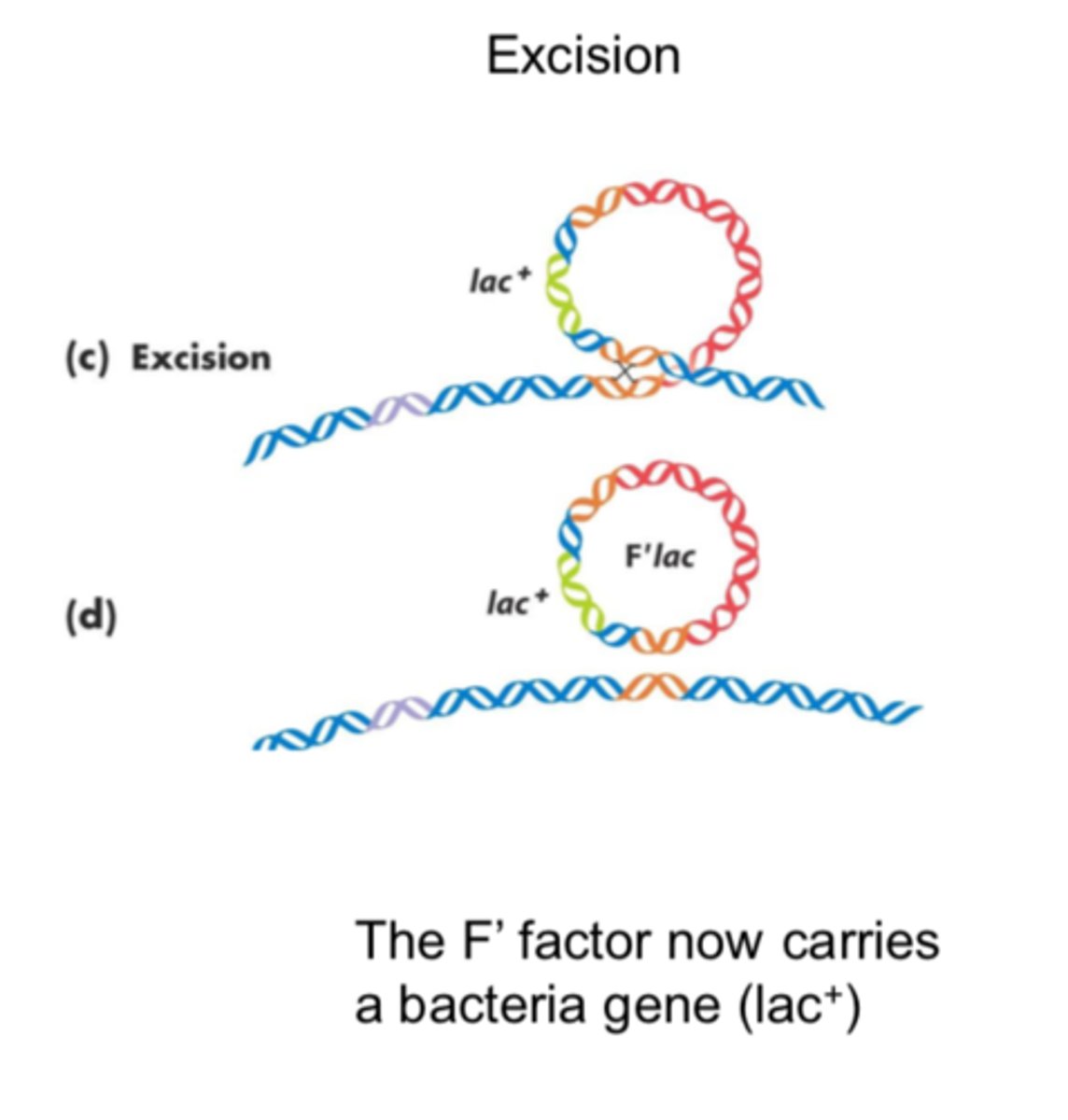
transfer of an F' factor during conjugation
carries some of the same genes already in the recipient's chromosome
merozygote or partial diploid
a partial diploid strain of bacteria containing F' factor genes
mutations that change the operator sequence so repressors cannot bind
- results in constitutive expression of the structural genes
- called operator-constitutive mutations (lacO^c)
- if partial diploid has a lacO^c and lacO+
>> only lacO^c will cause constitutive expression of struct genes
>> other copy expressed normally
>> called cis-dominant mutation
LacO^c changes the operator sequence so
repressor cannot bind
LacI- changes the regulator sequence so
repressor cannot bind operator
also causes constitutive operon expression
partial diploid with lacI and lacI-
- WT regulator gene makes WT repressor
- WT repressor can bind both copies of operator
- lac+ regulator sequence can overcome lacI- mutation
- lacI+ gene has trans-acting function to lacI- gene
superrepressor lacI^s changes the regulator sequence so
regulator can bind operator but not allolactose
once the lac^s repressor binds to operator
it cannot be induced to fall off
- results in basal transcription level even in presence of lactose
Superrepressor is trans-acting protein which means that
LacI^S dominant to LacI+
lacP changes the promoter so
RNA polymerase cannot bind
- -10 or -35 box mutation
- cis-acting function
LacI^d changes the regulator sequence so
subunits cannot form complete repressor
- repressor has homotetrameric structure
in partial diplods that are lac^-d/ lacI+
- if 1 or more subunits are lac^-d then, repressor will not form native confirmation
- repressor function will be lost
- ~12 repressor molecules in cell, so 1 mutant subunit blocks normal operator binding
- lacI^-d subunits trans-dominant to lacI+ subunit
trp operon
- contains the genes that code for enzymes that E. coli needs to make amino acid tryptophan
- negatively controlled by a repressor
Fundamental difference between lac and trp operons
- lac operon codes for catabolic enzyme (break down substance)
- such operons are turned ON by the substance (like lactose)
- trp operon codes operon codes for anabolic enzymes (builds up a substance)
- such operons are turned OFF by substance made (like tryptophan)
trp operon: genes
operon codes for 5 polypeptides that form 3 functional enzymes
- termed gene trpE, D, C, B, A
Trp E and trp D
code for polypeptides that make first enzyme
trp C
codes for single polypeptide enzyme that catalyzes intermediate steps
trp B and trp A
code for 2 polypeptides of enzyme that catalyzes last 2 steps
where does the trp operon lie
within the promoter region
the lac operon operator was where?
adjacent to the promoter
the presence of Trp means
the operon needs to shut down
- Trp acts like a corepressor
corepressor
it is half the functional repressor molecule
aporepressor
Trp can bind to a protein
aporepressors need whag\t
Trp to function
aporepressor + corepressor
binds operator and shuts off operon
low Trp concentration
inactive repressor allows transcription
high Trp concentration
active repressor complex binds to operator and prevents transcription
Trp bound to aporepressor forms active complex
causes conformational change in aporepressor to allow DNA binding
trp operon is
repressable
very common in prokaryotic a.a. synthesis pathway
what is the problem with this type of repression (repressable)?
- repressor/operator association is weak
- even at high [Trp], operon is still working at appreciable level
A second regulation mechanism:
ATTENUATION