Eukaryotic Gene Expression 1: Transcriptional Control
1/23
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
24 Terms
T/F: Different cell types synthesize different sets of RNAs
True
Many processes are common to all cells. Any two cells in an organism will have common gene products
Ex. B actin for cytoskeleton
What is an example of the level of expression of most genes varying by cell type?
Tyrosine aminotransferase gene is highly expressed in the liver only
What are the 4 key concepts of different cell types synthesizing different sets of proteins?
There are many proteins in common with any two cells [“housekeeping genes”]
Some proteins are more abundant in specialized cells [ex. hemoglobin only detected in RBCs]
Level of mRNA expression of every active gene varies from one cell type to another
Level of protein expression varies per cell type
T/F: A cell can change the expression of its genes in response to external signals
True
A cell can change the expression of its genes in response to external signals
Ex. glucocorticoid
How do different mammalian cells respond to glucocorticoid?
Liver cell - increase in production of glucose from amino acids [gluconeogenesis]
Fat cells - reduce production of certain proteins
Other cells - no response
Why do eukaryotic cells have more opportunity for regulation than bacterial cells?
Regulation of transcription in eukaryotic cells is VERY COMPLEX
Proteins can bind far from enhancers and still influence the promoter
RNA pol II. requires many GTFS and regulation steps (initiation, elongation, termination)
Packaging of eukaryotic DNA into chromatin introduces more regulation
Activators recruit coactivators to enhance gene transcription, repressors recruit corepressors to reduce gene transcription
What kind of structure does the eukaryotic activator protein have?
A modular structure. Activators are modular in design and have distinct DNA binding domains separate from the activation domain. Activation domains function to stimulate RNA pol II. transcription.
What two components do the activation domains interact with?
GTFs/RNA Pol II
Coactivator complexes
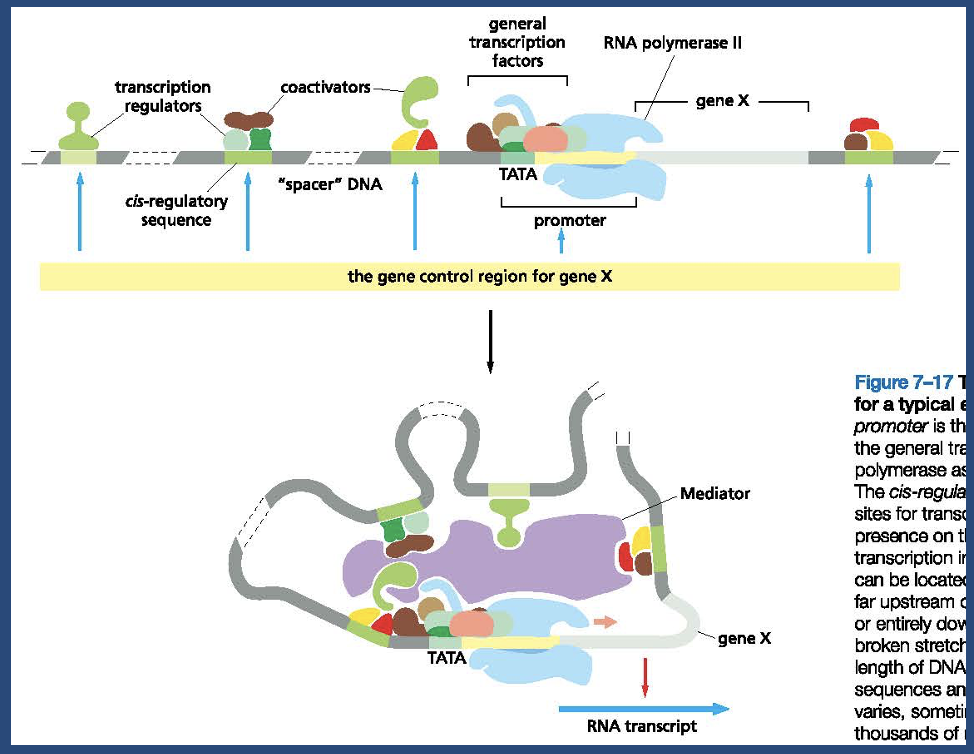
The control region for a typical eukaryotic gene
The main points about this figure
The promoter is where GTFs and polymerase assemble
The cis regulatory sequences are binding sites for transcription regulators
cis regulatory sequences will be adjacent to the promoter upstream or downstream
TATA box is the recognition sequence for TFIID
DNA looping allows transcription regulators to interact with the proteins at the promoter
T/F: Mediator and the GTFS are the same for all RNA pol II. transcribed genes
True
The locations of the binding sites relative to the promoter are different for each gene
What does acetylation do to the nucleosome?
Opens up the nucleosome
3 basic qualities of coactivators
Do NOT bind directly to DNA regulatory sequences, recruited to genes by binding a transcriptional activator
Often act in very large, multi subunit protein complexes
Can stimulate RNA Poll II transcription
What are the three ways coactivators can stimulate RNA pol II transcription?
Act as a bridge between the activator and GTFs/RNA poll II at the promoter (Mediator complex)
Having enzymatic activity that targets the histone tails in the nucleosome (ex. SWI/SNF chromatin remodeling complex)
Recruiting additional coactivators - acting as a scaffold (CBP recruiting another HAT called PCAF)
When would no coactivators be recruited?
If an activator protein only bound DNA minus its activation domain, no coactivators would be recruited and gene transcription would be minimal
How do eukaryotic transcription regulators work in groups?
2 TFs bind to DNA cooperatively
A protein dimer can create a surface that is recognized by a coregulator that carries either an activator or repressor function
eukaryotic genes are regulated by a committee of proteins
What kind of RNAs can act as scaffolds to stabilize a coregulator - transcription factor complex?
Long non coding RNAs
What is the main point to remember about transcription regulatory protein function?
Transcriptional regulatory proteins must be bound to DNA either directly (by activators) or indirectly (by coactivators) to influence target gene transcription
What does the rate of transcription of a gene ultimately depend on?
The spectrum of the regulatory proteins bound upstream and downstream of its transcription start site
What are the inputs that work together to influence overall transcriptional initiation rate at a given gene promoter?
Multiple sets of TFs
coactivators and corepressors
The nucleosome structure promotes what type of binding of transcription regulators?
Cooperative binding
Why do transcriptional regulators bind to DNA in nucleosomes with much lower affinity than they do DNA?
More accessibility
Cooperative binding
One transcription factor destabilizes nucleosome making it easier for next factor to bind
Transcription factors that do this are called pioneer factors
How to eurkaryotic transcription activator proteins direct local alterations in chromatin structure?
Activators can promote transcription initiation by recruiting factors that change chromatin structure and create histone post-translational modifications (“marks”) present on gene promoters
What are the three ways eukaryotic transcriptional activators modify local chromatin structure? What are some of the recruited coactivators?
Nucleosome sliding, removal, and replacement by ATP dependent chromatin remodeling complexes
Writers of covalent histone PTMs
Readers of covalent histone PTMs
An activator protein can recruit all of these activities, but in a temporal or kinetic fashion