Genetics Lab Midterm (From the powerpoints, and some math rules/tips)
1/100
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
101 Terms

I love you Pookie
Blondie


What is the gender of the flys shown on the left of each image?
Female. Female fruit flies are typically larger and have longer abdomens. There abdomens are light with smaller darker bands.

What is the gender of the flys shown on the right of each image?
Male. Male fruit flies are typically smaller, and have shorter abdomens. They have have darker bands, and have a darker bottom due to genital arch.
What is the wildtype of Drosophila melanogaster?
Brick-red eyes and normal wings
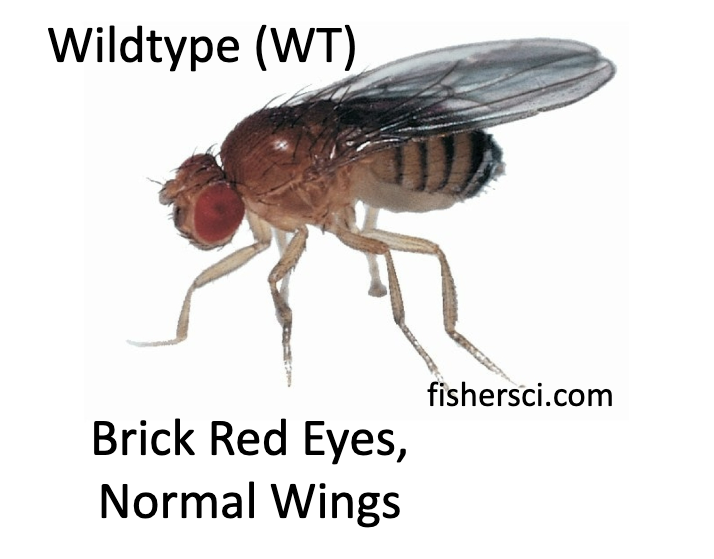
What are the two mutant-trait phenotypes Drosophila melanogaster can have for there eyes?
White eyes and Sepia (or brown eyes).

What does vestigial phenotype for Drosophila melanogaster?
It means the wings are crumpled and ineffective.

In the fly system, what symbol is used to denote the wild type allele?
A plus symbol, +.
For the wildtype phenotype, what is the genotype?
+/+
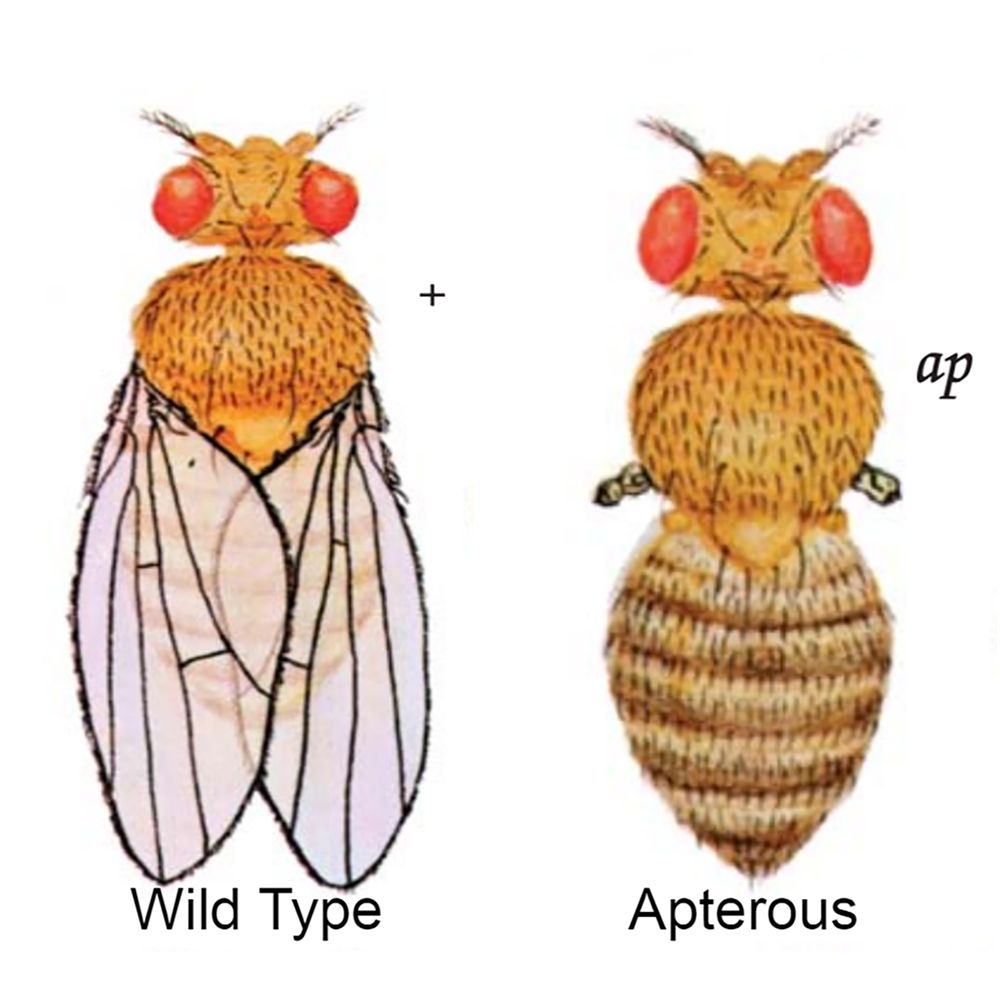
For the apterous phenotype, what is the genotype, part of the body effected, chromosome affected, and sex linkage?
Genotype - ap/ap
Body location - wing
Chromosome - chromosome 2
Sex linkage - not sex linked
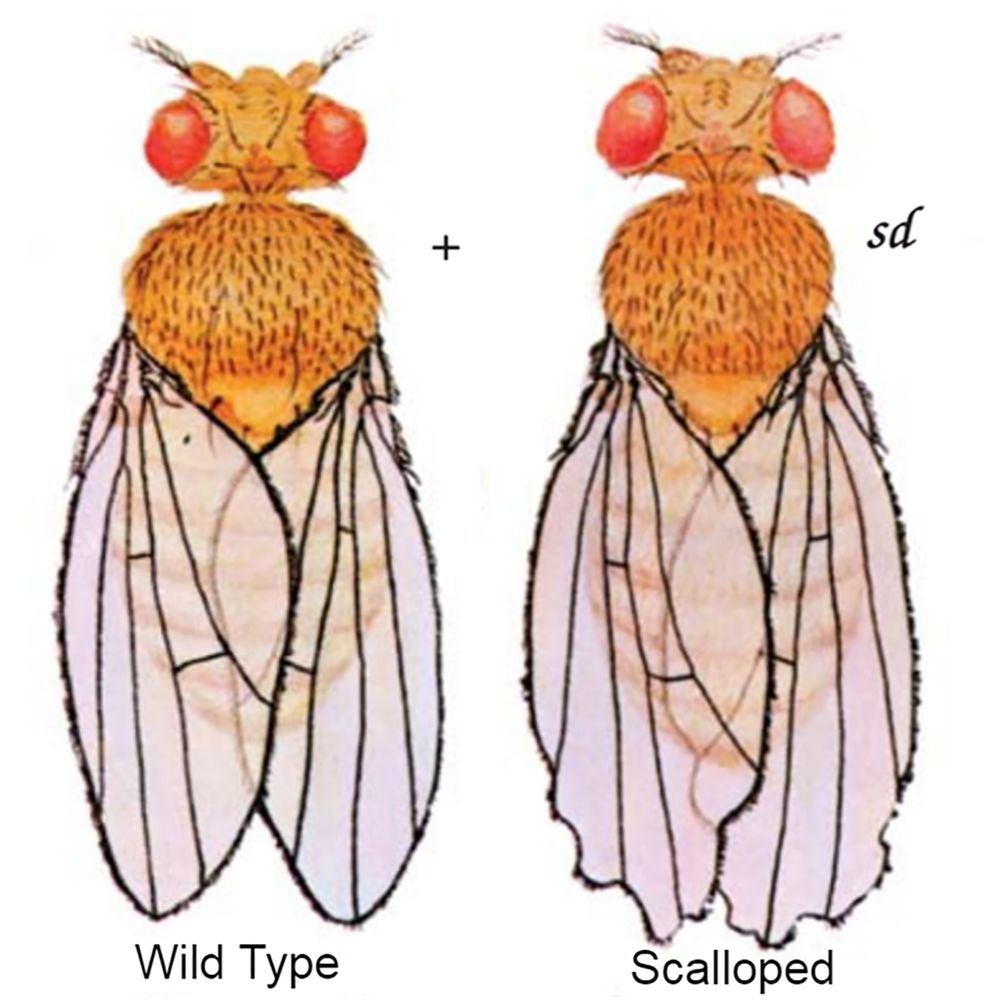
For the scalloped phenotype, what is the genotype, part of the body effected, chromosome affected, and sex linkage?
Genotype - sd/sd
Body location - wing
Chromosome - chromosome 1 (X)
Sex linkage - is sex linked, x chromosome
For the sepia phenotype, what is the genotype, part of the body effected, chromosome affected, and sex linkage?
Genotype - se/se
Body location - eye
Chromosome - chromosome 2
Sex linkage - not sex linked

For the vestigial phenotype, what is the genotype, part of the body effected, chromosome affected, and sex linkage?
Genotype - vg/vg
Body location - wing
Chromosome - chromosome 2
Sex linkage - not sex linked

For the white phenotype, what is the genotype, part of the body effected, chromosome affected, and sex linkage?
Genotype - w/w
Body location - eye
Chromosome - chromosome 1 (X)
Sex linkage - is sex linked, X chromosome

For the yellow phenotype, what is the genotype, part of the body effected, chromosome affected, and sex linkage?
Genotype - y/y
Body location - body
Chromosome - chromosome 1 (X)
Sex linkage - is sex linked, X chromosome

The probability of two or more independent events occurring at the same time (simultaneously) is?
The product of each their individual probabilities.
For two independent coin flips, the probability that they are both heads is ½ x ½ = ¼ .
The individual probability for a single coin to land on heads is 1/2, multiple the individual probabilities of both together, and you have the probability that both coins will be heads when flipped at the same time.
The probability of either one or the other, or two mutually exclusive events occurring is?
The sum of their individual probabilities.
An example is: “What is the chance when you flip a coin it will land on hears OR tails?”
The individual probabilities are ½, so the total probability is ½ + ½ =1.
Keyword is or.
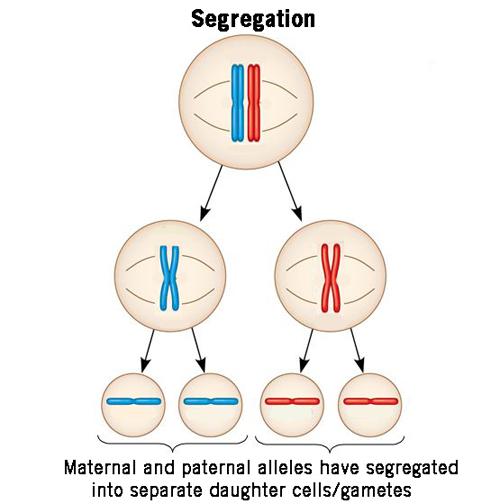
What does Mendel’s law of segregation state?
The law of segregation states that the two copies of a gene (the alleles) separate during meiosis, meaning the daughter gamete cells randomly receive only one of the alleles.
What does Mendel’s law of independent assortment state?
The law of independent assortment states that during meiosis (gamete formation), the alleles of two (or more) different genes segregate independently of each other. Essentially, the alleles of different genes get sorted into gamete cells independently of one another.
The gene pairs segregate independent during meiosis if they are not genetically linked by being near one another on the same chromosome.
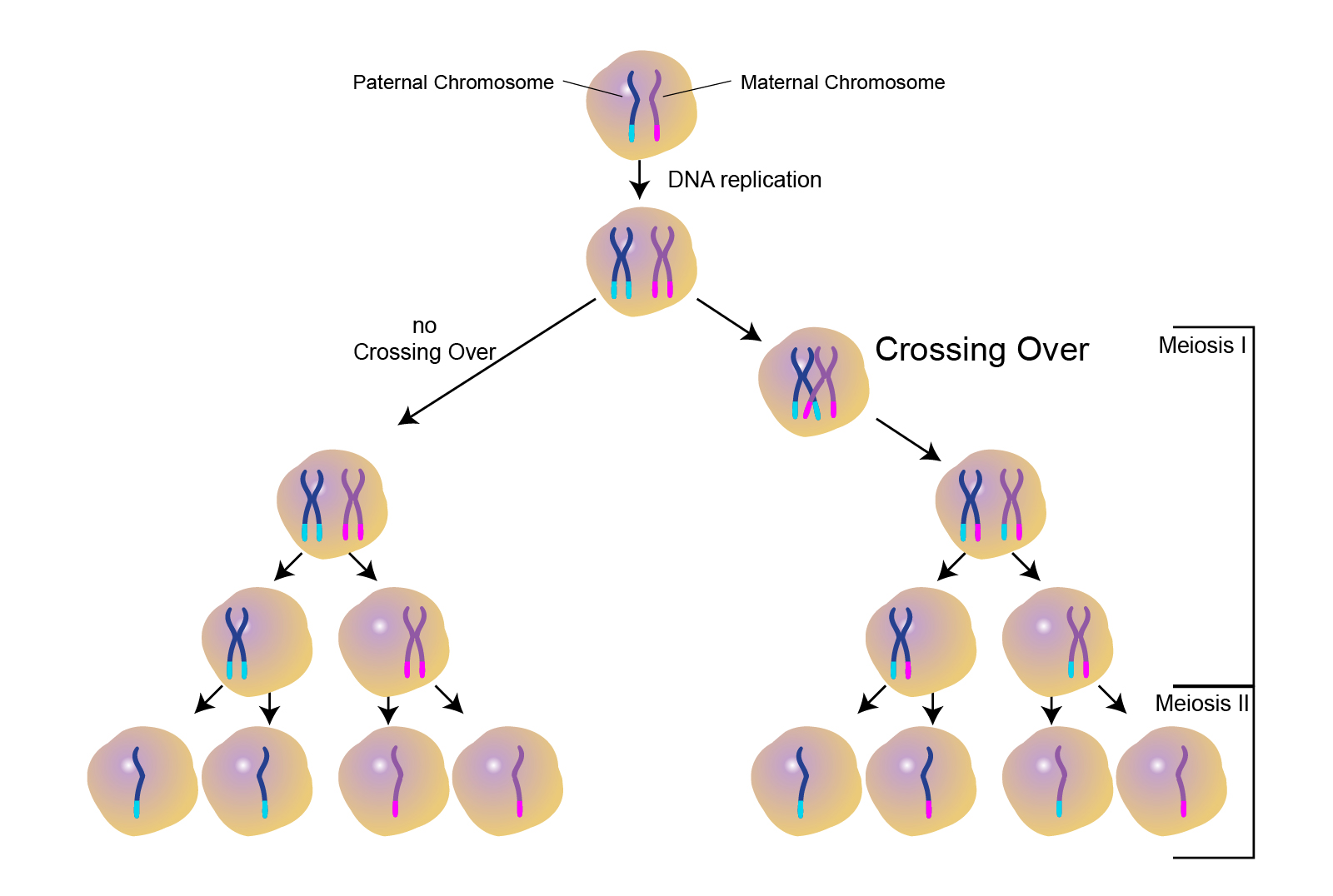
What is chromosomal crossover?
Crossover is the exchange of genetic material between two homologous chromosomes that “cross over”, touch, and exchange genetic material.
Crossover occurs during prophase 1 of meiosis.
Increases genetic variation.
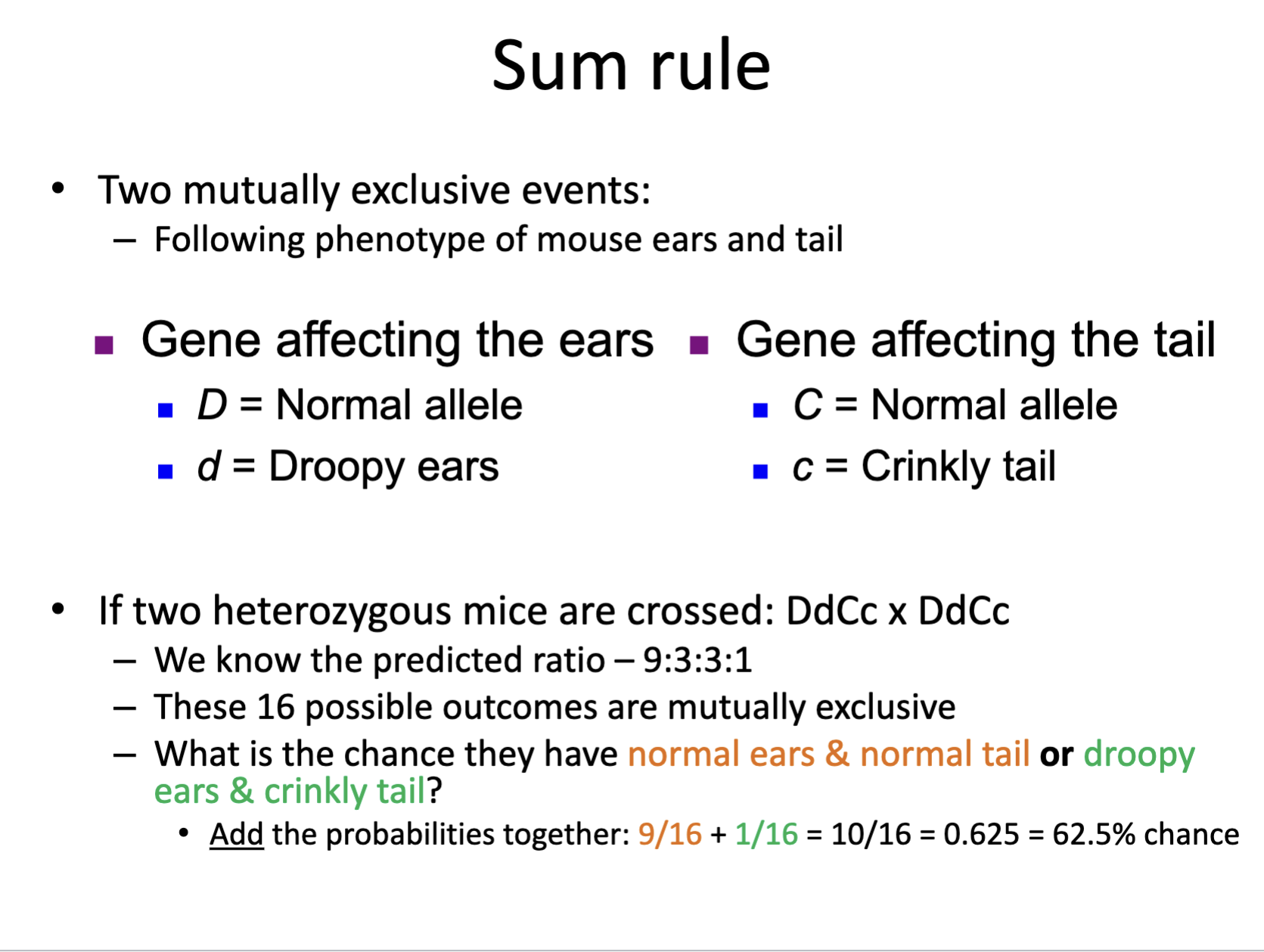
What is the OR rule? It is also called the Sum rule.
The OR/sum rule is used to determine the probability that two mutually exclusive events occur at the same time.
The sum rules sums (adds) together the independent probabilities of each mutually exclusive event:
½ + ½ = 1
What does mutually exclusive mean?
Mutually exclusive means that two or more events cannot happen simultaneously.
An example would be crossing two different genotypes for two different traits, and producing 16 offspring. The 16 offsprings combined genotypes are mutually exclusive, so the probability they would have both genotypes (both traits) would be the sum of the offspring that could have trait ones genotype plus the number of offspring that could have trait twos genotype.
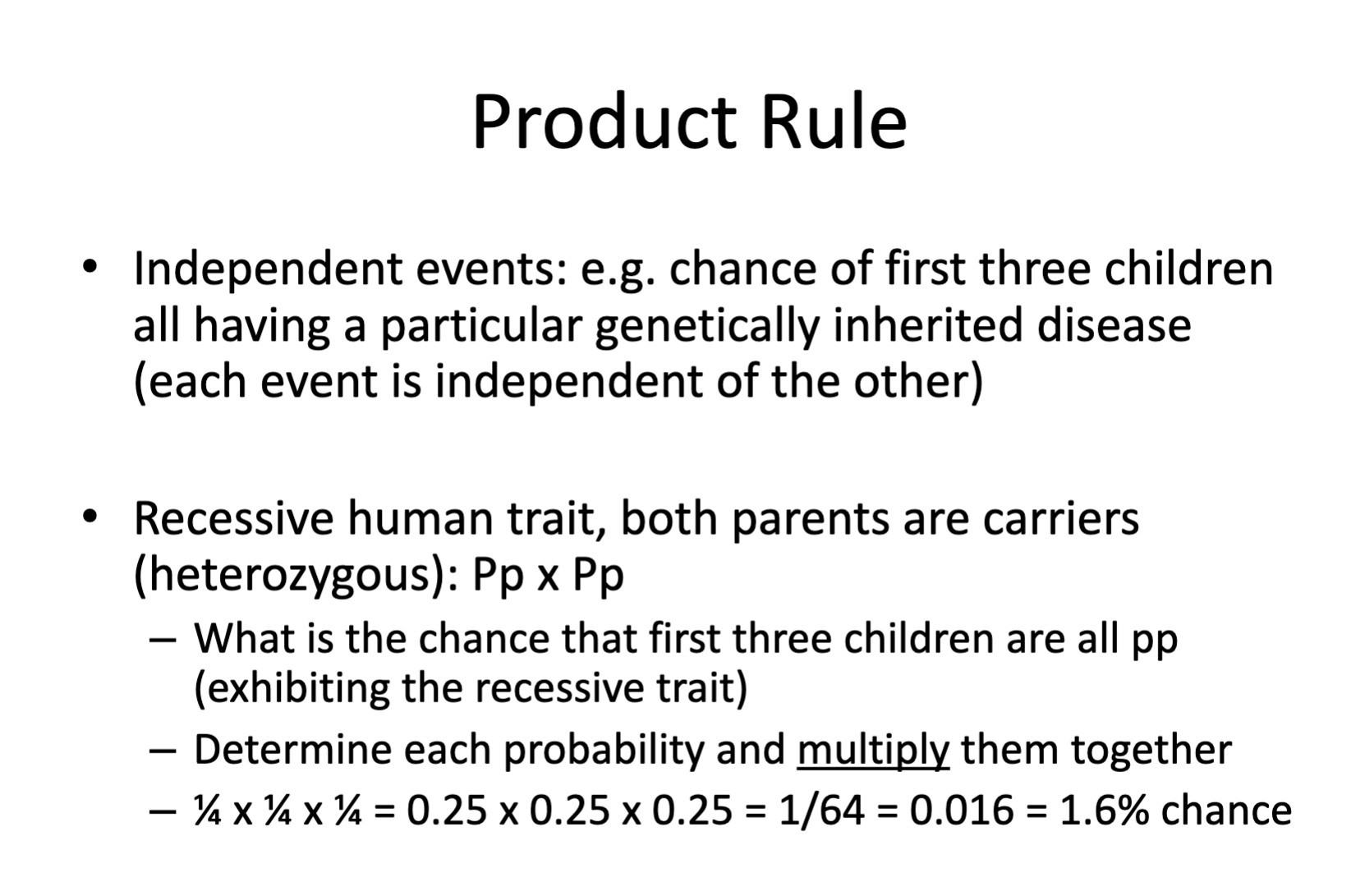
What is the AND rule? It is also called the Product rule.
The AND/Product rule is used to determine the probability that two independent events occur simultaneously at the same time. You multiple the individual probabilities of each independent event by each other, to determine the probability both events will have at the same time.
An example would be the first born child (event one) having a genotype of AA (event two).
½ x ½ = ¼
¼ is the probability both independent events will occur at the same time.
What is the major difference between the OR and AND rule?
The OR/sum rule is used to calculate the probability of one or more events are occurring, used when the events are mutually exclusive.
The OR/sum rule sums the individual probabilities together.
The AND/product rule is used to calculate the probability of two (both) independent events occurring at the same time.
The AND/product rule multiples the individual probabilities together.
In a potential test question, which buzzwords indicate what rule to use?
If the question says one and two, use the AND/product rule. If the question says one or two, use the OR/sum rule. And and or are the buzzwords.
And — use and rule
Or — use or rule
What is a gene locus?
A gene locus is the location of the gene in the organisms genome, or the specific location on a specific chromosome the gene is located.
For the vestigial locus, which allele would represent the non-mutated wild type allele?
vg+
The plus denotes the wild type
For the vestigial locus, what denotes the mutant vestigial allele (crumpled, nonfunctional wings).
vg
Not wild type, a mutant allele
Recessive allele
Which possible genotypes of the vestigial locus could result in a fruit fly having normal functional wings?
vg+/vg+ or vg+/vg
Wild type allele is dominant, the vestigial mutant allele (just vg) is recessive.
Which possible genotype could result in a fruit fly having nonfunctional vestigial wings?
vg/vg
In genetics, what is the goal of a Chi Square Analysis?
To test a genetic hypothesis
What does a Chi Square do in genetics?
It helps to determine if experimental results are consistent with a particular pattern of inheritance.
Does an observed number of offspring from a genetic cross fit well with the predicted outcomes according to the law of independent assortment and law of segregation?
A ___ ____ _____ can be used to determine goodness of fit for data.
Chi Square analysis
What value is used to asses the significance of the chi-square anaylsis result?
A p value. p is the probability that any difference seen between the predicted values and the outcome is due to random chance. p is the level of significance.
What is the Chi-sqaure formula?

What does an x² (square) test do?
It determines how well a set of data fits a pattern. The pattern is from a hypothesis that states: this data follows a particular pattern. The x² test sees how well the data fits the pattern the hypothesis predicted.
If the x² (Chi square) fit is good, is the hypothesis supported or rejected?
The hypothesis is supported if the x² fit is good.
What is the usually (most often used) level of significance value?
The level of significance is p. It is often p = 0.05
What is the critical value for a chi-sqaure test?
The critical value for a chi-square test is the value that is in the square table, and is the value you use to determine if the chi-square analysis fails to reject or rejects the null hypothesis.
How do you find the critical value for a chi-square test?
Determine your level of significance (p). Will usually be p = 0.05.
Determine your degrees of freedom, which is the number of phenotype classes (or just possible phenotypes) minus one.
I.e. #PC - 1 = Degrees of freedom
Then using the chi-square chart, find the row for the level of significance, and go to the column with the degree of freedom value.
Where they interest is the critical value for the chi-square analysis.
What is the null hypothesis in genetics?
The null hypothesis in genetics basically says that a crosses inheritance results is due to chance alone, and that their is not an underlying pattern of inheritance, like Mendelian inheritance.
The null hypothesis basically says there is no genetic linkage impacting the data.
In genetics, is the goal of an experiment to reject the null hypothesis or to fail to reject the null hypothesis?
You want to reject the null hypothesis. The null hypothesis is essentially the opposite of the hypothesis you want to prove.
In genetics, the hypothesis is usually, this cross undergoes mendelian inheritance.
The null hypothesis would state the cross does not have a mendelian inheritance genetic linkage, and is due to chance alone.
If the chi square (x²) value is less than the critical value for a level of significance of p = 0.05 for your degrees of freedom, does the data support the hypothesis?
Yes. If the chi-square value is less than the 5% (p < 0.05) critical value for however many degrees of freedom the experiment has, than the data supports the hypothesis
The data “rejects the null hypothesis”
What happens if the x² value is greater than the critical value for however many degree of freedom at a level of significance of p = 0.05?
You fail to reject the null hypothesis, meaning the data does not support the hypothesis.
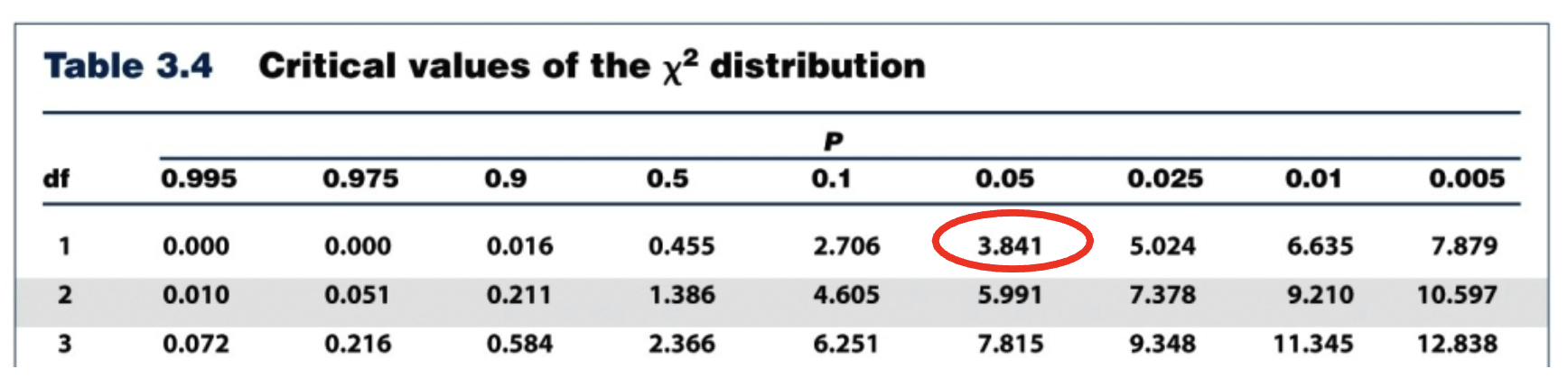
If your chi-square value is x² = 0.60, based on the chart below with a critical value of 3.841, does the data support the hypothesis?
Yes, the chi-square value (x² =0.60) is less than the critical value (3.841) so you reject the null hypothesis and the data supports the hypothesis.
0.06 < 3.841
Reject the null hypothesis, support the hypothesis.
Means that Mendelian patterns of inheritance has occurred, (if hypothesis accepted).
What are three restrictions of using a chi-square analysis in genetics?
Chi-square is not appropriate if the expected values are zero.
Not appropriate if any of the expected values are less than five, doesn’t work with small sample sizes.
Not appropriate if the observations are independent of one another, like one observation made per fly.
What are the general steps to solving Chi-square problems?
Establish the null-hypothesis and the expected phenotypic ratios.
Calculate the expected frequencies of each phenotype based on the sample size.
Use the observed frequencies, and the expected frequencies (they can be given to you also) and plug those into the chi-square formula.
Solve for the chi-square’s value.
Determine your degrees of freedom.
Consult the chi-square table, using your degrees of freedom and a 0.05 (5%) level of significance.
If the x² value is less than the critical value from the table, the data supports the hypothesis and is a good fit.
If the x² value is greater than the critical value from the table, the data does not support the hypothesis (reject the Null Ho).
How do you avoid contamination while using the pipettes?
Always use a new pipette tip for each pipetting action.
Ensure the pipette tip does not touch any surface when in use.
Keep the pipette upright.
The first stop (or press) of the pipette buttons does what?
Draws up the liquid. The second stop (or a second press) will dispense the liquid.
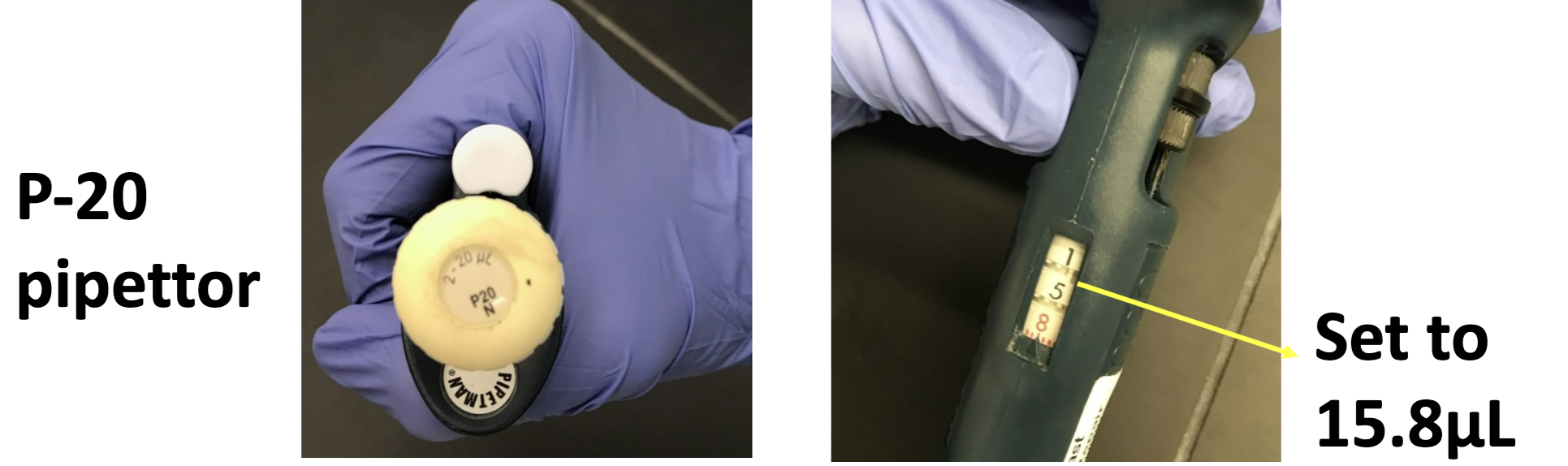
What microliter range (μl) is the P-1000 useful for?
200 to 1000 μl’s
What microliter range is the P-200 useful for?
20 to 200 μl’s
What microliter range is the P-20 useful for?
1 to 20 μl’s
What can happen if you lay the pipette on its side while liquid is drawn in the ti.
It can clog the pipette and lead to sample contamination.
When dispensing a liquid witha pipette into a tube or gel, where should you place the pipette tip to dispence?
Just below the surface, a few millimeters. Be sure not to touch the walls of the tube or the gel in the gel tray.
When pipetting and preparing solutions, what is a master mix?
A master mix is the combination of solutions that all samples will need. For instance, if each sample you want to test needs all three stock solutions, combining the thee stock solutions into a “master mix” to put in the samples instead can save you time.
Its one pipette dispense instead of individually dispensing each stock solution into each solution.
What equation can you use to determine how much of a specific reagent or stock solution (volume) you will need to achieve a specific concentration for your samples?
C1V1 = C2V2
For buffer one of your work manual, the stock concentration is 10x, but the working concentration is 1x. Calculate the initial volume you would need to use to achieve the 1x connection for a single reaction, where the total volume of all the reagents for a single reaction is 50 μl.
You would needed 5 μl.
C1V1 = C2V2
(10x)(V1) = (1x)(50μl)
V1 = 5 μl for a single reaction
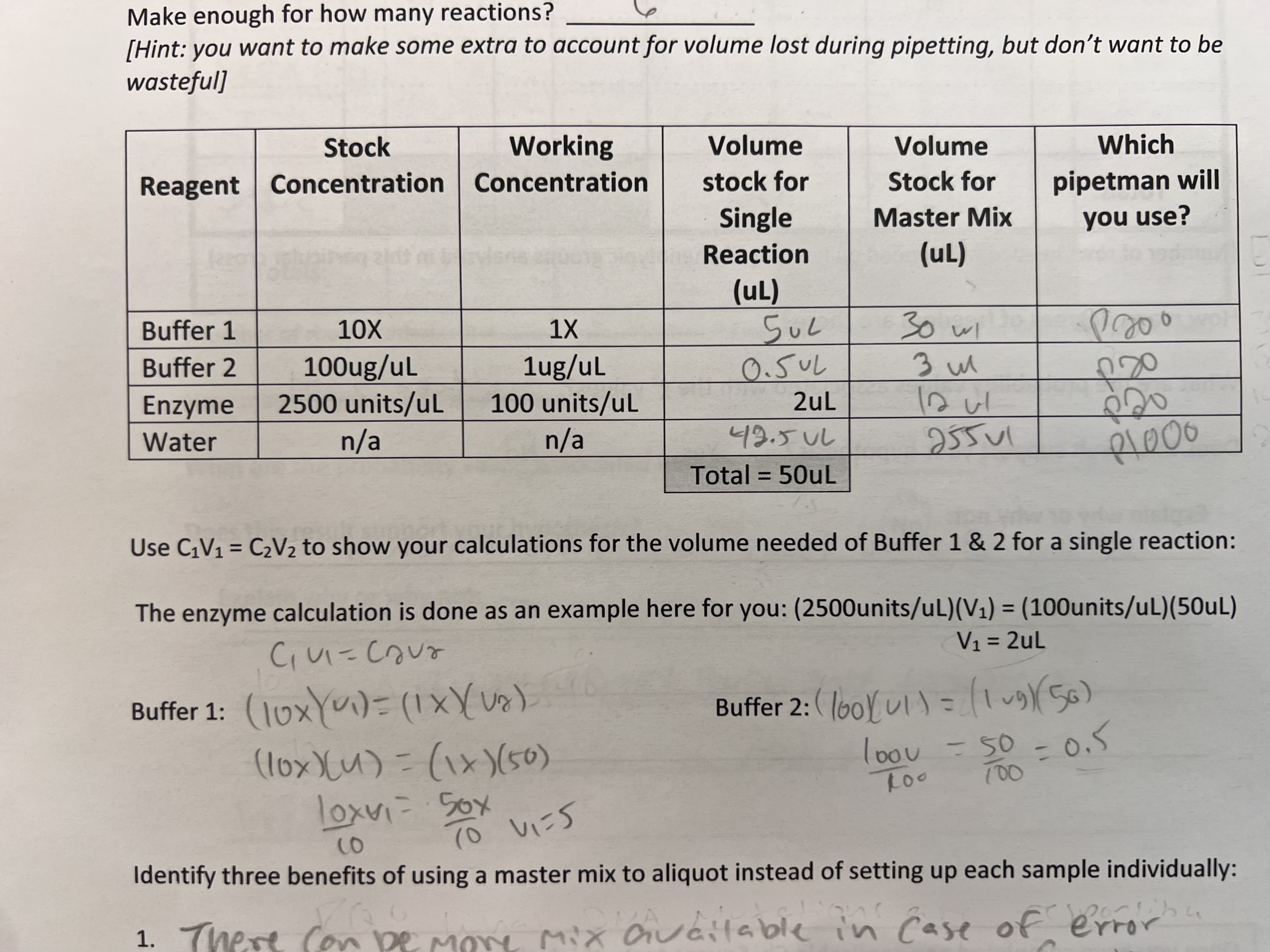
What is the goal of DNA gel electrophoresis?
To separate DNA fragments based on their size for analysis.
What gel is used in DNA gel electrophoresis?
Agarose gel
What is agarose? What is unique about agarose gel that allows it to be used for gel electrophoresis?
Agarose is a polysaccharide that can be melted down into a salt buffer to form a gel (agarose gel). When agarose gel is made, it produces a porous matrix, meaning the gel has holes throughout it. These holes allow for DNA to move through the gel.
What mechanism or property cause the DNA to move through the agarose gel?
DNA is negatively charged. By applying an electrical current, the negatively charged DNA is attracted to the positive end (terminal) of the gel apparatus. The holes of the agarose gel allow for DNA to move through these holes (move through the gel) towards the positive end.
When performing a DNA gel electrophoresis, what part and at which charged end do you place the processed DNA samples?
You place the processed DNA samples in the wells made when the agarose gel was cast. The wells are at the negative end (the anode) of the gel cast, so the negatively charged DNA will move to the positive end (the cathode).
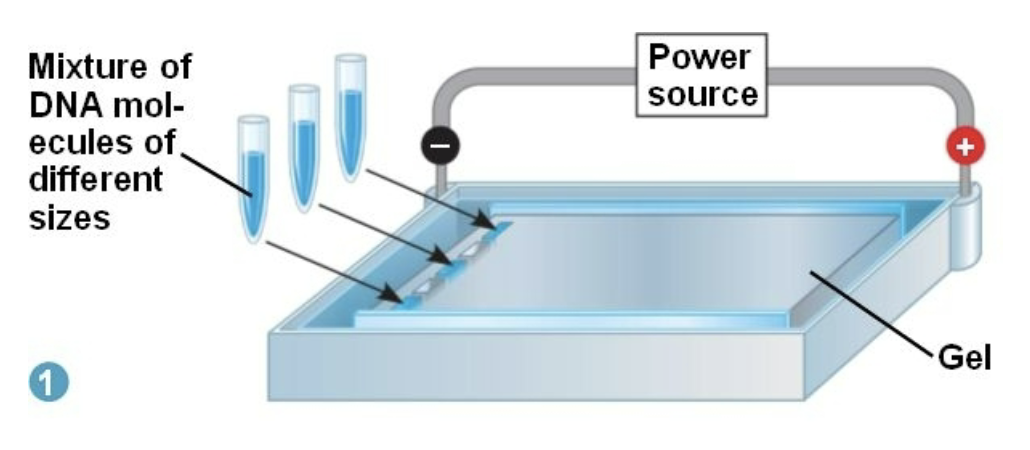
After a gel electrophoresis is performed, which sized DNA fragments will be closer to the positive end?
The smaller fragments. The smaller fragments of DNA migrate further and easier, as the larger DNA fragments get caught in the agarose gel holes.
The larger DNA fragments are closed to the anode (negative end).
The smaller DNA fragments are closer to the cathode (the positive end).
To make a 0.8% agarose gel, how many grams of agarose would you add to 150 milliliters of 1X conccentrateded of TBE solution?
You would add 1.2 grams of agarose to the 150 ml of 1x TBE buffer solution, to make a 0.8% agarose gel solution.
0.8% is 0.8/100 = 0.008
0.008 × 150 ml TBE = 1.2 grams
1.2 grams of agarose needed
If you were to change the concentration of agarose gel to more than 0.8%, say 1.5%, what would happen to the rate that the DNA moves?
The rate would slow down, because the agarose gel at a higher concentration (1.5%) or agarose, causes smaller pore sizes. Smaller pore sizes makes it harder for the DNA to migrate through the gel.
More agarose (higher concentration of it in the gel solution) means smaller pores. Smaller pores means the DNA will move through the gel at a slower rate.
Do smaller linear fragments or larger linear fragments of DNA migrate through the agarose gel faster?
Small linear fragments will move through the agarose gel quicker because they are easier able to move through the holes of the gel.
How does the conformation (shape) of the DNA fragment affect its migration through the agarose gel?
Supercoiled DNA (ie. a plasmid) will move through the agarose gel faster than a linear peace of DNA thats the same size.
A super coiled DNA (like a plasmid) has a conformation (shape) like a ball coiled up.
Which one migrates faster? Supercoiled DNA fragments or a relaxed open circle of DNA of the same size?
Supercoiled DNA will always move through the gel faster than relaxed open circles of DNA or linear strands of DNA.
Relaxed open circles of DNA could move faster or slower than linear DNA of the same size, depending on the concentration of the agarose gel.
What is the purpose off adding a tracker dye to the DNA samples?
To cause the samples to fall to the base of the plate (from within the wells) and to directly visualize the progress while the gel electrophoresis is running.
Which tracker dye does the lab manual mention?
Bromophenol blue
Which two reagents do you combine to make an agarose gel?
Agarose and 1x TBE solution.
The TBE is dissolved in water
The agarose is a polysaccharide (sugar)
What reagent is 1x TBE? What does TBE stand for?
TBE stands for Tris-Borate-EDTA. It is made of:
Tris-HCl
0.08 M Boric Acid
1.0 mM EDTA
Its pH is 8.0
What reagent dye is used to indirectly visualize and track the DNA?
Ethidium bromide
How does Ethidium bromide work to visualize DNA?
Ethidium bromide is an intercalating agent, meaning it binds between the nitrogenous bases. The Ethidium bromide will bind between the bases of the DNA fragments, and when exposed to UV light, Ethidium bromide fluoresces very strongly.
This causes the DNA fragments to appear as bands, which can be photographed.
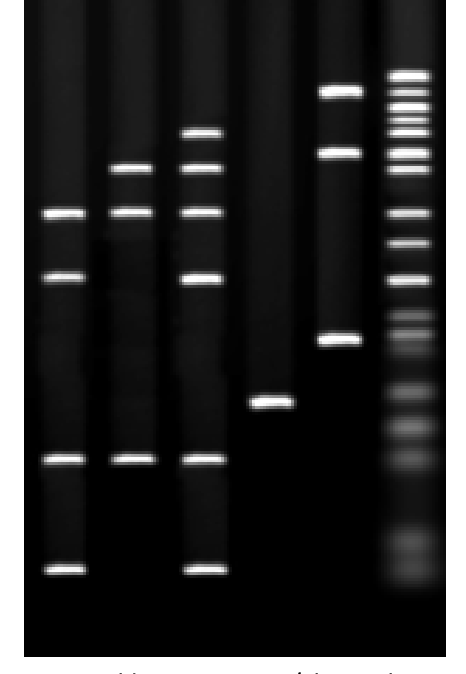
1 kb (kilobase) corresponds to how many base pairs?
1 kb = ~1000 base pairs
0.8 kb is about 800 base pairs
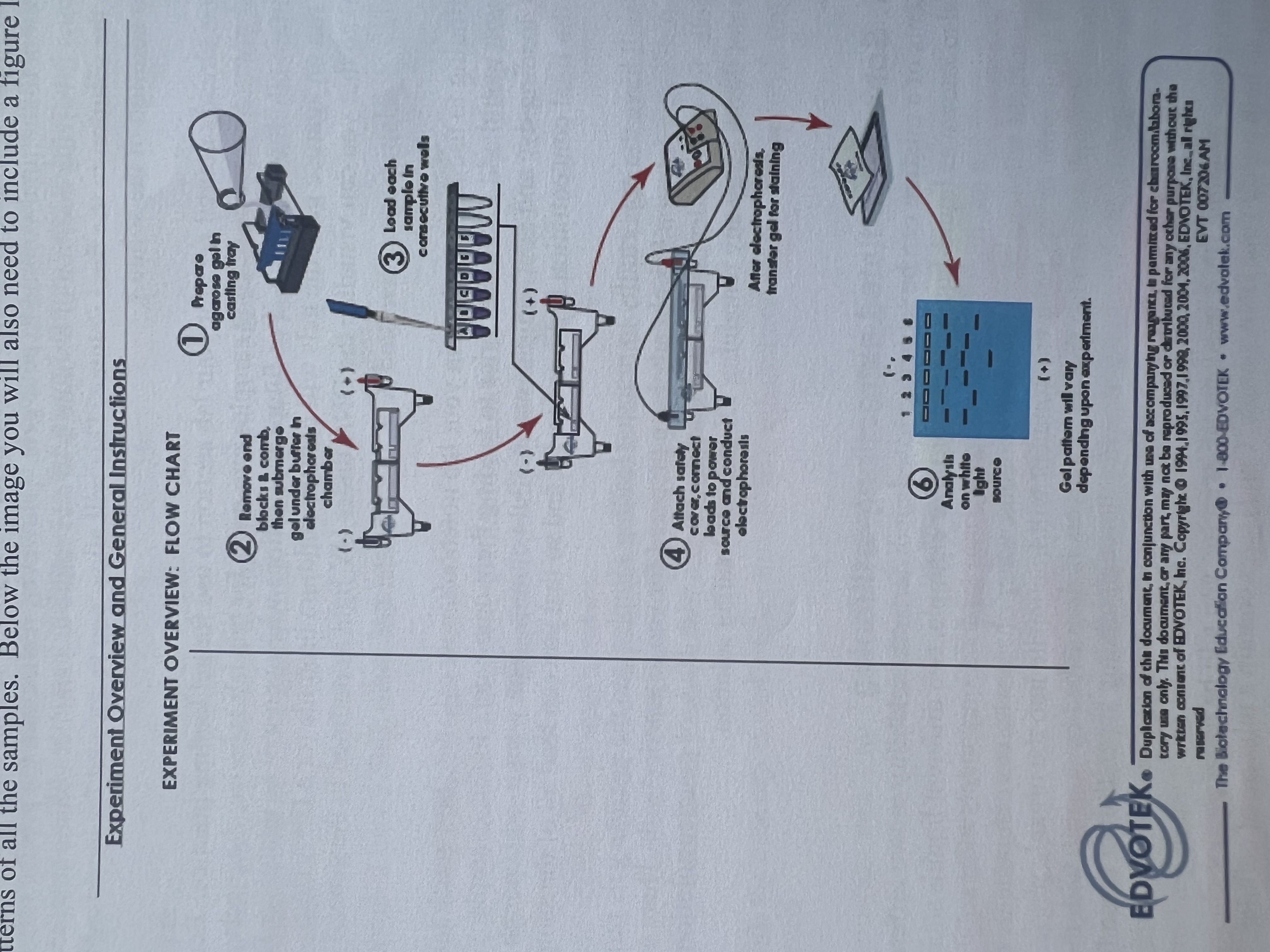
What are the general steps (from the lab manual) in pouring and running a 1.0% agarose gel electrophoresis.
Add the appropriate amount of agarose to the 1X TBE buffer, to make a 1.0% agarose solution, and put the flask into the microwave oven.
Heat the solution until it is gently boiling to dissolve the agarose without it spilling. (about a minute or two)
Using hand protection, remove the flask and allow the solution to cool to about 60 degrees Celsius, swirling occasionally.
The TA then comes and adds the Ethidium bromide at a 50ug/ml concentration to the gel solution.
Prepare a clean gel tray by turning it sideways within the gel box to seal the edges.
Choose the right well comb for the experiment and place it into the slots of the gel tray. Pour in the hot agarose solution and allow it to cool/set for 15-20 minutes.
When the gel is fully set, remove the comb and place the gel and tray into the electrophoresis box (the one with the wires)
Once the gel is set, fil the gel electrophoresis box with 1X TBE buffer to that the gel is fully covered with the buffer.
Mix the samples with the appropriate concentration and amount of a tracker dyes.
Carefully load the samples into the wells using a pipette by putting the pipette tip below the surface of the TBE buffer but not touching the bottom of the well.
Put the lid on the gel bock so that the electrodes make contact. Ensure the electrodes are on the right end, and turn on the power supply with the appropriate voltage.
Let it run for however long specified.
Once the gel has run, using gloves, place the gel on a UV Lightbox, put the plastic shield down over the light box, and turn on the switch.
Photograph the florescent (due to the ethidium bromide) results.
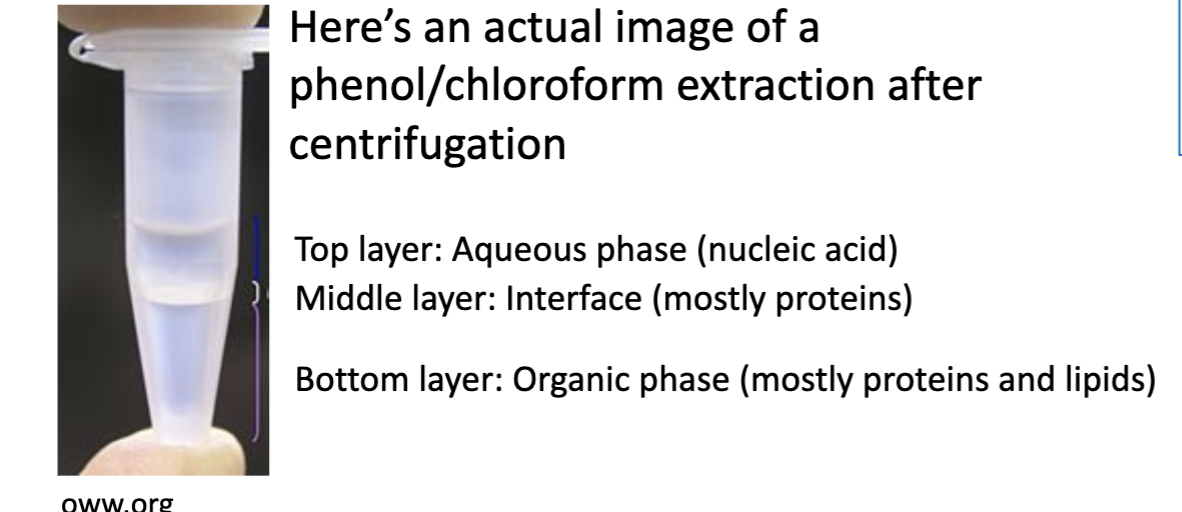
What is liquid-liquid extraction?
Separating components of a mixture based on solubility.
An example is separating two immiscible liquids, like water and an organic solvent separating into their layers.
In liquid-liquid extraction, what phase do polar molecules fractionate (seperate) into?
The aqueous phase. The polar molecules separate into the polar water.
In liquid-liquid extraction, what phase do non-polar moleulces fractionate (separate) into?
The organic phase. The non-polar molecules separate into the non-polar organic solvent.
What determines the separating of the aqueous and organic solvent layers on top or bottom?
Density
Protein removal by phenol-chloroform extraction is a common step performed for?
DNA extraction and purification.
What is the mechanism behind phenol-chloroform extraction?
When the phenol is mixed with the aqueous cell lysate solution (just broken down cells in solution):
The negatively charge nucleic acids (DNA & RNA) stay in the polar aqueous phase (in the water).
The phenol-soluble proteins are extracted out of the aqueous cell lysate and go into the non-polar phenol phase (the organic solvent phase).
Phenol is denser than water, so after centrifugation, the phenol with the proteins goes to the bottom, and the top is the aqueous solution with the DNA and RNA.
The top aqueous phase can be harvest and used for further tests/analysis.
Chloroform is often used with phenol to improve the solubility of the proteins in the organic solvent (organic phase).
In a phenol-chloroform DNA extraction, where do the nucleic acids settle (top or bottom)?
The nucleic acids (DNA and RNA) settle on the top in the aqueous solution. The proteins settle on the bottom in the phenol and chloroform organic solvents.
What is the general idea/steps to extract genomic DNA (gDNA) from the Drosphilia melanogastor?
Grind the flies in a small volume of lysis buffer that contains sodium dodecyl sulfate (SDS).
Centrifuge the crushed flies and lysis buffer. The sample will contain proteins, DNA, and RNA.
Using the phenol-chloroform liquid liquid extraction, use equal parts of phenol and chloroform to extract the proteins (in the bottom).
Transfer the aqueous phase (the top layer) containing DNA and RNA into a new sterile/clean tube.
Remove the RNA by adding RNase A to the sample, an enzyme that degrades (digests) RNA.
Then the DNA can be used for analysis (i,e, gel electrophoresis).
What does sodium dodecyl sulfate (SDS) due in extracting genomic DNA from the Drosphilia?
SDS lyses the cells, allowing the materials, like the proteins, DNA, and RNA to freely move in the solution, for the extraction.
What is the purpose of RNase A in the genomic DNA extraction from the Drosphilia?
To digest the RNA in extracted aqueous solution, so only DNA is left.
What chemical species are in the lysis buffer?
The lysis buffer has 0.1% sodium dodecyl sulfate (SDS), 10 mM of Tris-pH 8.0, and 1 mM of EDTA pH 8.0.
What is the chemical formula of chloroform?
CHCl3
The lab manual kept putting the formula instead of just the name for some reason.
What does PCR stand for?
Polymerase chain reaction.
What is the main goal of PCR?
To amplify specific sequences of nucleic acids, like the double-stranded gDNA.
What is the mechanism behind the PCR used in the lab?
Two oligonucleotides hybridize, each to opposite strands, at each end of the specific DNA region (segment) to be amplified.
The reaction mixture contains the oligonucltodiesj DNA polymerase, primers, nucleotides, salt, and other critical buffers.
This is all mixed together, and then the reaction is heated to 94-96 degrees celsius to denature the double-stranded DNA into single strands. This allows the primers access to individual template strands.
Next the temperature is lowered to allow for primer annealing to the complementary sequence in the template DNA. The optimal annealing temperature is 50-62 degrees Celsius.
The temperature is then raised to 72 degrees Celsius for extension, which is the amplification of DNA synthesis by the DNA polymerase.
The cycles are repeated about 25-40 times to produce enough of the DNA for analysis (like gel electrophoresis).
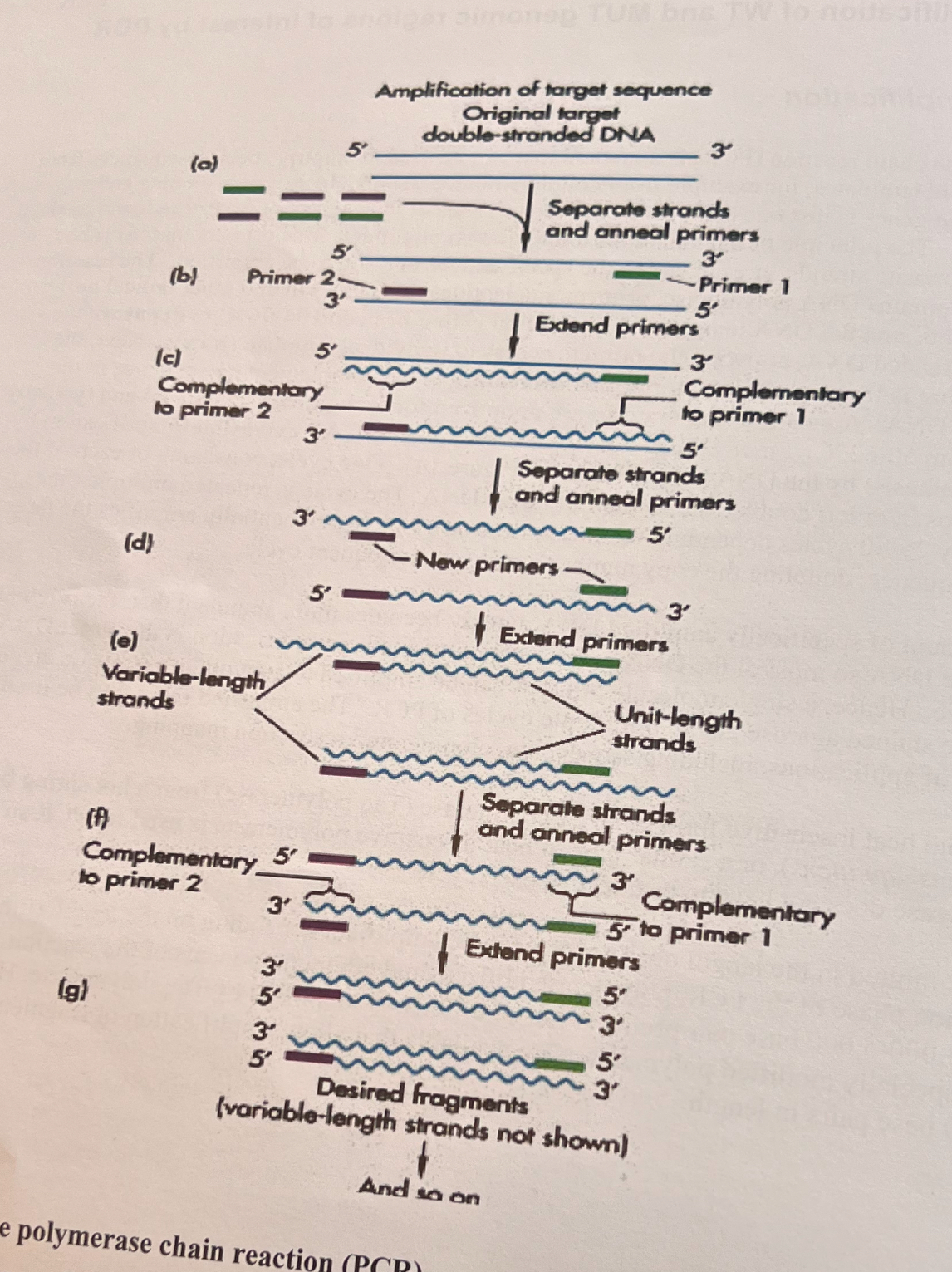
What is annealing in PCR?
Annealing is when the primers (very short DNA sequences) bind to their complimentary regions in the single-stranded DNA sequences.
The DNA becomes single stranded due to the temperature denaturing the double-helix.
What is extension in PCR?
Extension is the amplification of DNA synthesis by DNA polymerase. Its when the DNA polymerase copies the single strands of DNA by using the primers as extensions.
What is DNA polymerase?
DNA polymerase is an enzyme that copies DNA.
What is the purpose of dNTP’s in the PCR?
They are the nucleotides that are used by the DNA polymerase to build new strands of DNA. It stands for deoxynulcotide triphosphates.
What is the name of the special heat insensitive (heat resistant) form of DNA polymerase?
Taq polymerase. It is heat insensitive, which means that DNA polymerase doesn’t have to be added for every cycle if Taq polymerase is used.
Where does the forward primer bind to on the DNA in PCR?
The forward primer binds to the antisense end, also called the minus end (-) of the DNA strand. It runs from the 3’ to the 5’ end direction.
Where does the reverse primer bind to on the DNA in PCR?
The reverse primer binds to the sense end, also called the positive end (+) of the DNA strand. It runs from the 5’ end to the 3’ end direction.
The master mix you used in lab contained what?
The components needed for PCR. The only thing it did not have was the template DNA, which is what you added to the individual samples by extracting the gDNA from the Drosphilia.
List the components of the master mix.
dNTPs - the nucleotides to build new DNA
Taq polymerase
Various buffer components at a 5X concentration (not named). This was diluted down to 1X working concentration.
The labs (your) own primers that you add.