Biochem Lec 9- Enzymes 1: Thermodynamics and Equilibria
1/25
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
26 Terms
What is K’eq?
-It relates the concentration of substrates (reactants) and products
-Same as chemical equilibrium but assumes aqueous conditions ([H2O] = 55.5 M) and a pH of 7
K’eq=[products]/[reactants]
What is ΔGº’?
The standard biological free energy change for the reaction assuming [substrates] and [products] are 1 M, [H2O] = 55.5 M and pH=7
How are ΔGº’ and K’eq related?
Related by ∆Gº′ = − RT ln K′eq
R=gas constant= 8.315 X10-3 kJ/mol∙deg
T=temperature= 298 K for ambient temperature (25 C), 310 K for mammalian tissues (37 C)
RT= 2.48 kJ/mol at 25 C or 2.58 kJ/mol at 37 C
What is an example of an unfavorable (+∆Gº′) biological reaction?
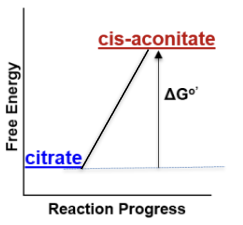
Citric acid cycle: +8.4 kJ/mol

What is an example of a favorable (-∆Gº′) biological reaction?
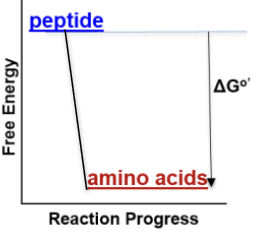
Proteolysis: -10 kJ/mol
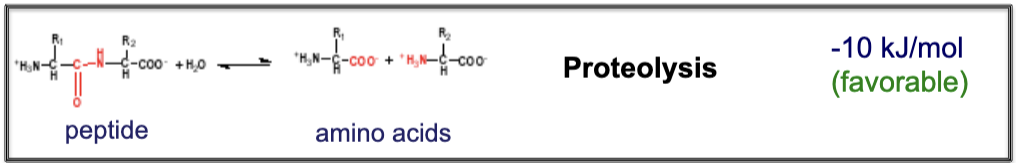
What is an example of a biological reaction that occurs in equilibrium (∆Gº′=0) ?
Nucleotide synthesis: 0 kJ/mol

How does the free energy change throughout an unfavorable reaction (ex. citric acid cycle)?
Reactants lower energy than products (+∆Gº′)
How does the free energy change throughout a favorable reaction (ex. proteolysis)?
Reactants higher energy than products (-∆Gº′)
How does the free energy change throughout an equilibrium reaction (ex. nucleotide synthesis)?
Reactants have same energy as products (∆Gº′=0)
If [cis-aconitate]=0.8 mM (product) and [citrate]=1 mM (reactant), would the reaction proceed? K’eq=0.034
∆Gº′= -2.48*ln(0.034)=+8.4 kJ/mol
∆G=8.4 + 2.48 ln(0.8)= +7.85 kJ/mol
Would not proceed
In respiring mitochondria, [cis-aconitate]=0.05 mM (product) and [citrate]=2 mM (reactant). Would this proceed? K’eq=0.034
∆Gº′= -2.48*ln(0.034)=+8.4 kJ/mol
∆G=8.4 + 2.48 ln(0.025)= -0.75 kJ/mol
Would proceed
How do cells solve the issue of unfavorable endergonic reactions?
Energy coupling: couple an unfavorable endergonic reaction with a favorable exergonic reaction
Can also regulate the amount of substrate and product present in order to lower the [P]/[S] ratio→ lower [P] or increase [S]
Reactions using energy coupling often involve the _____ of ______
Hydrolysis, adenosine triphosphate (ATP)
How does the hydrolysis of ATP make reactions exergonic?
The cleavage of the terminal phosphoanhydride bond of ATP releases energy and the products are more stable than ATP, so it’s favorable
Do thermodynamics influence the rate of a reaction? Provide an example to support your answer.
NO thermodynamics do not influence reaction rate.
Ex. Hydrolysis of peptide bond is exergonic, but takes a very long time (about 31 years)
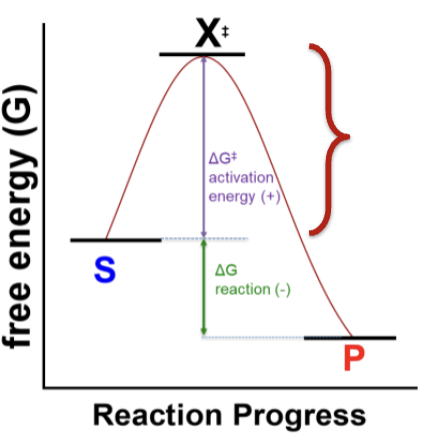
What is the rate of the reaction dependent on? (Kinetics)
The difference in energy between the substrates and the transition state X‡. This is the activation energy ΔG‡.
ΔG‡ = GX‡ – GS
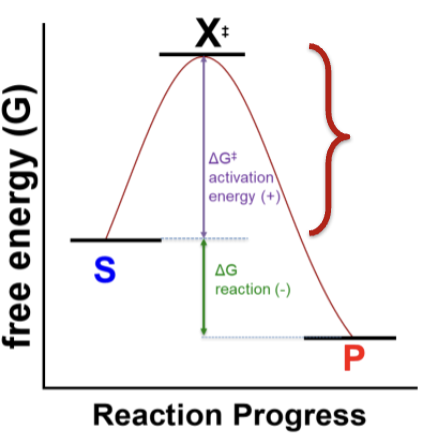
How is ΔG for a reaction found?
It’s the difference between the free energy of the substrates and products
ΔG = GP – GS
What is the rate of an uncatalyzed reaction dependent on?
It is dependent on a first order rate constant kuncat and the concentration of substrate [S]
Vº=kuncat[S]
Vº=measured rate of production of P from a solution of substrate (S) per unit of time
S→(kuncat)P
![<p>It is dependent on a first order rate constant k<sub>uncat</sub> and the concentration of substrate [S]</p><p>V<sub>º</sub>=k<sub>uncat</sub>[S]</p><p>V<sub>º</sub>=measured rate of production of P from a solution of substrate (S) per unit of time</p><p>S→(k<sub>uncat</sub>)P</p>](https://knowt-user-attachments.s3.amazonaws.com/98f9f311-b979-45a3-95e0-b56db0e1b4df.png)
What is the rate of a catalyzed reaction dependent on?
It is dependent on a first order rate constant kcat and the concentration of substrate [ES]
Vº=kcat[ES]
Vº=rate of production of P per unit of time when starting with a solution of substrate (S) and adding an enzyme catalyst that binds the substrate in a complex (ES)
ES→(kcat)E+P
![<p>It is dependent on a first order rate constant k<sub>cat</sub> and the concentration of substrate [ES]</p><p>V<sub>º</sub>=k<sub>cat</sub>[ES]</p><p>V<sub>º</sub>=rate of production of P per unit of time when starting with a solution of substrate (S) and adding an enzyme catalyst that binds the substrate in a complex (ES)</p><p>ES→(k<sub>cat</sub>)E+P</p>](https://knowt-user-attachments.s3.amazonaws.com/90a031ff-e8fe-472b-b78d-962d23544b64.png)
How do enzymes lower activation energy/speed up reaction?
Enzymes bind to substrate and form ES complex. The geometry of the enzyme binding site stabilizes/lowers the energy for the formation of transition state (X‡).
This substantially lowers the activation energy ΔG‡ and increases the rate of reaction Vo
What is an example of an enzyme catalyzed reaction? Compare the rates both cat and uncat.
Hydrolysis of the following peptide bond by the Pancreatic enzyme “chymotrypsin”
Uncatalyzed rate: kuncat= 10-9 sec-1
Chymotrypsin enzyme catalyzed rate: kcat= 100 sec-1
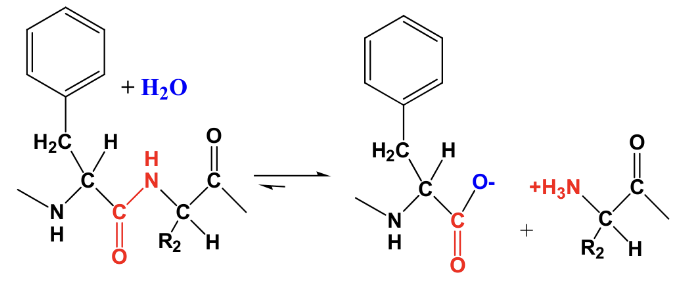
Are enzymes specific for their substrates?
-Enzymes are highly specific for their substrates
-Enzyme active sites have “chemical complementarity” with substrates
-Active site residues have the proper shape and stereochemistry that allows binding of substrate and stabilization by noncovalent interactions with key amino acid R-groups and other cofactors
What is the active site?
A small pocket in the native 3D tertiary structure of a protein where substrate molecules bind reversibly.
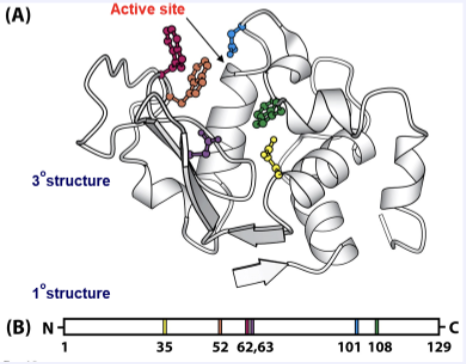
Briefly describe the Lock and Key Hypothesis.
The lock and key hypothesis states that an enzyme’s active site has a specific, rigid shape that fits the substrate exactly, like a key fitting into a lock. This precise fit explains the enzyme’s specificity, since only the correct substrate can bind and undergo the reaction. This is partly correct but not fully.
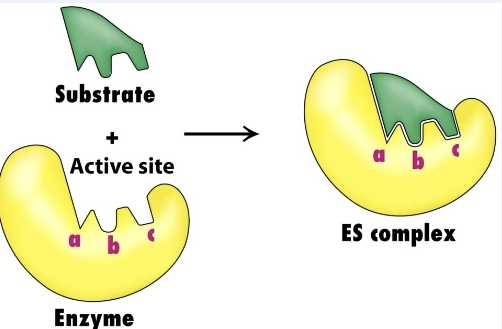
Briefly describe the Induced Fit Hypothesis.
The induced fit hypothesis states that when a substrate binds to an enzyme, the enzyme’s active site changes shape slightly to fit the substrate more snugly. This flexibility positions key amino acids for catalysis, lowers the activation energy, and makes the reaction more efficient. This is more accurate than lock and key.
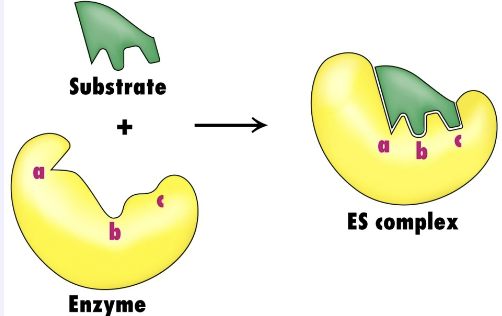
What are the properties of the active site?
Nonaqueous environment (affects dielectric constant)
High specificity for substrates due to chemical complementarity
High binding affinity for substrate binding
Even higher affinity for the transition state which lowers activation energy ΔG‡