BIOS 300 Lecture 15: Eukaryotic Transcription
1/122
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
123 Terms
Eukaryotic RNA Synthesis = Complex
Eukaryotic RNA Synthesis = Complex
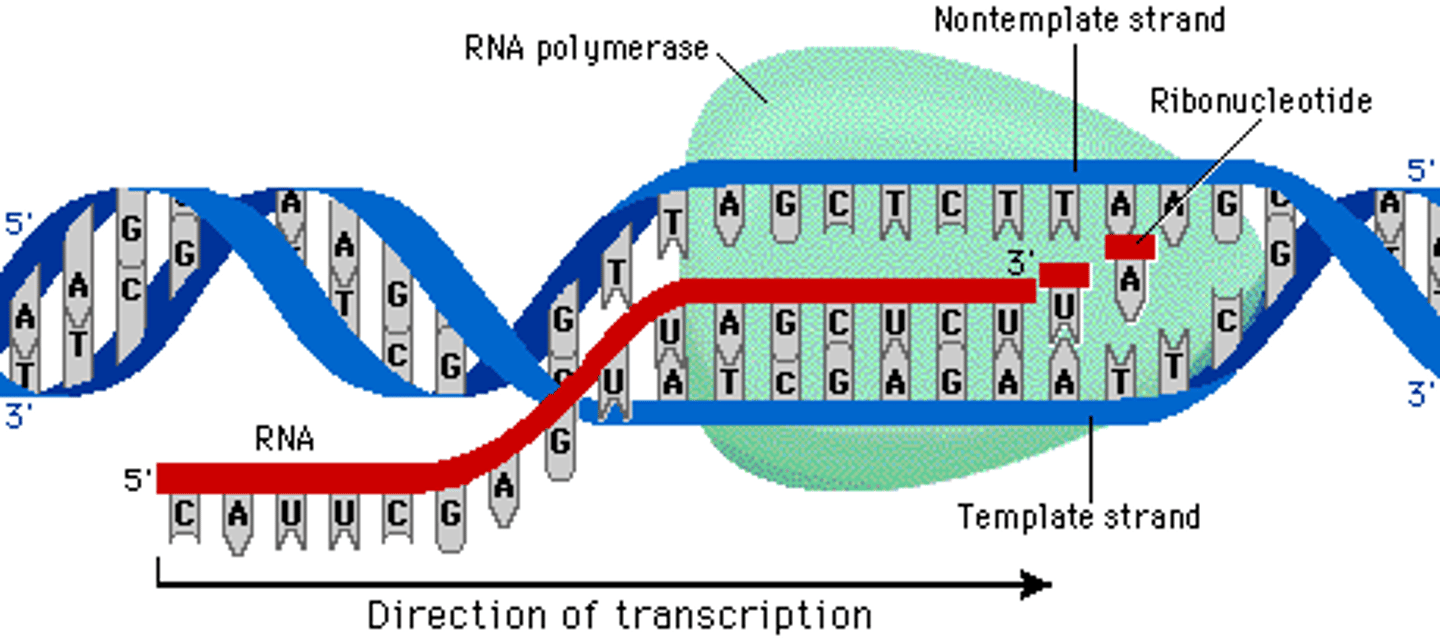
How many different RNA polymerases are needed for eukaryotic transcription?
Three (RNA Pol I, RNA Pol II, RNA Pol III)
True or False?: Eukaryotic RNA polymerases function similarly to prokaryotic RNA polymerases.
True
RNA Polymerase I (Pol I)
synthesizes ribosomal RNA (rRNA) precursors)
RNA Polymerase II (Pol II)
transcribes protein-coding genes to produce messenger RNA (mRNA) and some small nuclear RNAs (snRNAs)
Which RNA Polymerase produces mRNA?
RNA Polymerase II
RNA Polymerase III (Pol III)
responsible for producing transfer RNA (tRNA), 5S rRNA, and other small RNAs
Eukaryotic transcription requires multiple cofactors, known as _________________________________ _________________________.
transcription factors
True or False?: Just like prokaryotic RNA polymerase, eukaryotic RNA polymerases can initiate transcription on their own (de novo).
False; Unlike prokaryotic RNA polymerase, eukaryotic RNA polymerases cannot initiate transcription on their own and require the assistance of various protein factors, including general transcription factors
Eukaryotic RNA must copy DNA in nucleosomes
histone traffic -> histones present! -> have to get histones out of the way so RNA Pol can do its job
What components are involved in the Basal Transcription Machinery?
Core promoter, proximal enhancers, and RNA Polymerase II
What is the function of the Basal Transcription Machinery?
Forms the basic complex required for transcription initiation
Core Promoter
A DNA sequence located near the transcription start site where RNA Polymerase II (RNAP) and the basal transcription machinery assemble to begin transcription
Proximal Enhancers
DNA sequences located close to the core promoter that have binding sites for regulatory transcription factors.
These factors help to increase the rate of transcription.
Proximal enhancers help to ______________________ the rate of transcription
increase
Coactivators for Transcription
These are proteins that bind to transcription factors and help to activate transcription.
Transcription coactivators can also modify ____________________ _____________________ to make the DNA more accessible for transcription.
chromatin structure
What does the complex regulatory machinery in eukaryotic transcription involve?
Distal enhancers, coactivators, and other regulatory proteins.
What is the role of the complex regulatory machinery in eukaryotic transcription?
It modulates the transcription process.
How does the complex regulatory machinery facilitate transcription?
By looping the DNA to allow interaction between distal enhancers and the core promoter.
In eukaryotic transcription, complex regulatory machinery facilitates the __________________ of the DNA to allow for the interaction between distal enhancers and the core promoter
looping
What are distal enhancers?
DNA sequences located far from the gene they regulate.
How far can distal enhancers be located from the gene they regulate?
They can be located thousands of base pairs away.
What do distal enhancers bind to in order to regulate gene expression?
Transcription factors.
How do distal enhancers interact with the core promoter region?
The DNA loops to bring these distal enhancers into proximity with the core promoter region.
Eukaryotic transcriptional regulation
The overall process shown is a highly coordinated activity involving various transcription factors, RNA polymerase, and chromatin regulators to control gene expression.
Where does eukaryotic RNA synthesis begin?
promoter 'TATA box'
Transcription factors and TATA-binding protein (TBP)
regulate access to the promoter
Do eukaryotes have a sigma factor?
No; eukaryotes do not have sigma factor - instead, they have the TATA box!!
Eukaryotic transcription -> dealing with nucleosome structure
Chromatin-modifying proteins
Histone modifying enzymes (e.g., kinases)
Transcriptional activators and repressors
What occurs at the core promoter region of a gene?
Various transcription factors (such as TFIID (transcription factor IID), TFIIA, TFIIB, etc.) and RNA polymerase II (Pol II) assemble to initiate transcription
Gene Expression Control in Eukaryotic Transcription
Regulatory elements determine which proteins are produced, in what quantities, in which cells, and at what times
Transcription is regulated by . . .
chromatin-modifying proteins
histone-modifying enzymes
transcriptional activators and repressors
What is the role of Mediator in eukaryotes?
Transcriptional coactivator for RNA polymerase in all eukaryotes
How many subunits does Mediator contain?
More than 25 subunits
What is the molecular weight of Mediator?
Approximately 1.2 million Da (Daltons)
What does Mediator bind to?
RNA polymerase holoenzyme (RNAPII)
Why does the Mediator have a large surface area?
To interact with other proteins
Activators
Repressors
What is the function of the Mediator complex?
helps regulate the initiation of transcription by recruiting RNAPII and other factors to the promoter region of a gene
Why do prokaryotes lack a protein like Mediator?
Prokaryotes lack a protein like Mediator because their gene expression is less complex than that of eukaryotes
Are the complexities of eukaryotic transcription offset by opportunities created by these complexities?
Yes; the complex regulatory machinery of eukaryotes, including the Mediator complex, allows for precise control over gene expression.
The Mediator is involved in the assembly of the pre-initiation complex at promoters
TATA binding protein
Core transcription factors
Why does the Mediator undergo a large conformational change in binding?
Associates with RNA Pol II and other general transcription factors (bridge role?)
True or False?: The Mediator can activate transcription on its own.
True
Unwinding DNA for transcription creates supercoiling -> complicated by nucleosomal structure in eukaryotes
Unwinding DNA for transcription creates supercoiling -> complicated by nucleosomal structure in eukaryotes
Supercoiling
The twisting of DNA that occurs when the helix is unwound for transcription
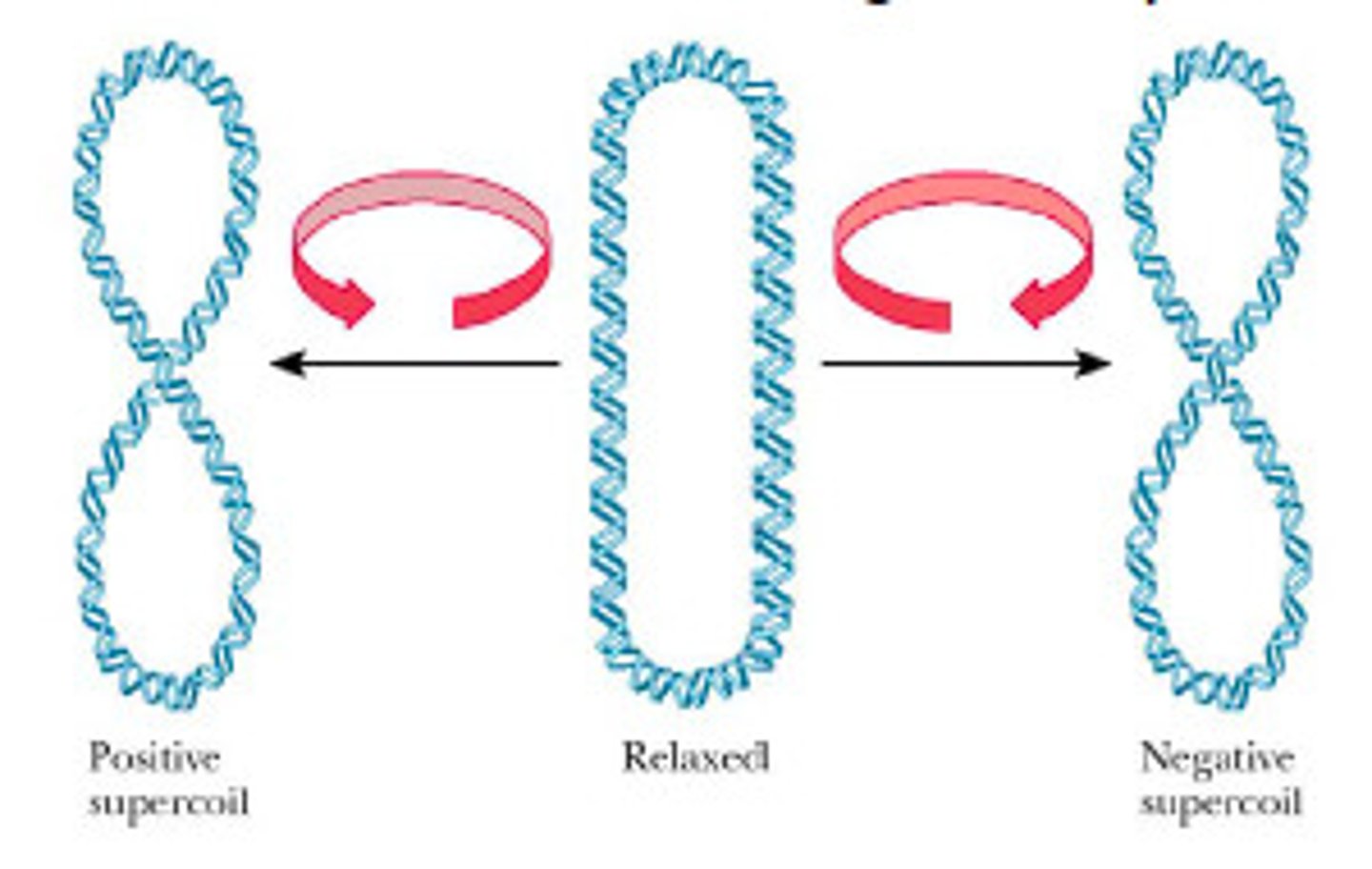
Unwinding the DNA at one end of the RNA polymerase creates _____________________ _________________________ (overwound) ahead of the enzyme
positive supercoils
Rewinding the DNA behind the enzyme creates ___________________ ______________________ (underwound)
negative supercoils
Topoisomerase unwinding relaxes ____________________ supercoils
positive
Topoisomerase rewinding relaxes ________________________ supercoils
negative
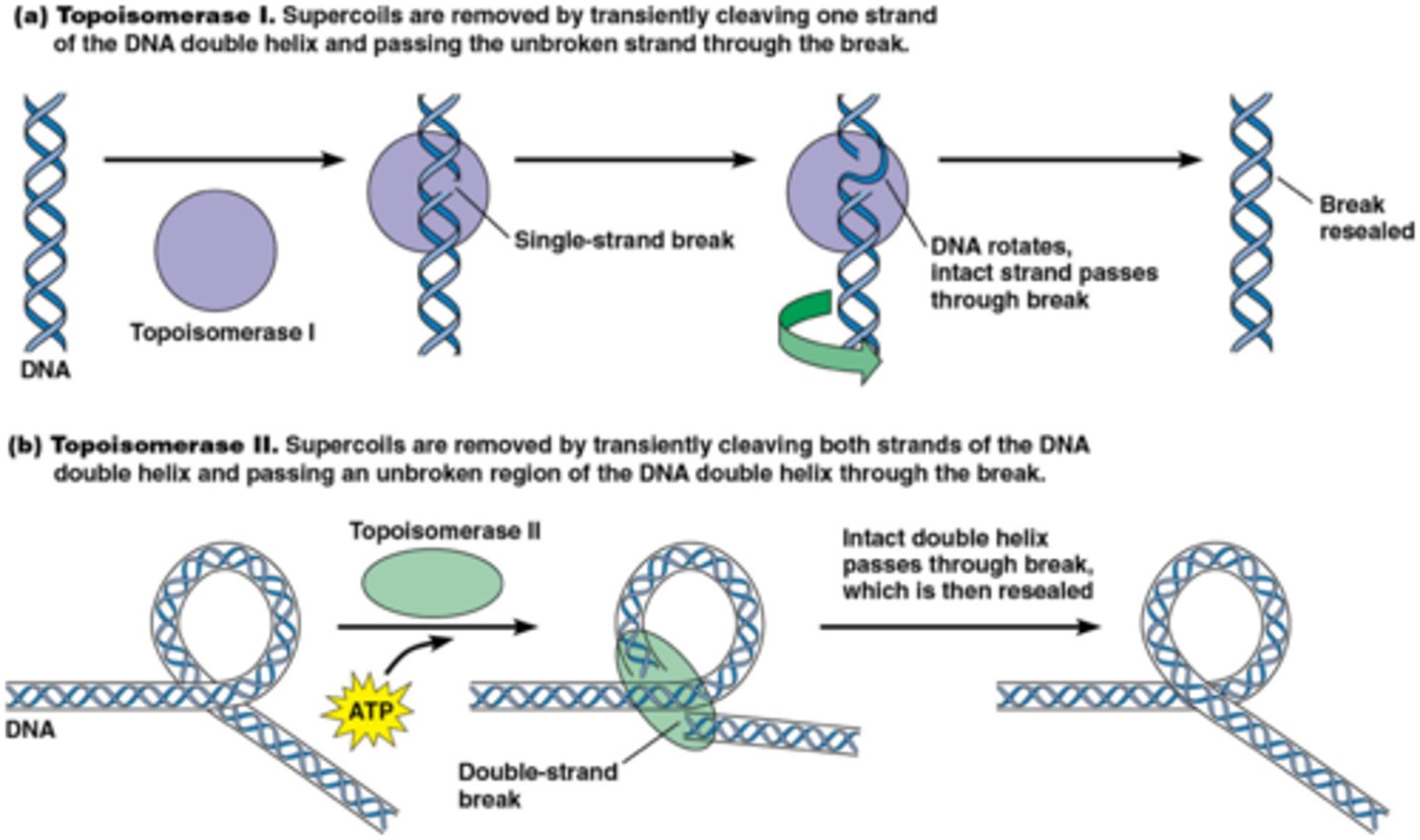
Type I topoisomerases
create a single-strand break
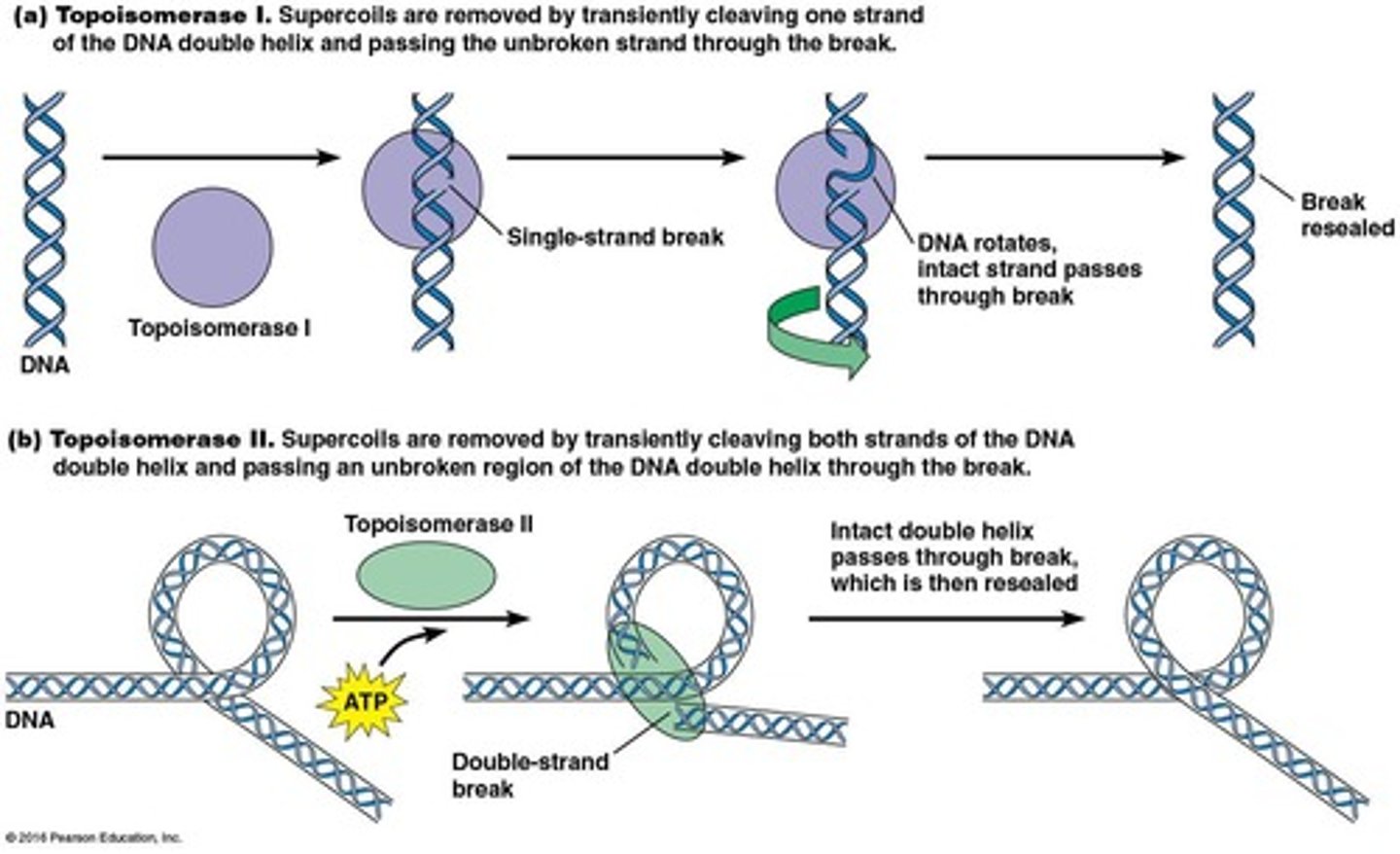
Type II topoisomerases
create a double-strand break
If topoisomerase activity is insufficient, the accumulation of supercoils can inhibit transcription, especially for long genes.
It can also lead to the formation of DNA knots and R-loops, which are harmful to the cell.
List the 3 steps of mRNA processing in eukaryotes.
1) Capping (5' cap)
2) Splicing
3) Polyadenylation (Poly-A tail)
5'-end of pre-mRNA is modified with addition of
5' cap (m⁷G cap)
m⁷G cap
7-methylguanosine cap (modified guanine nucleotide)
What does the 5' cap do?
Protect the mRNA from being degraded by enzymes
What else does the 5' cap do?
Export of mRNA from nucleus
Helps ribosomes recognize the mRNA to begin translation
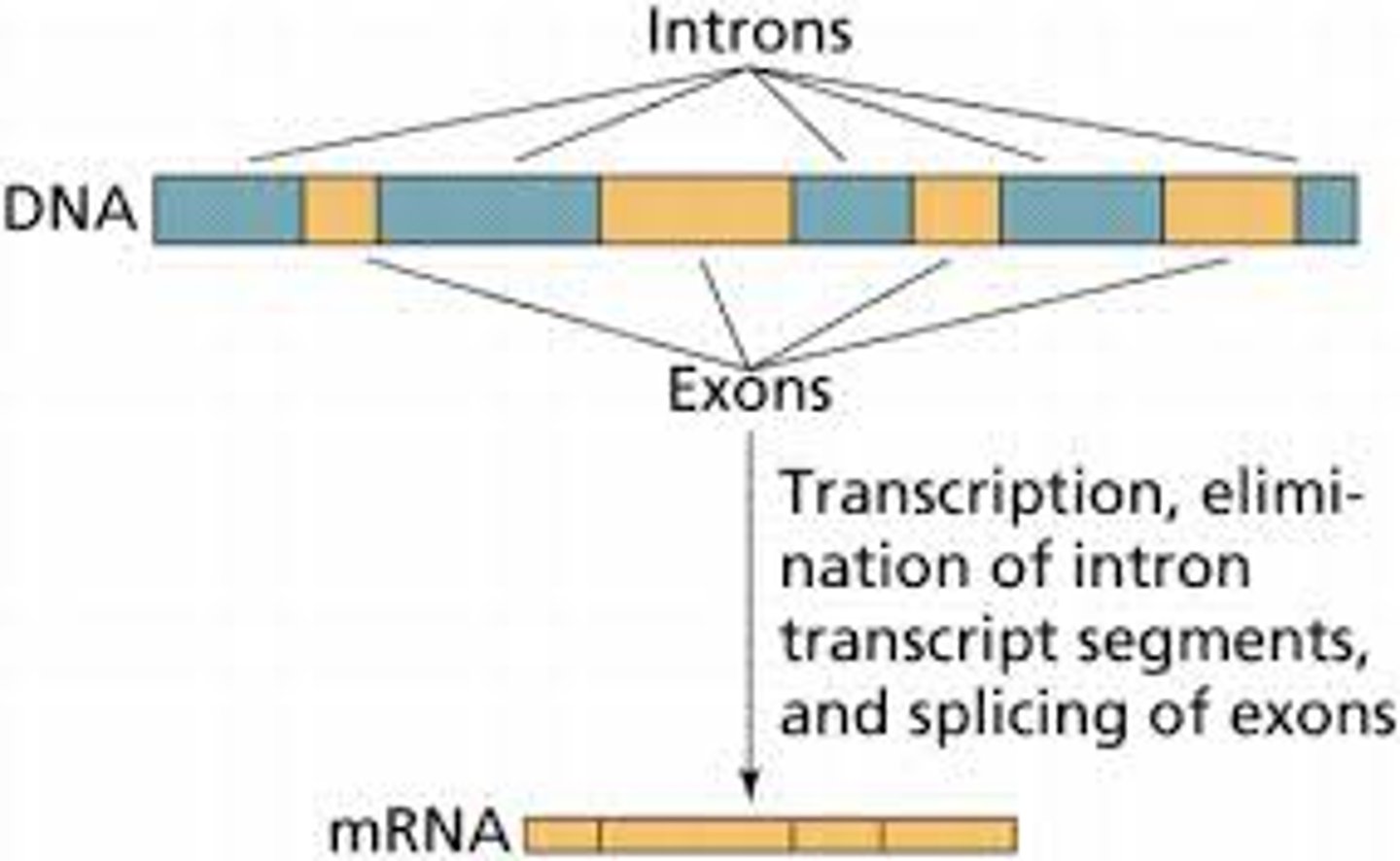
RNA splicing removes/excises . . .
non-coding regions (introns)
RNA splicing also joins/splices together . . .
coding regions (exons)
Where is RNA splicing carried out?
Spliceosome
Poly-A tail
string of adenine nucleotides
The Poly-A tail is added to the ________-end of the mRNA
3'
What does the Poly-A tail do?
Extends the life of an mRNA by protecting it from degradation
UTR
untranslated regions at 5' and 3' ends of mRNA
Parts of exons -> not always coding
Important for gene expression
What is the primary RNA polymerase involved in transcriptional termination for mRNA in eukaryotes?
RNA polymerase II
What do termination factors bind to during transcriptional termination in eukaryotes?
Specific sites
What do termination factors promote in relation to RNA polymerase II?
Dissociation from DNA within 300 bp
What happens to the 300 bp region during processing after transcriptional termination?
It is removed
What is added to the 5' end of eukaryotic mRNA during processing?
A 7-methylguanosine cap
How is the 7-methylguanosine cap linked to the mRNA?
Through a unique 5' to 5' triphosphate bridge
What is the function of the 5' cap on mRNA?
To protect the mRNA from degradation by enzymes
What type of methylation occurs in eukaryotic mRNA processing?
Methylation on 2'-OH of 5' bases and sometimes on the 5'-base (often adenine)
Can you imagine alternative ways to achieve the goals of these modifications provided at the 5'-end of the eukaryotic mRNA (besides capping, splicing, and export)?
Alternative capping mechanisms
Circularization of mRNA
Internal ribosome entry sites (IRES)
Chemical modifications
Alternative capping mechanisms
Instead of a 7-methylguanosine cap, other non-canonical caps like NAD+, FAD, or dpCoA could be used to protect the mRNA and regulate translation
Circularization of mRNA
Creating circular molecules would eliminate the 5' and 3' ends, making them resistant to exonuclease degradation. This approach has been explored for therapeutic applications
Internal ribosome entry sites (IRES)
Some viruses and cellular mRNAs use IRES to initiate translation without a 5' cap. This would allow for cap-independent translation initiation.
Chemical modifications (alternative mRNA processing)
Various chemical modifications to the nucleotides themselves could increase mRNA stability and translation efficiency without a traditional cap
Spliceosome consists of
snRNAs (small nuclear RNAs) and specific proteins, such as small nuclear ribonucleoprotein particles (snRNPs)
Energy for mRNA splicing comes from . . .
ATP hydrolysis
What is mRNA splicing?
A crucial step in gene expression where non-coding regions are removed from a precursor messenger RNA molecule.
What are introns?
Non-coding regions that are removed during mRNA splicing.
What are exons?
Coding regions that are joined together during mRNA splicing.
What is the final product of mRNA splicing?
a mature messenger RNA (mRNA) that can be translated into a protein
Which complex carries out mRNA splicing?
spliceosome, which is composed of small nuclear ribonucleoproteins (snRNPs) such as U1, U2, U4, and U6
The spliceosome recognizes specific sequence signals at the ______1________ (___1____) and _________2___________ (____2____) ends of the intron, as well as a branch site within the intron.
1) donor (5')
2) acceptor (3')
The intron is removed in a ______1__________ structure, and exons are ________2_________ together.
1) lariat
2) ligated
Where does mRNA splicing occur?
Nucleus (nuclear membrane)
What opportunities does mRNA splicing offer for eukaryotes?
Alternative splicing
Regulation of gene expression
What is alternative splicing?
A process that allows a single gene to code for multiple proteins.
What does alternative splicing increase?
The diversity of the proteome without needing a larger number of genes.
Regulation of gene expression (mRNA splicing)
Splicing can be regulated to control the amount or type of protein produced
What complexities and/or problems accompany the requirement for mRNA splicing?
Splicing errors
Energetic cost
Regulation complexity
What does the Poly-A tail consist of?
Long sequence of (~200) A (adenine) residues is added to create the tail of the "mature RNA"
When is the Poly-A tail created?
During the polyadenylation process
Which proteins are involved in the polyadenylation process?
CPSF (Cleavage and Polyadenylation Specificity Factor)
PAP (Poly-A Polymerase)
What is RNA Editing?
Direct alteration of one or more nucleotides in an mRNA that has already been synthesized.
What do enzymes do during RNA Editing?
They modify just the base, changing a single base.
Alterations in sequence can change meaning
alter amino acid encoded or signal early termination
What is a specific type of RNA editing?
The conversion of adenosine to inosine.