RNA transcription
0.0(0)
0.0(0)
Card Sorting
1/57
Earn XP
Description and Tags
Study Analytics
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
58 Terms
1
New cards
Molecular definition of a gene
Entire nucleic acid sequence that is necessary for synthesis of proteins OR rna.
\
Genes are segments of DNA that are transcribed into RNA
\
1. Resulting RNA encodes a protein (e.g., mRNA)
2. The resulting RNA functions as RNA and ay not be translated into protein (e.g., tRNA and rRNA)
\
Genes are segments of DNA that are transcribed into RNA
\
1. Resulting RNA encodes a protein (e.g., mRNA)
2. The resulting RNA functions as RNA and ay not be translated into protein (e.g., tRNA and rRNA)
2
New cards
More RNA means…
More protein
3
New cards
Transcription is for…
Creating RNA
4
New cards
1st rule: RNA is made \[….\] and \[….\] to DNA
Parallel, complementary
5
New cards
2nd rule: RNA is made in the \[..\] direction
5’ → 3’
6
New cards
The DNA template is read… direction
3’ → 5’
7
New cards
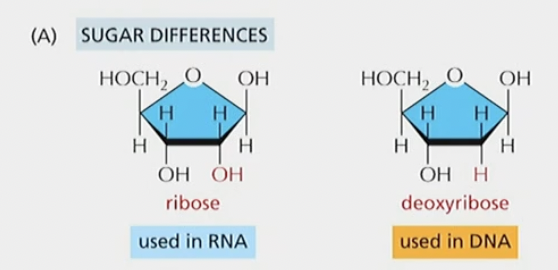
Sugar differences
Ribose has an OH group, deoxyribose does not
8
New cards
Is Uracil enough to determine whether something is RNA?
No, you can have dimers. you need to look for sugar differences.
9
New cards
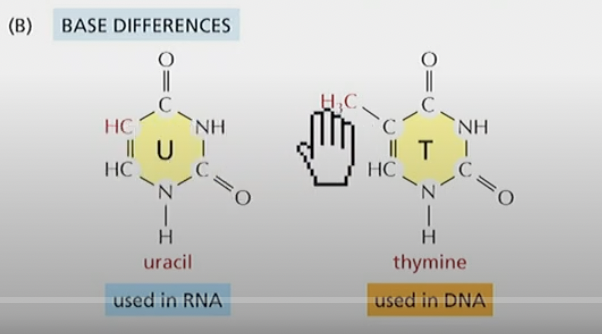
Difference between Uracil and Thymine
Uracil is missing a Methyl group
10
New cards
An important difference between RNA and DNA
Ribonucleoside triphosphates used (ATP, UTP, CTP, GTP)
11
New cards
RNA Pol. Holoenzyme
Sigma factor and core enzyme
12
New cards
Transcription cycle (bacterial)
Steps:
Sigma factor binds to RNAP and finds promoter sequence
* Holoenzyme localized unwinding of DNA, abortive transcription (a few short RNAs synthesized initially) and RNAP clamps down, sigma factor released
* elongation (processive) phase
* Termination & release of RNA
* Bacteria have different sigma factors
Sigma factor binds to RNAP and finds promoter sequence
* Holoenzyme localized unwinding of DNA, abortive transcription (a few short RNAs synthesized initially) and RNAP clamps down, sigma factor released
* elongation (processive) phase
* Termination & release of RNA
* Bacteria have different sigma factors
13
New cards
Does RNA polymerase require a primer?
No
14
New cards
Is RNA polymerase accurate? Why?
No, because it has exonuclease activity (the backspace)
15
New cards
How is the promoter sequence numbered?
Upstream is the more negative number, downstream is more positive.
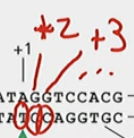
16
New cards
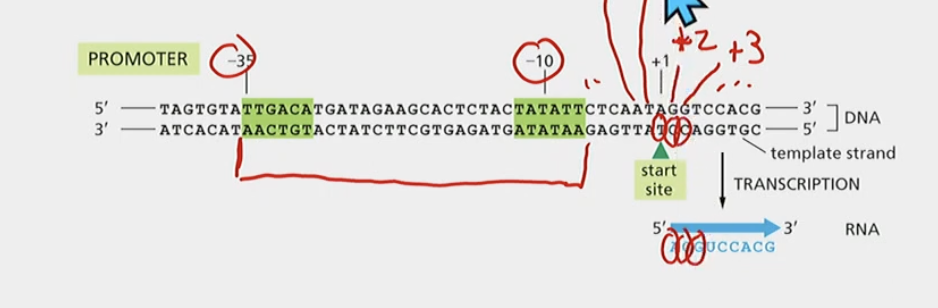
Where is the bacterial promoter
TTGACA to TATA box
17
New cards
Why does the sigma bean bound to the green sequences?
So that the active site is positioned over +1, right after the promoter consensus sequences
18
New cards
Where are the promoter consensus sequences shown and what are they?
The green sequences
* most common/average sequences
* -10, -35
* most common/average sequences
* -10, -35
19
New cards
Significance of the promoter
Positions RNA polymerase and determines whicg strand is template strand (3’ → 5’)
20
New cards
Hairpin loops
Formed due to base pairing with itself when the RNA transcript leaves RNAP
21
New cards
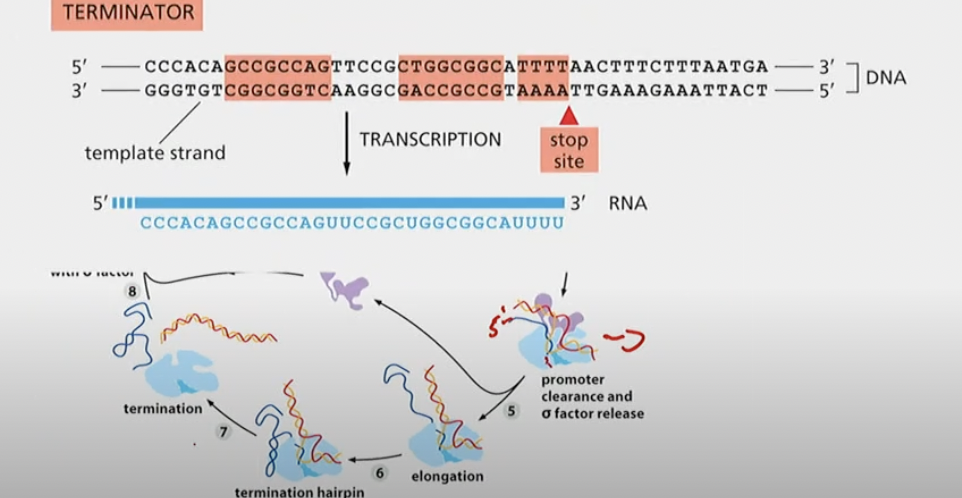
Terminator sequences
G’s and C’s followed b y A’s and T’s.
When the G’s and C’s bond, since they have 3 bonds they’re much stronger and form the hairpin formation. The DNA has weaker A’s, U’s, and T’s but since the hairpin is so strong it pries itself off of it
When the G’s and C’s bond, since they have 3 bonds they’re much stronger and form the hairpin formation. The DNA has weaker A’s, U’s, and T’s but since the hairpin is so strong it pries itself off of it
22
New cards
Key points about the transcription cycle
1) initial steps of RNA synthesis are relatively inefficient
2) elongation mode of RNAP is highly processive (moves very quickly)
2) elongation mode of RNAP is highly processive (moves very quickly)
23
New cards
What are some characteristics of RNA termination signals?
* Hairpin structure formed as a result of GC rich sequences
* AT rich DNA sequences following hairpin sequences (weak A-U bonding in the DNA/RNA duplex)
* AT rich DNA sequences following hairpin sequences (weak A-U bonding in the DNA/RNA duplex)
24
New cards
How do termination signals help to dissociate the RNA transcript from the polymerase
Disrupts H-bonding of new mRNA transcript with DNA template
25
New cards
@@Difference between sigma factor mode and elongation mode@@
\
26
New cards
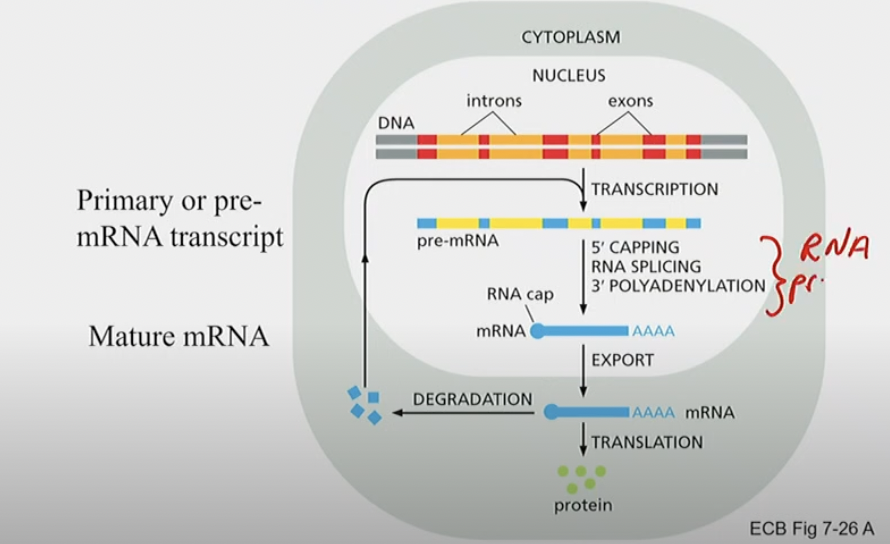
Key difference between prokaryotic gene expression and eukaryotic gene expression
Translation can begin before Transcription is complete because of the absence of a nucleus.
Eukaryotes have introns and exons, creating pre-mRNA transcript before translation begins.
5’ capping, RNA splicing and 3’ polyadenylation removes introns before mRNA leaves nucleus for translation.
Eukaryotes have introns and exons, creating pre-mRNA transcript before translation begins.
5’ capping, RNA splicing and 3’ polyadenylation removes introns before mRNA leaves nucleus for translation.
27
New cards
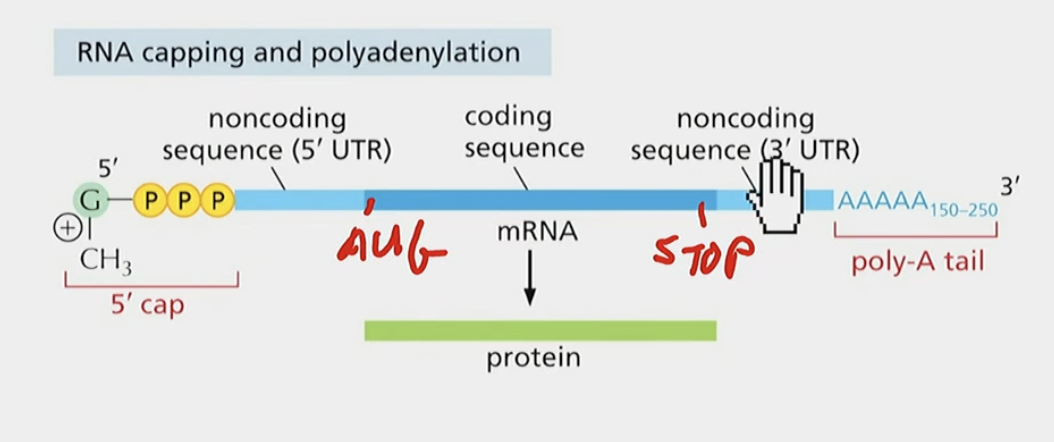
Do exons have to be coding?
No, because EVERYTHING left over after transcription is exons, (5’ UTR and 3’ UTR) and that includes noncoding sequence. They are essential or else ribosomes do not work.
UTR stands for UNTRANSLATED (i.e., non-coding)
UTR stands for UNTRANSLATED (i.e., non-coding)
28
New cards
RNA types present in both Eukaryotes and Prokaryotes
mRNA rRNA tRNA
29
New cards
Do all RNAs help create proteins?
No
30
New cards
RNAP I
transcribes most RNA genes
31
New cards
RNA II
transcribes all protein-coding genes
32
New cards
RNA Pol III
transcribes tRNA genes
33
New cards
What are protein-coding genes
mRNAs
34
New cards
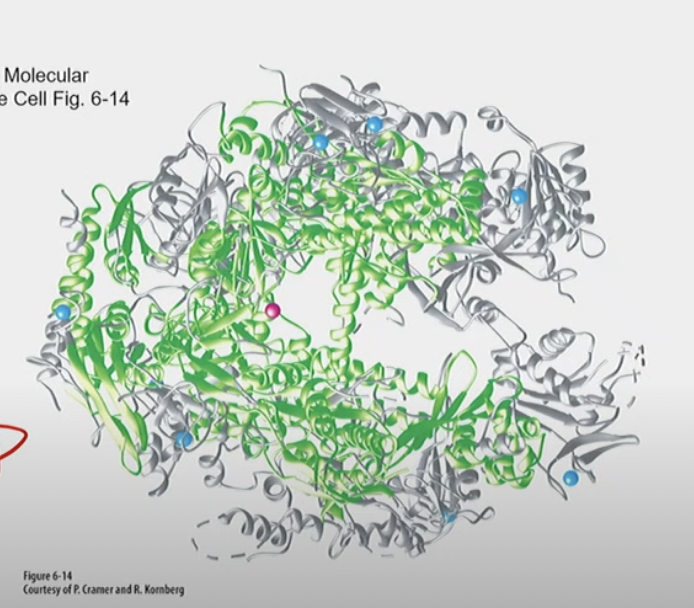
Eukaryotic RNAP II vs. Bacterial RNAP structure
Bacterial RNAP has 5 subunits, eukaryotic RNA Pol II has 12
* RNA Pol II has a %%SPECIAL carboxyl terminal domain (CTD)%% not found in bacterial or other eukaryotic RNAPs
* RNA Pol II has a %%SPECIAL carboxyl terminal domain (CTD)%% not found in bacterial or other eukaryotic RNAPs
35
New cards
Eukaryotic vs. bacterial RNA polymerases
Eukaryotic RNAPs require transcription factors
and also need to deal with chromosomal structures, like histones
and also need to deal with chromosomal structures, like histones
36
New cards
Transcription factors
* proteins to help position RNAP at the promoter
* *similar* to the sigma subunit to bacterial RNA polymerases
* *similar* to the sigma subunit to bacterial RNA polymerases
37
New cards
Sigma subunit
bean-shaped protein behind active site of BACTERIAL RNAP
38
New cards
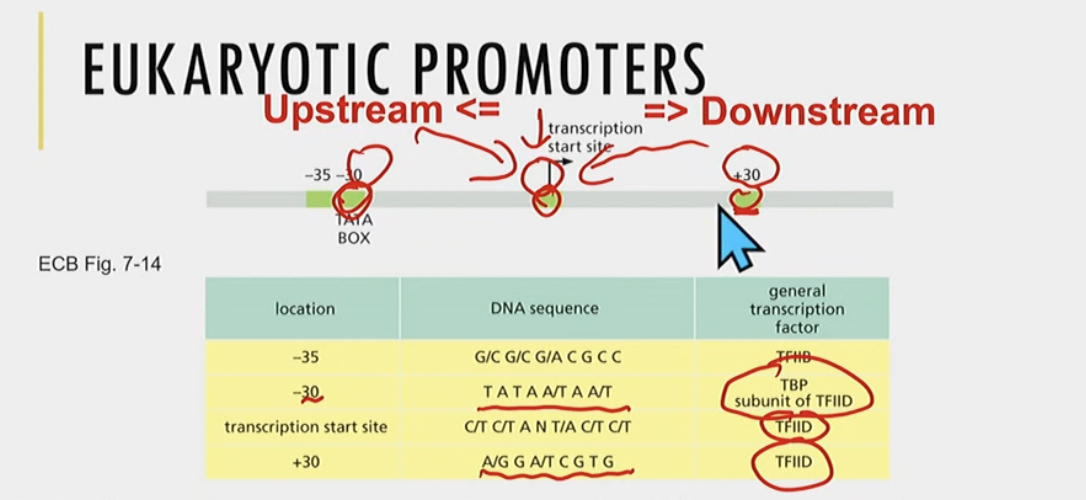
Eukaryotic promoters
* more variable than bacterial promoters
* 1 or more specific sequences called elements
* specific elements are at specific locations
* elements are recognized by specific general transcription factors, which in turn, help position the RNAP relative to the transcription start site
* 1 or more specific sequences called elements
* specific elements are at specific locations
* elements are recognized by specific general transcription factors, which in turn, help position the RNAP relative to the transcription start site
39
New cards
Elements
40
New cards
TATA box
sequence TATA is highly (evolutioarily) conserved, found \~30 bp upstream from start site for transcription (-30)
* Helps position RNAP II ANDDD general transcriptiion factors
* TATA box common, but many other types of promoter sequences (elements)
* Helps position RNAP II ANDDD general transcriptiion factors
* TATA box common, but many other types of promoter sequences (elements)
41
New cards
Steps in the initiation of transcription
Binding of TBP (TATA Binding Protein) subunit of TFIID (Transcription Factor (RNA pol II)) near TATA box sequences
* mobilizes the binding of TFIIB complex adjacent to TATA box
* Other transcription factors (TFs) bind
* RNAP II and other TFs will be able to bind in correct orientation at transcription start site
* SPECIAL: Helicase activity & Phosphorylation (Kinase activity) of C-Terminal Domain (CTD) of RNAP II -- both jobs performed by TFIIH
* Helicase activity separates strands, RNAP II is abortive until phosphyrlation happens, then it keeps on going
* OTHER RNAPs DO NOT HAVE THE CTD TAIL
\
4 steps summarized:
1) TBP, a subunit of TFIID, binds to the TATA box promoter in the minor groove, bending and distorting DNA
2) this attracts other TPs, which help to orient and bind RNAP II to the DNA
3) the helicase activity of TFIIH uses ATP to pry apart DNA strands at transcription start site
4) TFIIH ALSO phosphorylates the C-terminal domain of RNAP II, activating it so that transcription can begin
* mobilizes the binding of TFIIB complex adjacent to TATA box
* Other transcription factors (TFs) bind
* RNAP II and other TFs will be able to bind in correct orientation at transcription start site
* SPECIAL: Helicase activity & Phosphorylation (Kinase activity) of C-Terminal Domain (CTD) of RNAP II -- both jobs performed by TFIIH
* Helicase activity separates strands, RNAP II is abortive until phosphyrlation happens, then it keeps on going
* OTHER RNAPs DO NOT HAVE THE CTD TAIL
\
4 steps summarized:
1) TBP, a subunit of TFIID, binds to the TATA box promoter in the minor groove, bending and distorting DNA
2) this attracts other TPs, which help to orient and bind RNAP II to the DNA
3) the helicase activity of TFIIH uses ATP to pry apart DNA strands at transcription start site
4) TFIIH ALSO phosphorylates the C-terminal domain of RNAP II, activating it so that transcription can begin
42
New cards
Activator protein
binds to enhancer DNA sequences, folds over and kisses mediator, telling RNAP to go faster, getting out of abortive phase
43
New cards
RNAP II c-terminal domain
the carboxyl terminal domain of largest subunit has a stretch of 7 amino acids that is repeated multiple times (tandem repeats)
1 reapt = tyr-ser-pro-thr-ser-pro-ser
yeast enzyme has 26 repeats
human enzyme has 52 repeats
C-TERMINAL DOMAIN IS ESSENTIAL FOR VARIABILITY
1 reapt = tyr-ser-pro-thr-ser-pro-ser
yeast enzyme has 26 repeats
human enzyme has 52 repeats
C-TERMINAL DOMAIN IS ESSENTIAL FOR VARIABILITY
44
New cards
How is RNAP II activated?
Phosphorylation
45
New cards
What is phosphorylated in RNAP II acitvation?
adding phosphate groups on S (Ser) located on the CTD
46
New cards
How many proteins are involved in initiating eukaryotic transcription?
>100 subunits of many proteins
47
New cards
mRNA Processing: Phosphorylation of C-terminal tail of _____ results in…
RNAP II
\
binding of RNA processing proteins
Additional phosphorylation of CTD and phosphorylation pattern changes
\
YOU RARELY GET THE FULL UNPROCESSED PRE-MRNA
\
binding of RNA processing proteins
Additional phosphorylation of CTD and phosphorylation pattern changes
\
YOU RARELY GET THE FULL UNPROCESSED PRE-MRNA
48
New cards
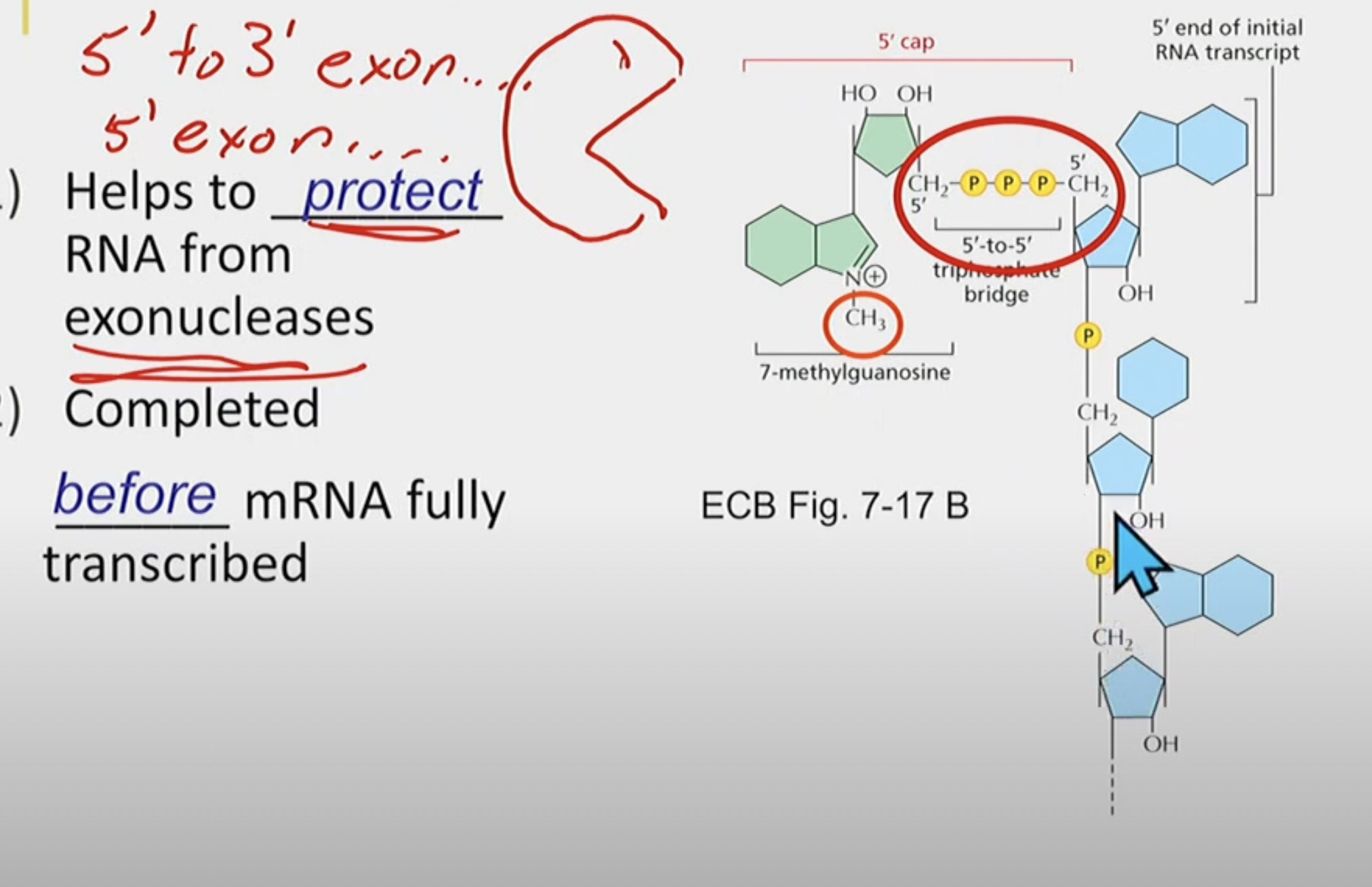
5’ pre-mRNA capping
Helps to protect RNA from 5’ → 3’ exonucleases
completed BEFORE mRNA fully transcribed
completed BEFORE mRNA fully transcribed
49
New cards
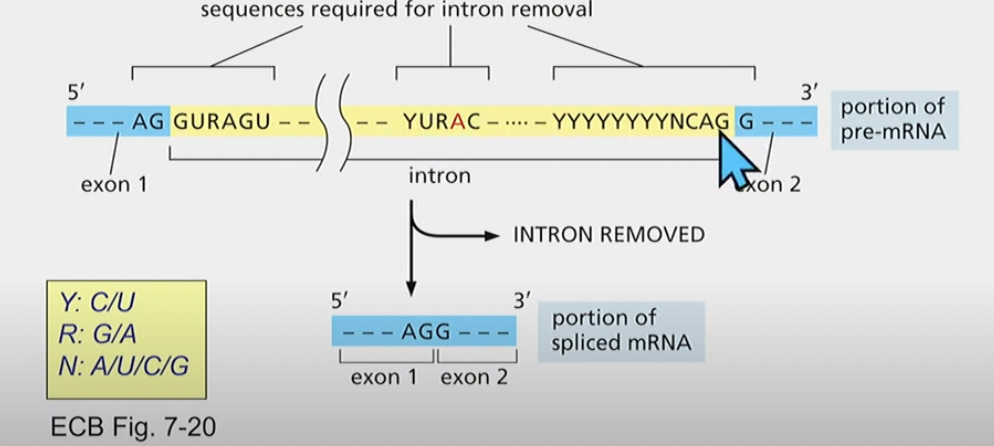
Removal of introns from pre-mrna
1) branch-point A attacks the 5’ (OH) splice site
2) 3’ of on exon reacts with 5’ of next exon to release intron
THIS ALL HAPPENS THROUGH A SPLICESOME COMPLEX
2) 3’ of on exon reacts with 5’ of next exon to release intron
THIS ALL HAPPENS THROUGH A SPLICESOME COMPLEX
50
New cards
Catalytic mechanism is RNA dependent (significance of what functional group?)
3) 2’OH group of the ribose sugar is not present in deoxyribose
4) this group is necessary for the formation of the lariat structure in intron splicing
4) this group is necessary for the formation of the lariat structure in intron splicing
51
New cards
snRNPs
pre-mRNAs not able to self-splice
Splicesosomes contain snRNAs bound to protein -- (snRNPs) plus other associated proteins
Spliceosomes assemble on mRNA to remove introns
When splicing complete, exon junction complex added to signify completion of splicing
Splicesosomes contain snRNAs bound to protein -- (snRNPs) plus other associated proteins
Spliceosomes assemble on mRNA to remove introns
When splicing complete, exon junction complex added to signify completion of splicing
52
New cards
Correct sequences required for accurate mRNA splicing
53
New cards
Why does alternative RNA splicing increases the number of gene products?
Because different kinds of splicing results in different kinds of gene expression (different kinds of mRNA)
54
New cards
Abnormal splicing
You can have exon skipping, cryptic (incomplete) splice sites, or new splice sites created causing new exons to be incorporated
55
New cards
Termination in eukaryotes
3’-end modifying proteins
Gu-rich or U-rich sequences are encoded in the genome
but after they are transcribed, the 3’ end processing proteins recognize them on the mRNA and are recruited to the mRNA
note that the poly-A sequence is NOT encoded in the genome
Gu-rich or U-rich sequences are encoded in the genome
but after they are transcribed, the 3’ end processing proteins recognize them on the mRNA and are recruited to the mRNA
note that the poly-A sequence is NOT encoded in the genome
56
New cards
mRNA processing (CTD phosphorylation)
CTD phosphorylation happens because, each time, it allows a different protein to bind to the tail.
change in phosphorylation happens each time for:
1. move capping proteins from CTD to 5’ end of growing mRNA from RNAP
2. move spliceosome over to splice
3. move 3’ end processing proteins
change in phosphorylation happens each time for:
1. move capping proteins from CTD to 5’ end of growing mRNA from RNAP
2. move spliceosome over to splice
3. move 3’ end processing proteins
57
New cards
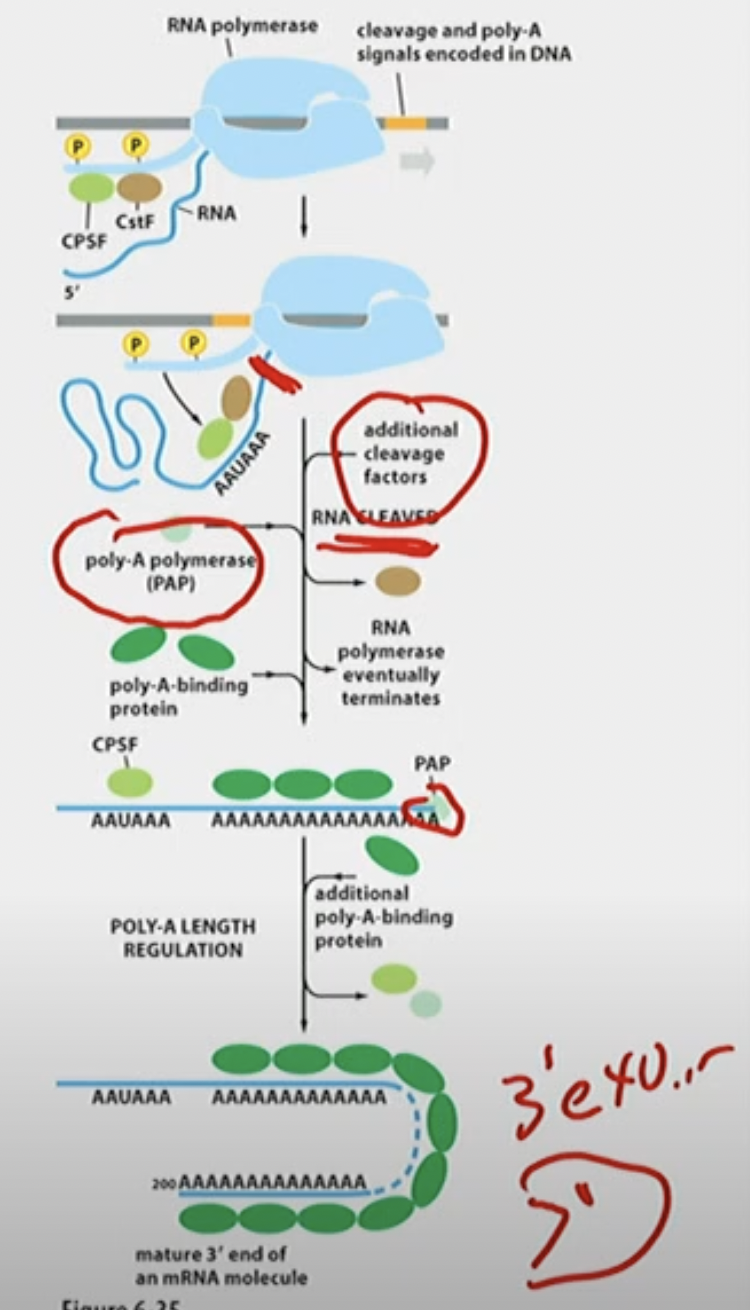
3’ end processing
CPSF (Cleavage (meaning cuts RNA) and Polyadenylation specificity factor) and CstF (Cleavage Stimulation Factor)
poly-A signals (Ts) transcribed to AAUAAA
CPSF and CstF transferred over to RNA
CPSF cleaves RNA CPSF is still binded
poly-A-polymerase (PAP) directly adds (binds) a bunch of A sequences to RNA after AAUAAA, using ATP. CPSF is still binded.
Length is regulated when PAP terminates, poly-A-binding proteins protect 3’ end from 3’ → 5’ exonucleases on mature 3’ end of an mRNA molecule
\
in summary:
\
→ consensus sequences (yellow portion in picture) direct cleavage and polyadenlyation of the 3’ end.
→ 3’ end processing proteins move from CTD to MRNA
→ Cleavage and addition of a poly-A 3’ tail along with Poly A-binding proteins result in the mature mRNA
poly-A signals (Ts) transcribed to AAUAAA
CPSF and CstF transferred over to RNA
CPSF cleaves RNA CPSF is still binded
poly-A-polymerase (PAP) directly adds (binds) a bunch of A sequences to RNA after AAUAAA, using ATP. CPSF is still binded.
Length is regulated when PAP terminates, poly-A-binding proteins protect 3’ end from 3’ → 5’ exonucleases on mature 3’ end of an mRNA molecule
\
in summary:
\
→ consensus sequences (yellow portion in picture) direct cleavage and polyadenlyation of the 3’ end.
→ 3’ end processing proteins move from CTD to MRNA
→ Cleavage and addition of a poly-A 3’ tail along with Poly A-binding proteins result in the mature mRNA
58
New cards
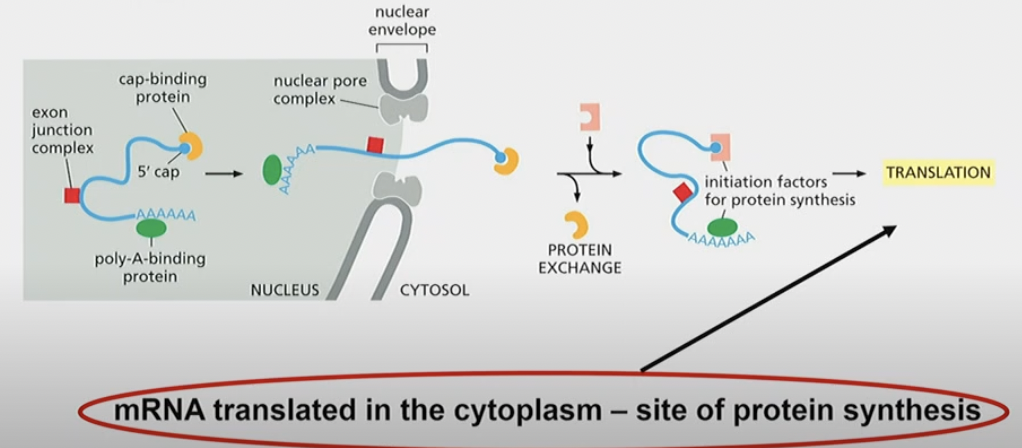
mRNA export
* exported from nucleus to cytosol
* cap-binded protein binds to 5’ end
* exon junction complex also binds to where introns used to be
* note: they are not binding the exons, they’re just.. there
* poly-A-binding protein and sequence at 3’ end
* exits through nuclear pore complex
* In cytosol, cap binding complexes can be removed, and initiation factors for protein synthesis bind, leading to translation
* cap-binded protein binds to 5’ end
* exon junction complex also binds to where introns used to be
* note: they are not binding the exons, they’re just.. there
* poly-A-binding protein and sequence at 3’ end
* exits through nuclear pore complex
* In cytosol, cap binding complexes can be removed, and initiation factors for protein synthesis bind, leading to translation