cancer genetics part 1
1/137
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
138 Terms
tumor suppressor examples
p53, Rb, APC, BRCA1
oncogene
accelerator
mutated genes that cause cancer
genome maintenance mutations lead to cancer because _____
mutations in DNA cause oncogenes
chromosome loss
more mutations = more likely to hit consequential genes contributing to cancer
BRCA1 repairs damaged DNA
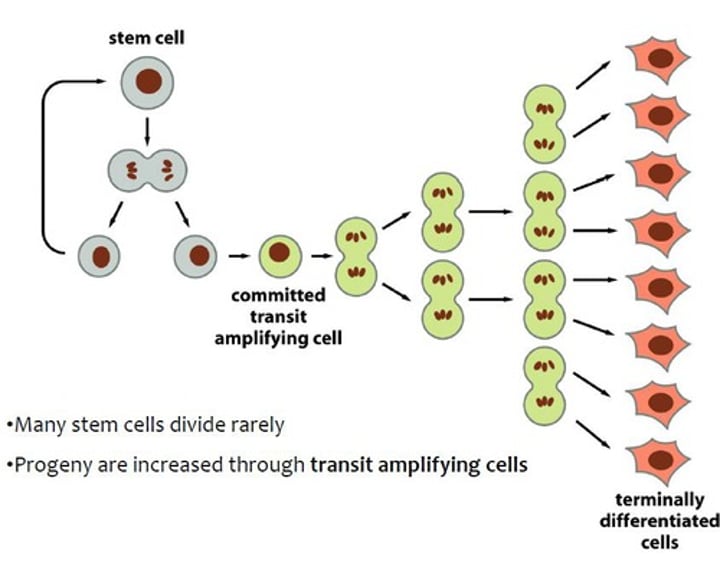
cell fate mutations lead to cancer because _____
defects in differentiation cause uncontrolled growth
stem cells trapped in transit amplifying state
cell growth and survival genes lead to cancer because _____
signals that say divide are unleashed when they shouldnt be
no apoptosis
p53 signaling hub - normally inhibits cell cycle
cdk1 - normally promotes cell cycle
braf - pro mitogenic growth
passenger mutation
not directly contributing to cancer
silent mutation (no change in AA)
unrelated, benign
not going to be listed on cards
driver mutation
consequential in inducing cancer
common/important pathways
listed on cards
different combinations of mutations can ________
contribute to the same type of cancer
mutations in the same gene can be associated with ________
different types of cancers
how do normal cells become cancer cells
cancer relevant genes → altered function
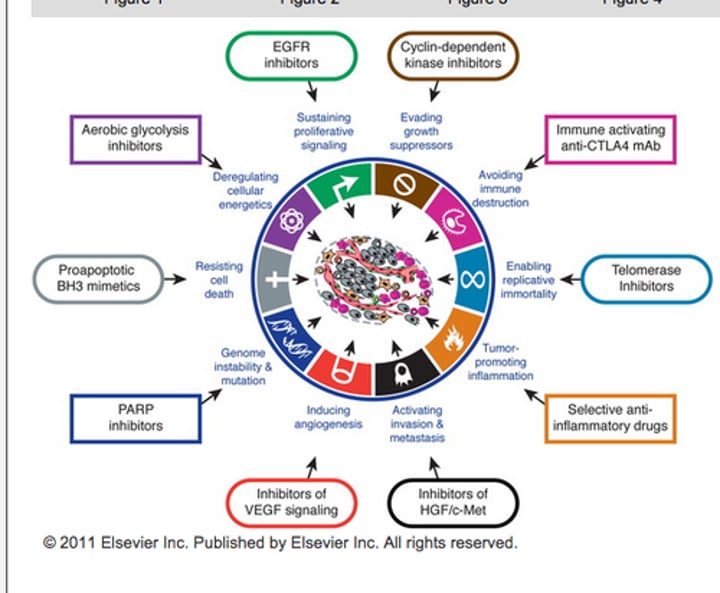
hallmarks of cancer
Sustaining proliferative signaling
Evading growth suppressors
Resisting cell death
Enabling replicative immortality
Inducing angiogenesis
Activating invasion and metastasis
how is cancer a genetic disease
normal cell → (mutations, epigenetics) → cancer cell
mutations - direct changes to nucleotide
epigenetics - changing what genes are on/off
genetic changes - sequencing + genetic studies
2 things that cancer cells do are ____
1. grow/divide
2. spread (metastasis, goes through basement membrane)
cancer is caused by ___
dysregulation of genes
Two major ways in which genes become dysregulated
1. genetic - mutations
2. epigenetic - histone mods, DNA methylation
genetic causes of cancer
inherited/somatic
induced by mutagens (UV radiation, chemical, viruses, smoking, cells dividing)
epigenetic causes of cancer
histone modification, DNA methylation
mutagens
cause mutations
always carcinogenic
carcinogens
cause cancer
not always mutagens (could be epigenetic)
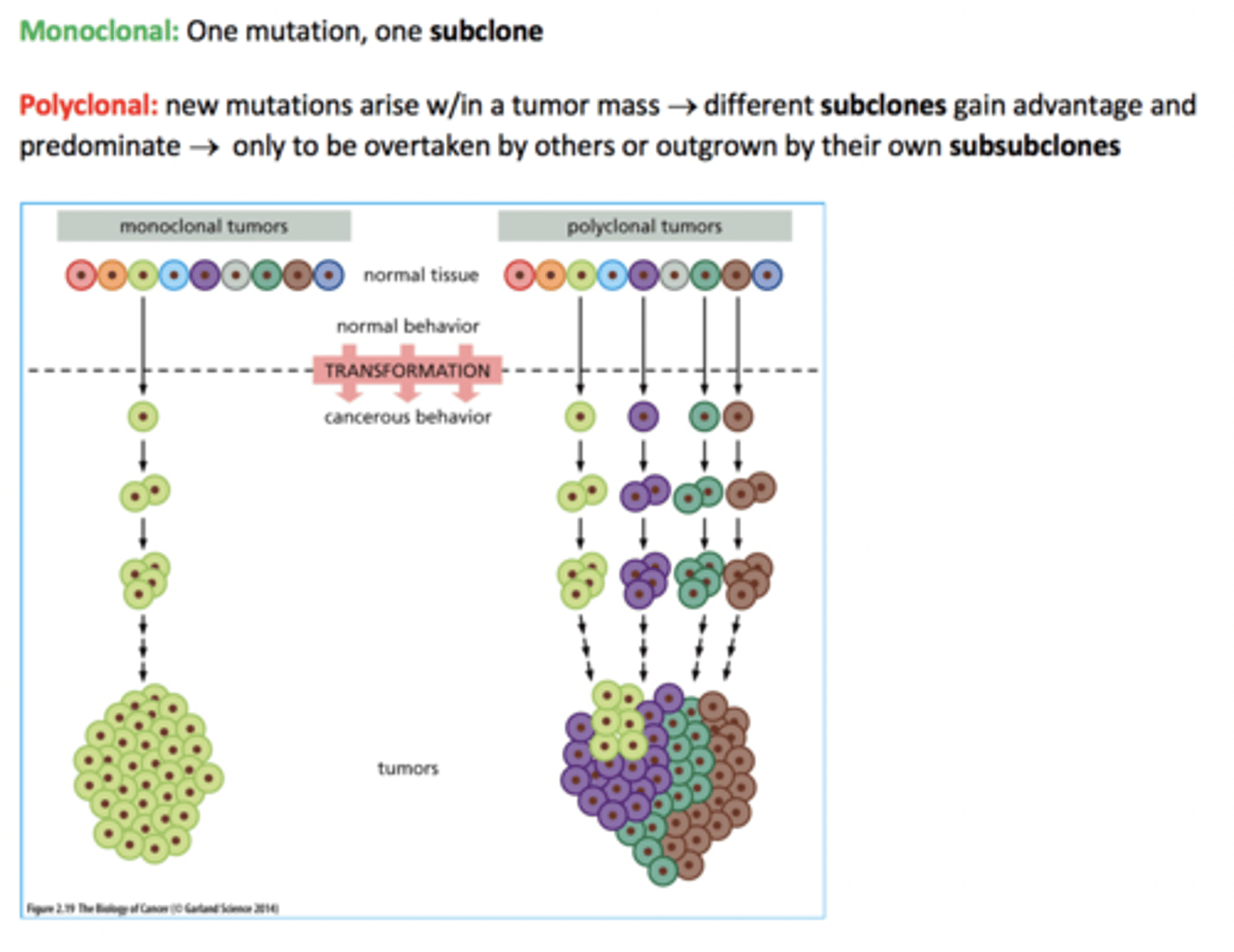
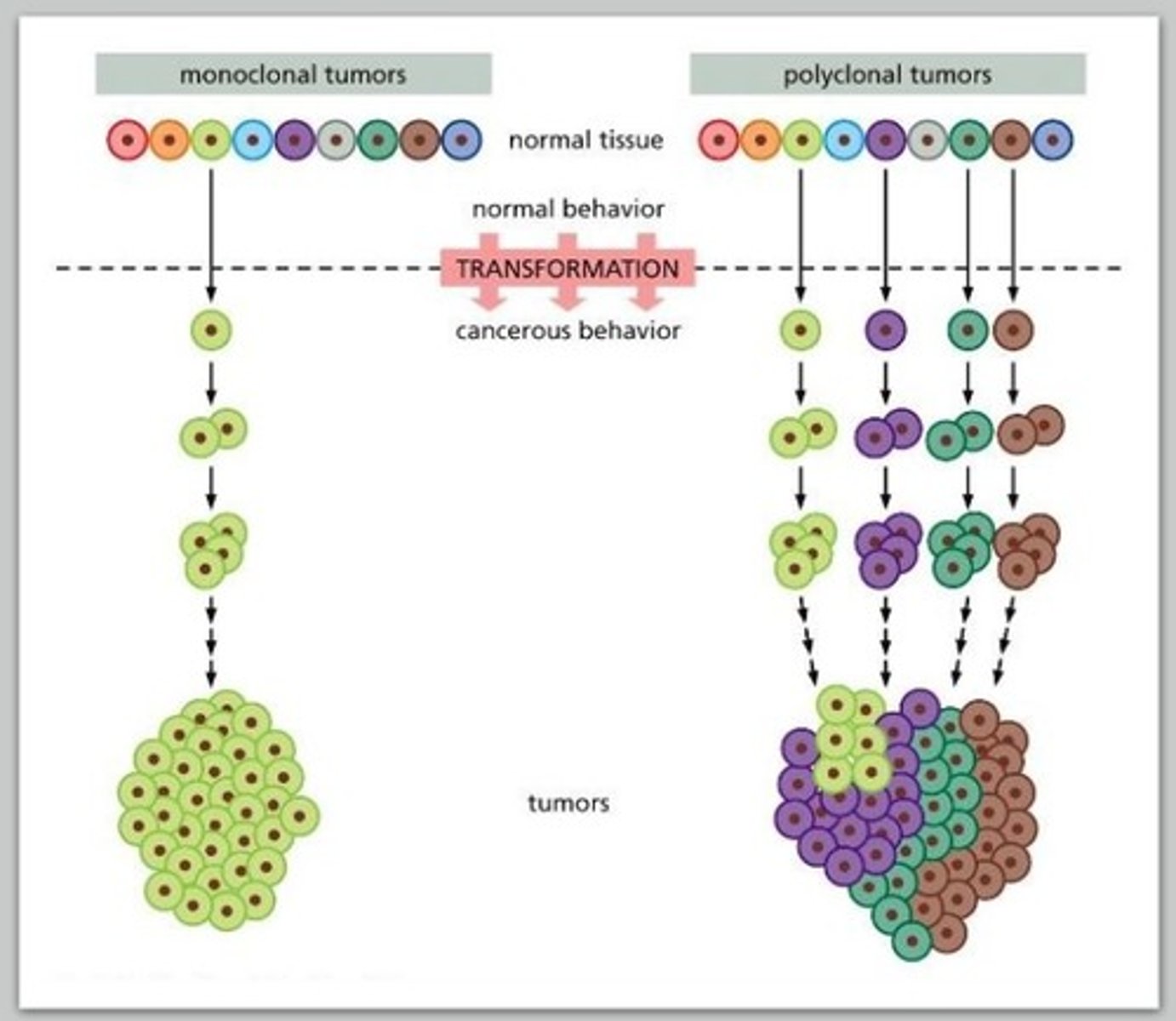
cancer cells are _____clonal in origin
monoclonal
1 cell was the source
2 pieces of evidence for monoclonal origin
1. myeloma: tumor of B cell precursors (antibody producing cells)
2. chromosomal aberrations
evidence for monoclonal origin:
myeloma
each B cell produces a different antibody
Antibody diversity is produced by VDJ recombination
a monoclonal myeloma cancer cell will express that antibody in high amounts
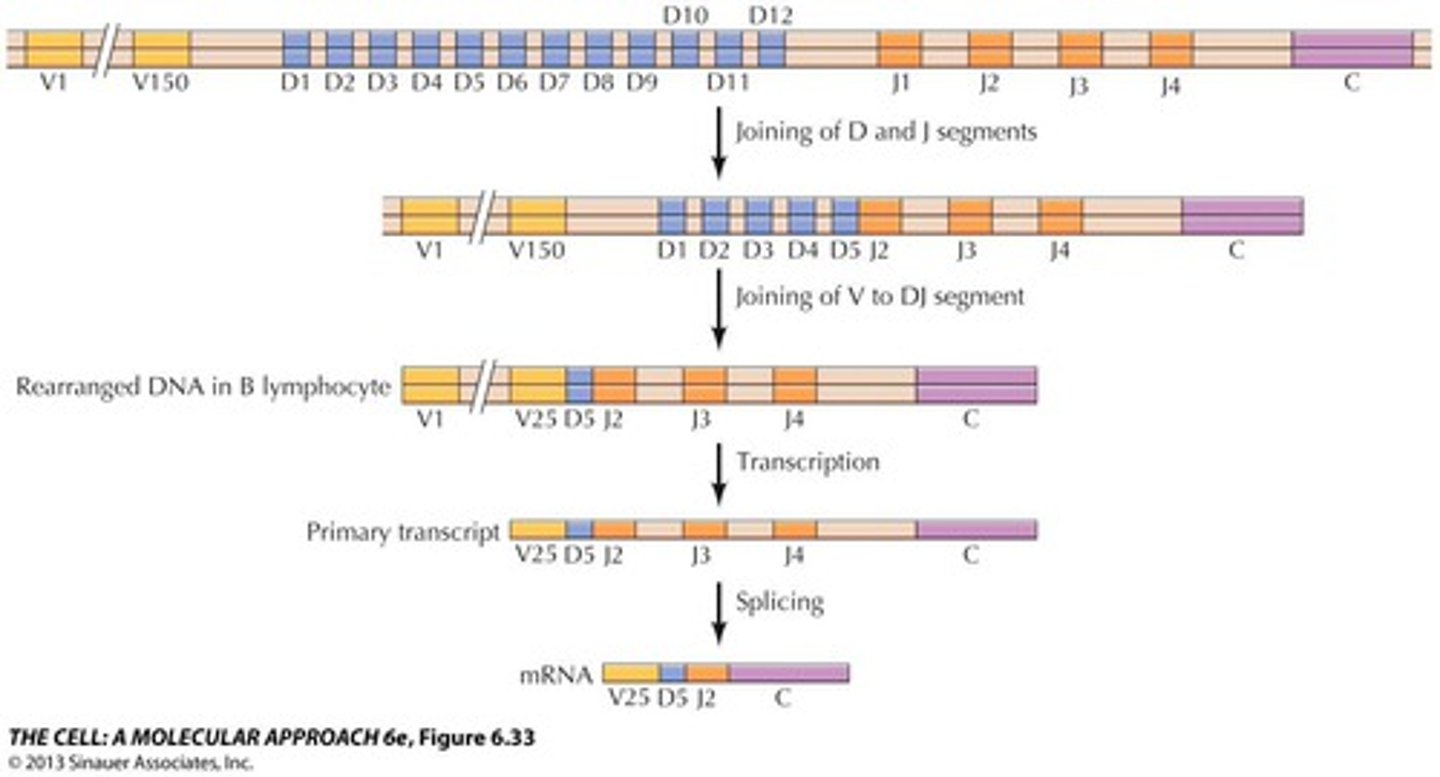
evidence for monoclonal origin:
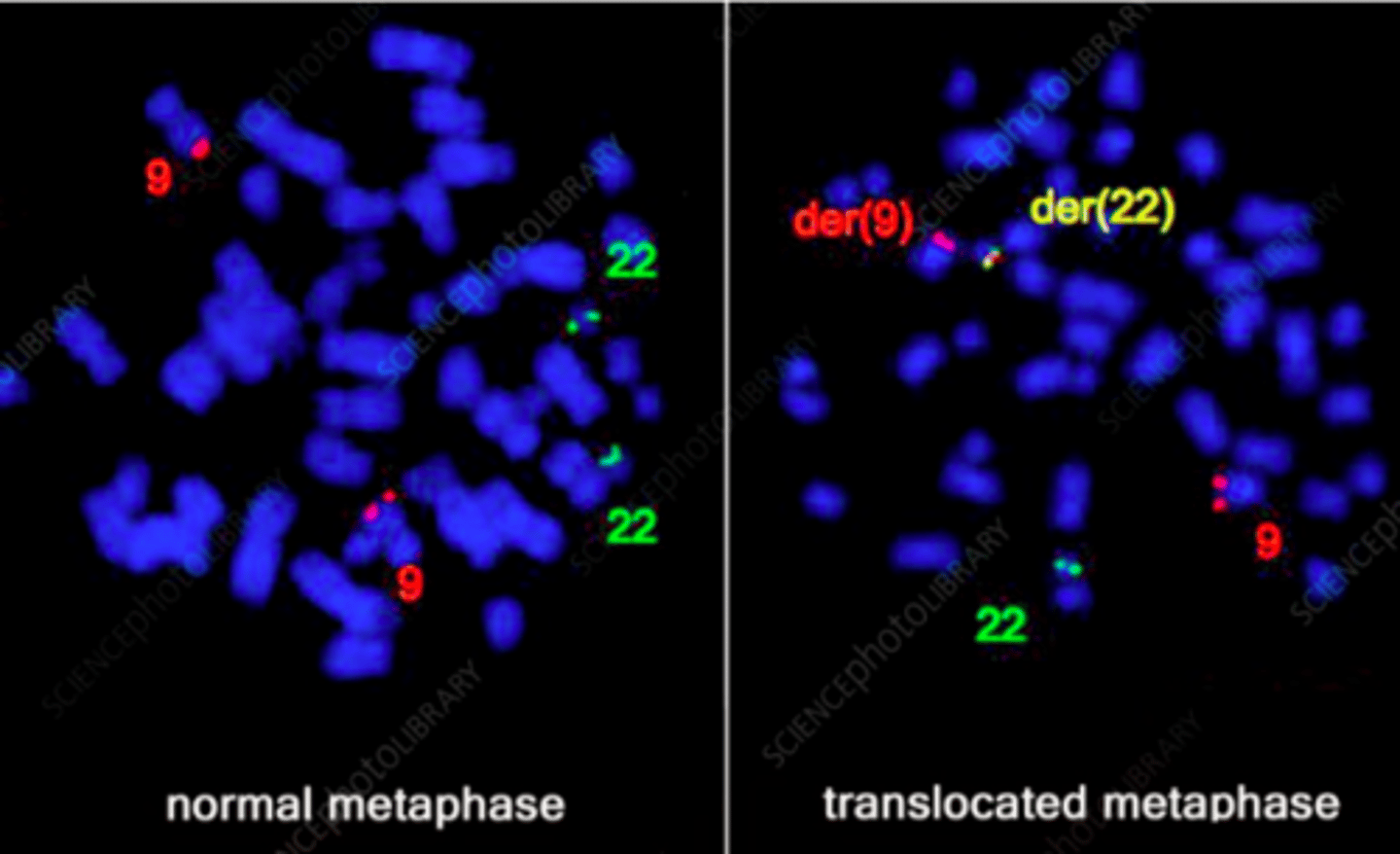
chromosomal aberrations
translocations
all cells in tumor tissue will have the same translocation
stain using FISH
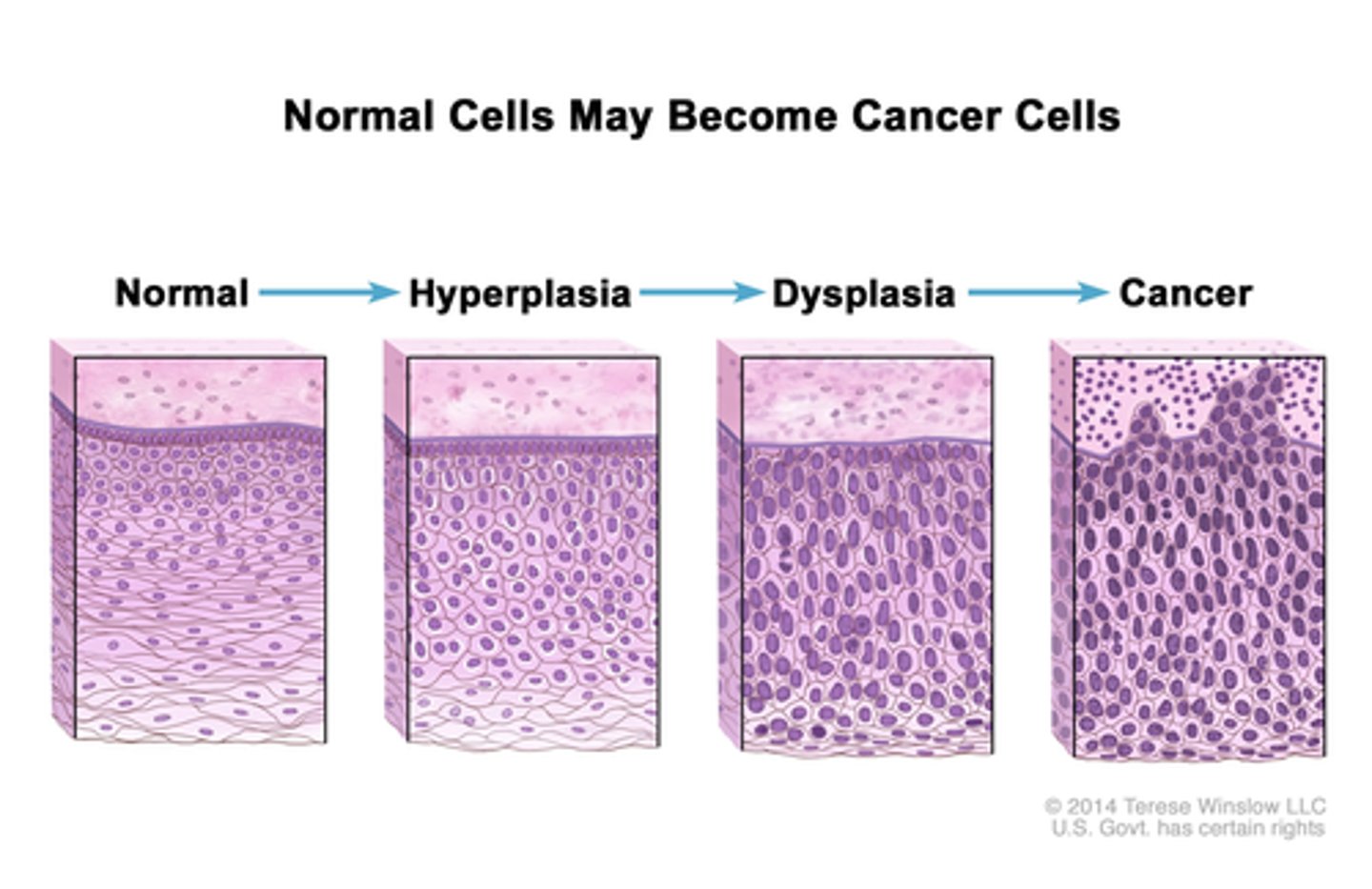
multistep process of cancer:
histopathological
normal
hyperplasia - excess # of normal cells
dysplasia - abnormal size/shape
^^^BENIGN^^^
neoplastic - breaks basement membrane
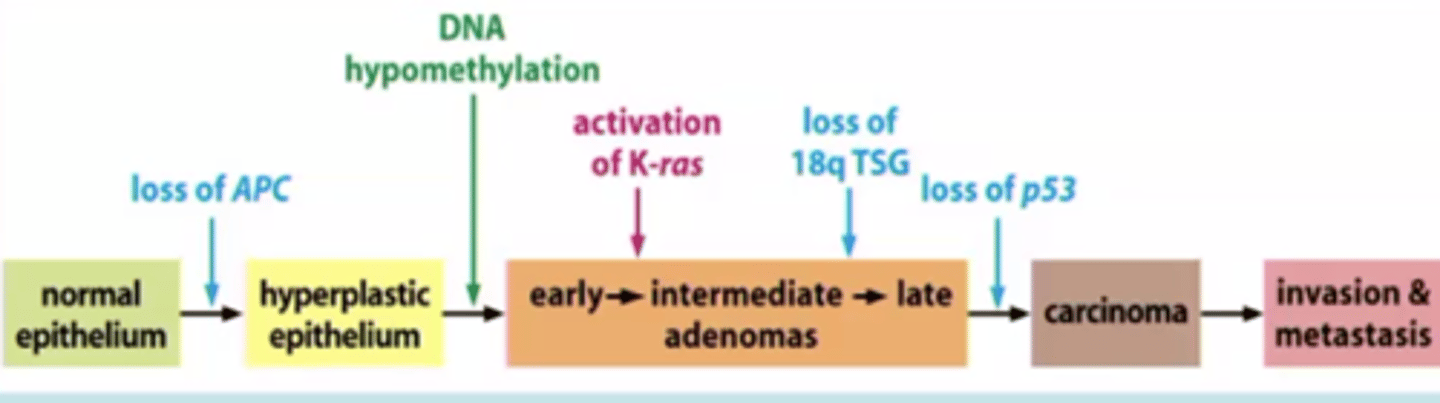
multistep process of cancer:
genetic
increasingly neoplastic
increasing # altered genetic loci
normal → loss APC → hyperplastic → hypomethalyation/activation → carcinoma
tumor progression takes ____
time
could be years after initiated to develop into cancer
tumor evolution
evolving units are cells
leads to clonal expansions
high mutation rates =
clonal diversification
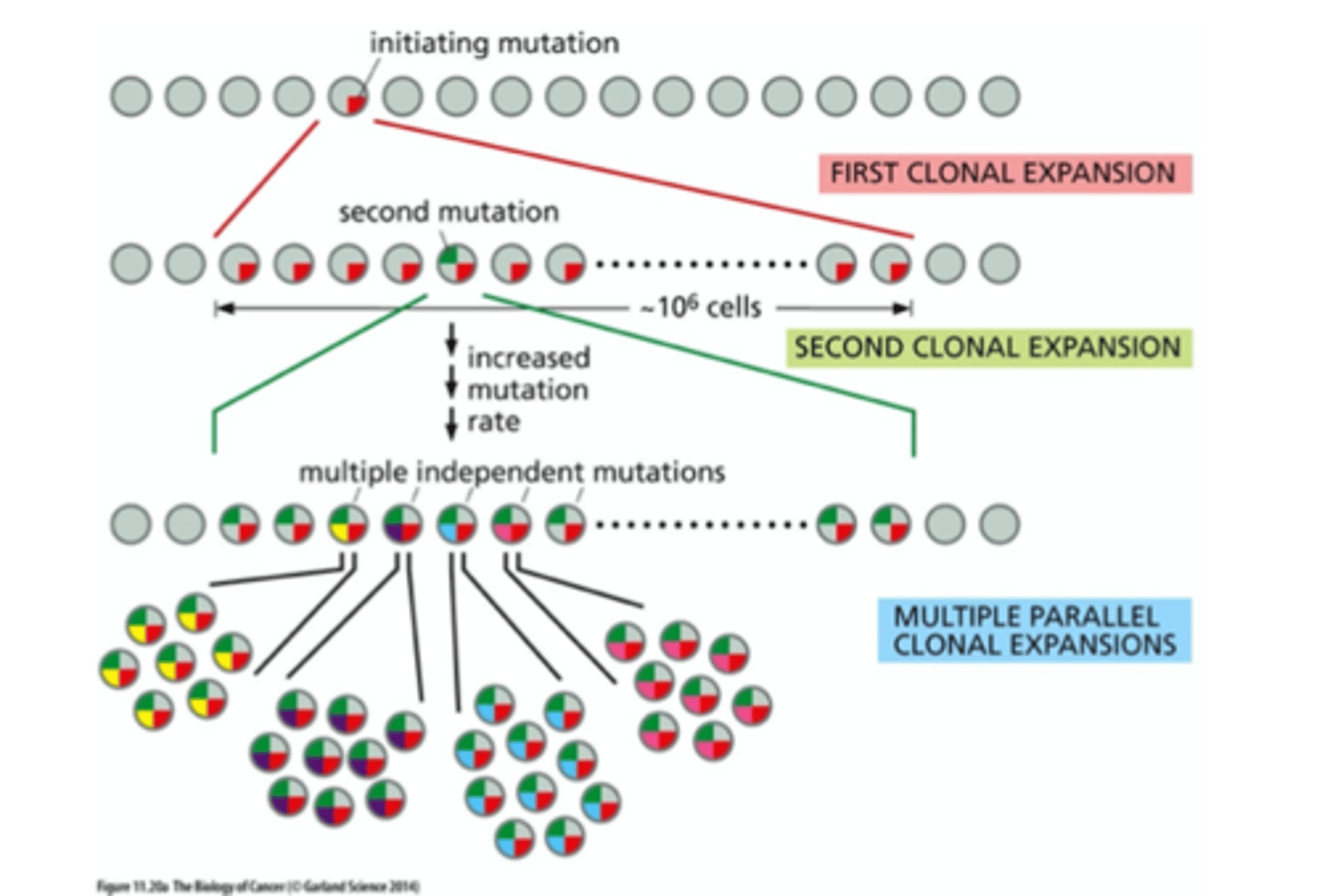
monoclonal origin, but subsequent ____ can lead to ______
mutations can lead to heterogeneity
Where do cancer causing mutations come from?
inherited pre-disposition
knudson's two hit hypothesis
mutation terminology based on phenotypic effects
- Null, loss-of-function, gain-of-function, reduction-of-function
- hypermorphic (increase gene function), hypomorphic (decreased), neomorphic (new)
- Recessive
- Dominant
- dominant negative
- Haploinsufficient
- Autosomal -vs- X-linked
- Lethal, sterile
- Conditional (WT phenotype under one condition, mutant phenotype under other envi condition)
mutation terminology based on phenotypic effects:
hypermorphic
hypomorphic
neomorphic
increase gene function
decrease gene function
new gene function
mutation terminology based on molecular lesion
- Point mutation: change of 1 base
- missense: change aa
- nonsense: change to stop
- silent: no change in aa
- neutral: occurs in intergenic region
Indel (insertion/deletion)
Sometimes called DIPs (deletion/insertion) polymorphism
Point mutations
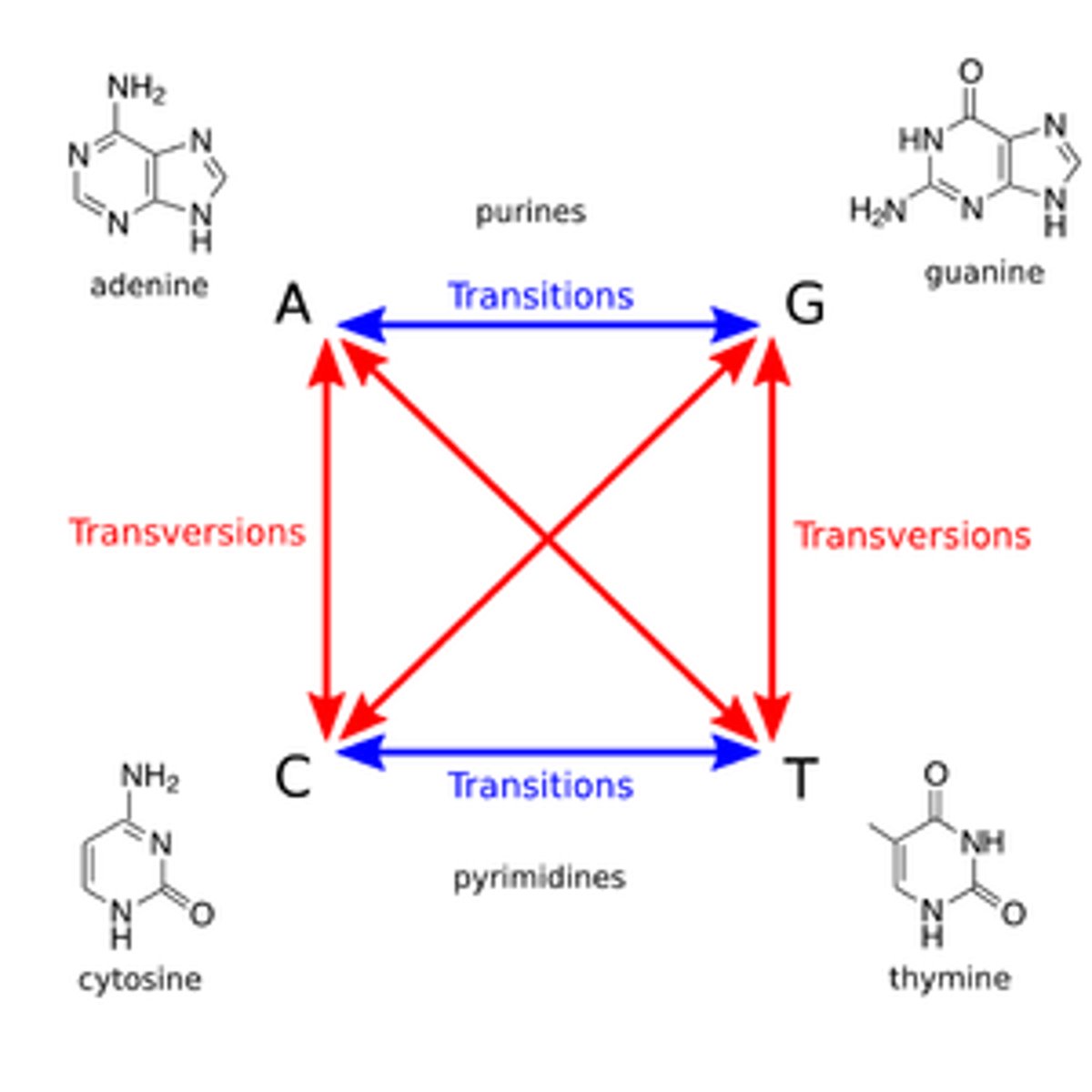
transitions: purine->purine, pyrimidine->pyrimidine
transversions: purine
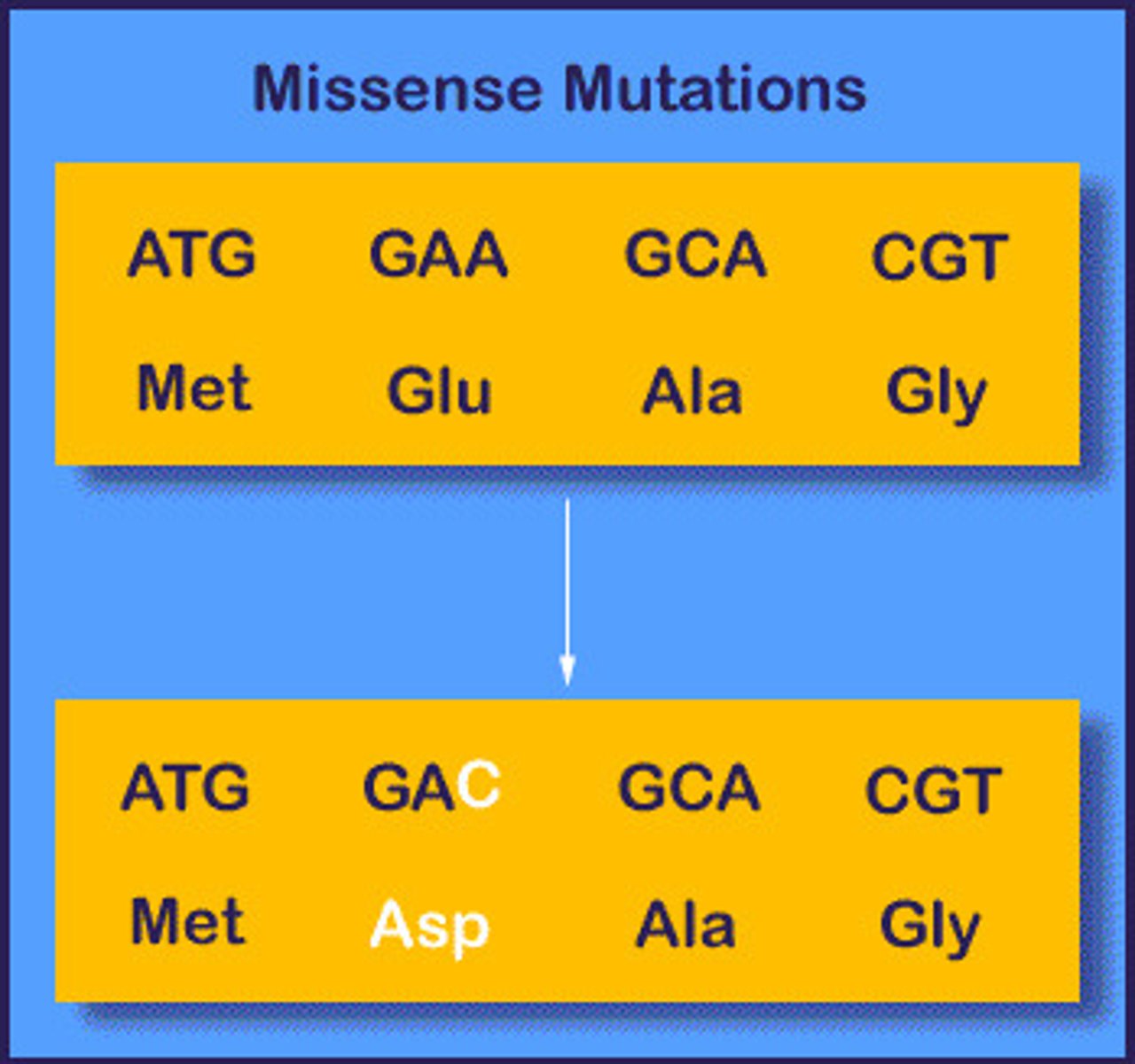
missense mutation
based on molecular lesion
point mutation
change aa
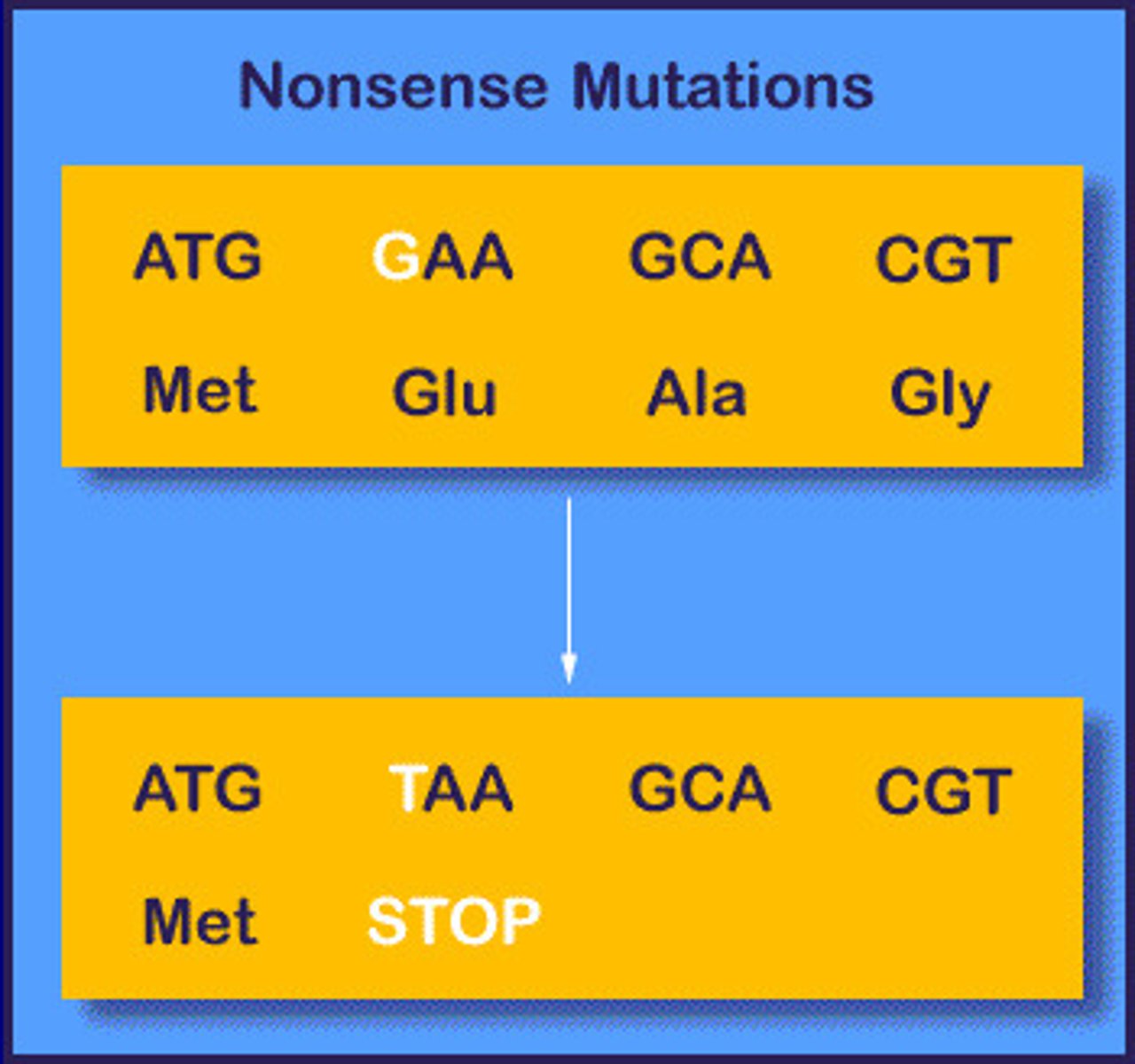
nonsense mutation
based on molecular lesion
point mutation
change to stop
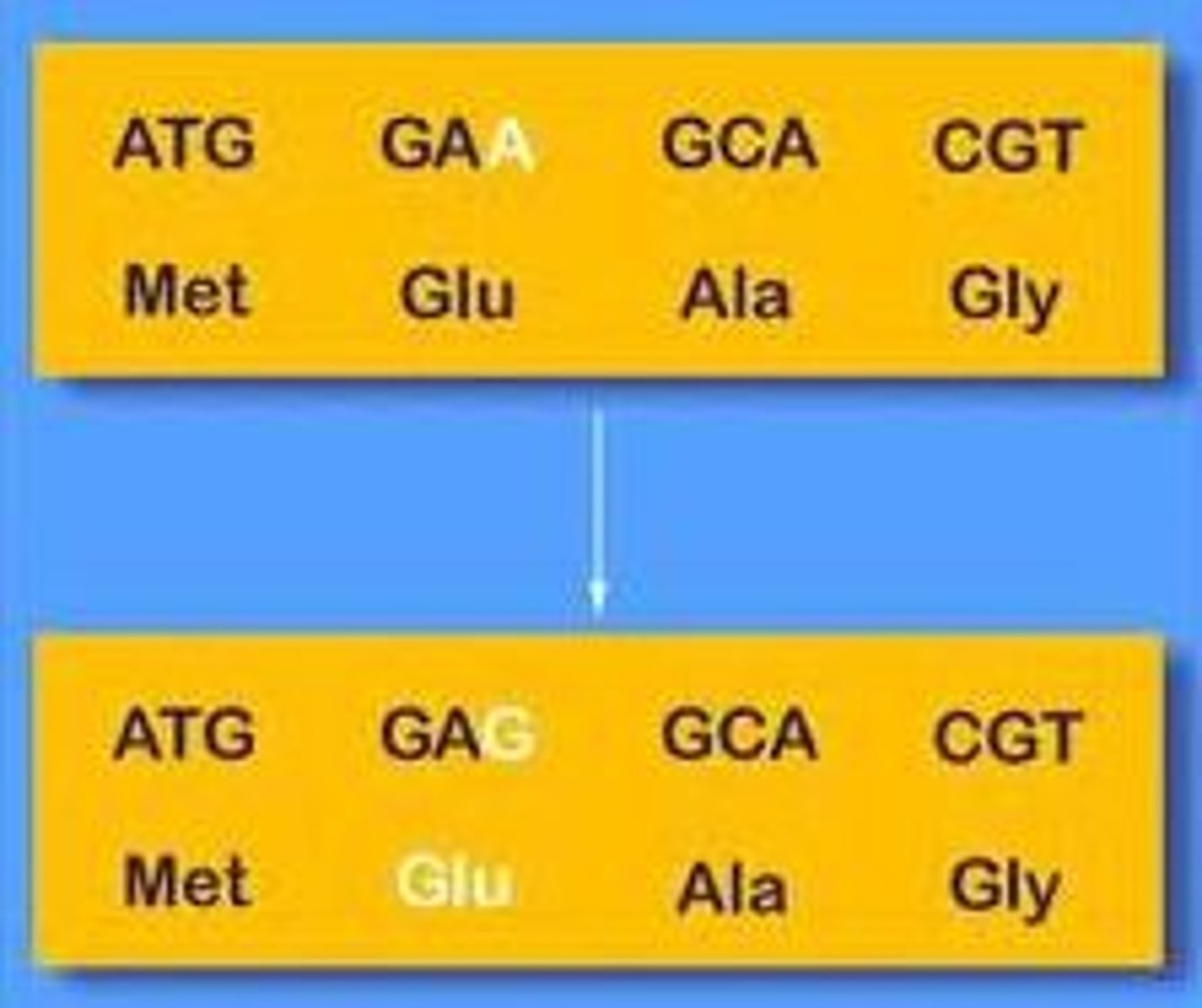
silent mutation
based on molecular lesion
point mutation
no change in aa
neutral mutation
based on molecular lesion
point mutation
occurs in intergenic (non-coding) region
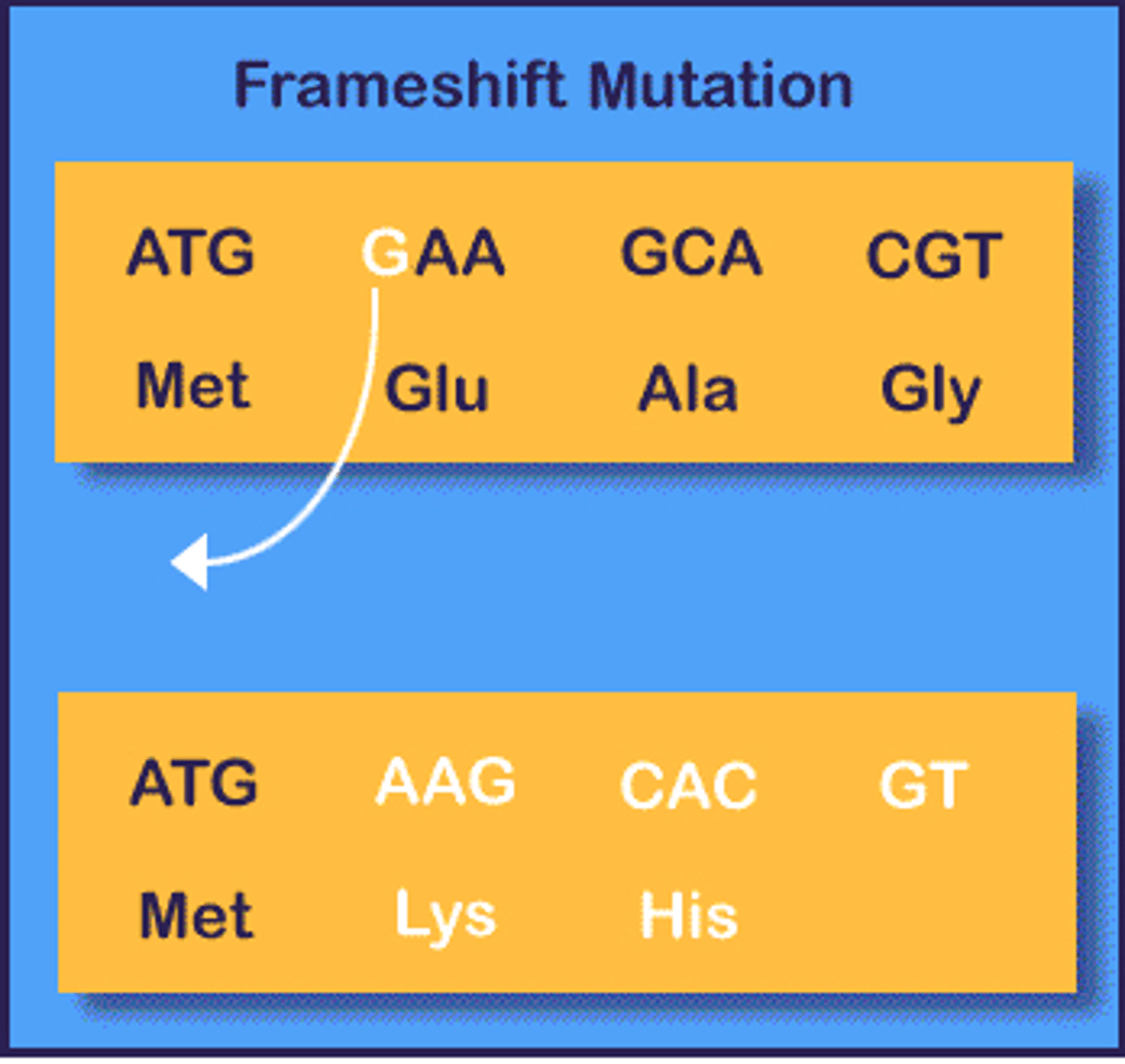
indel mutation
based on molecular lesion
point mutation
insertion/deletion
Sometimes called DIPs (deletion/insertion) polymorphism
transition mutation
purine → purine
pyrimidine → pyrimidine
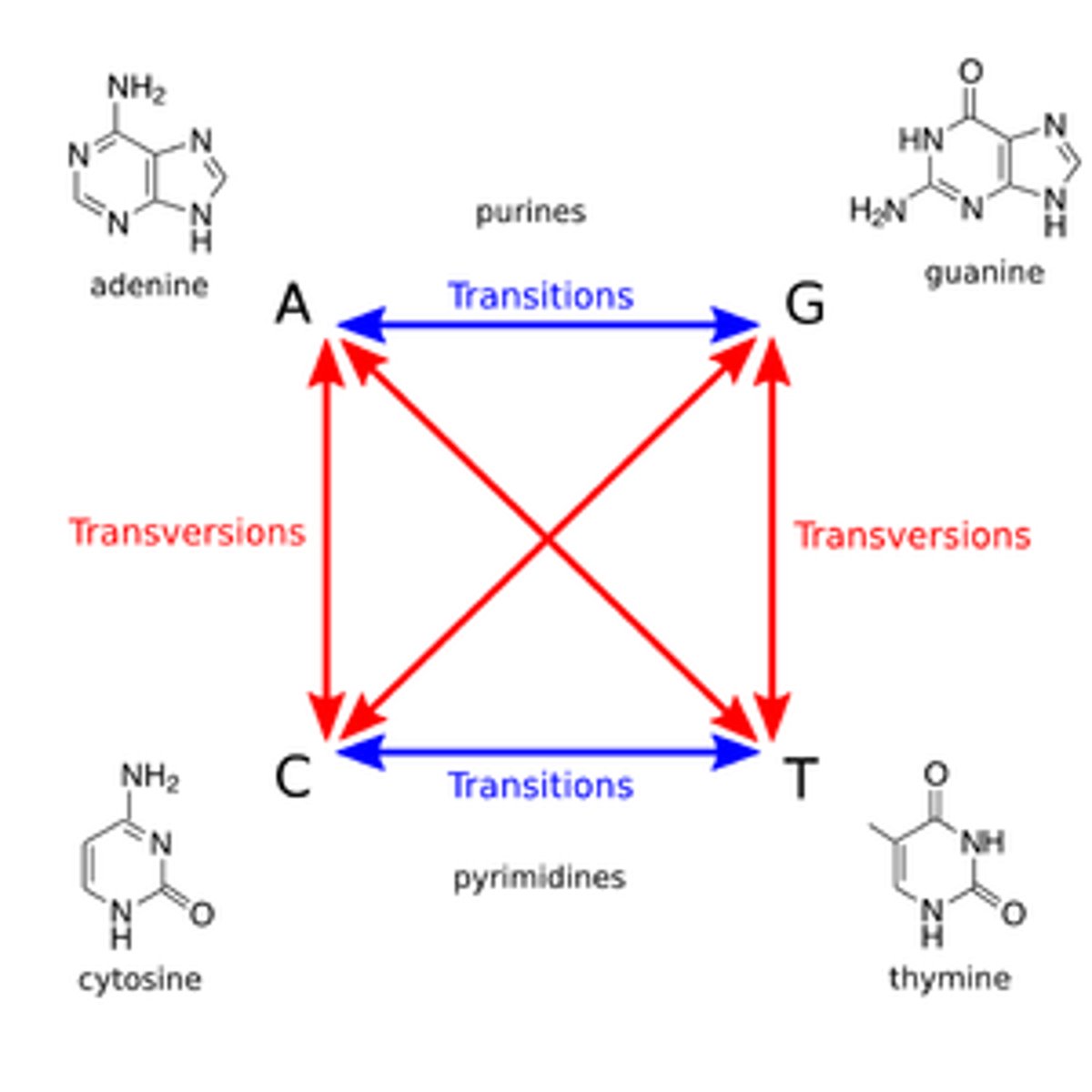
transversion mutation
purine → pyrimidine
vice versa
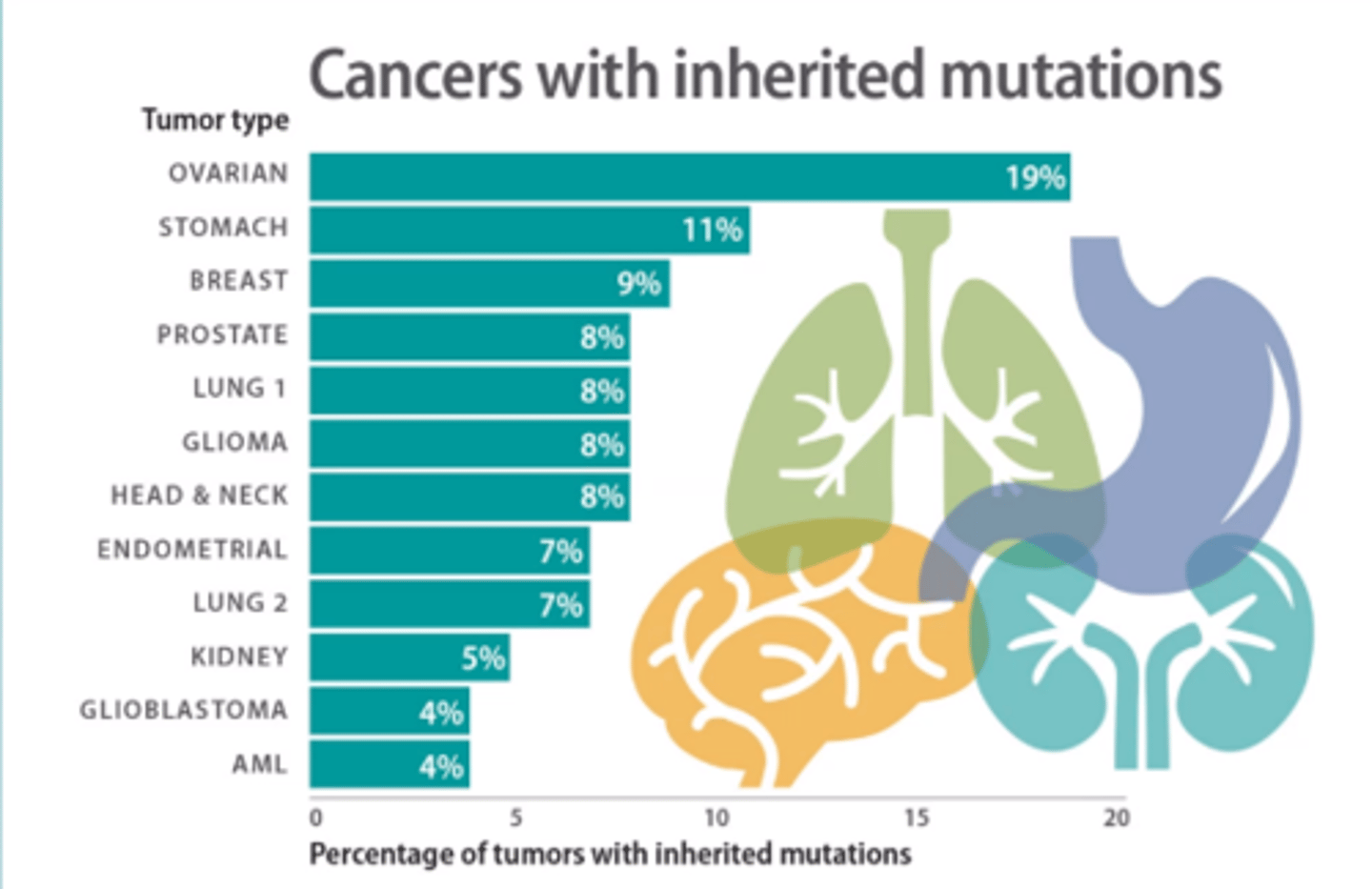
Where do cancer causing mutations come from?
Inherited Pre-disposition
ovarian has highest %
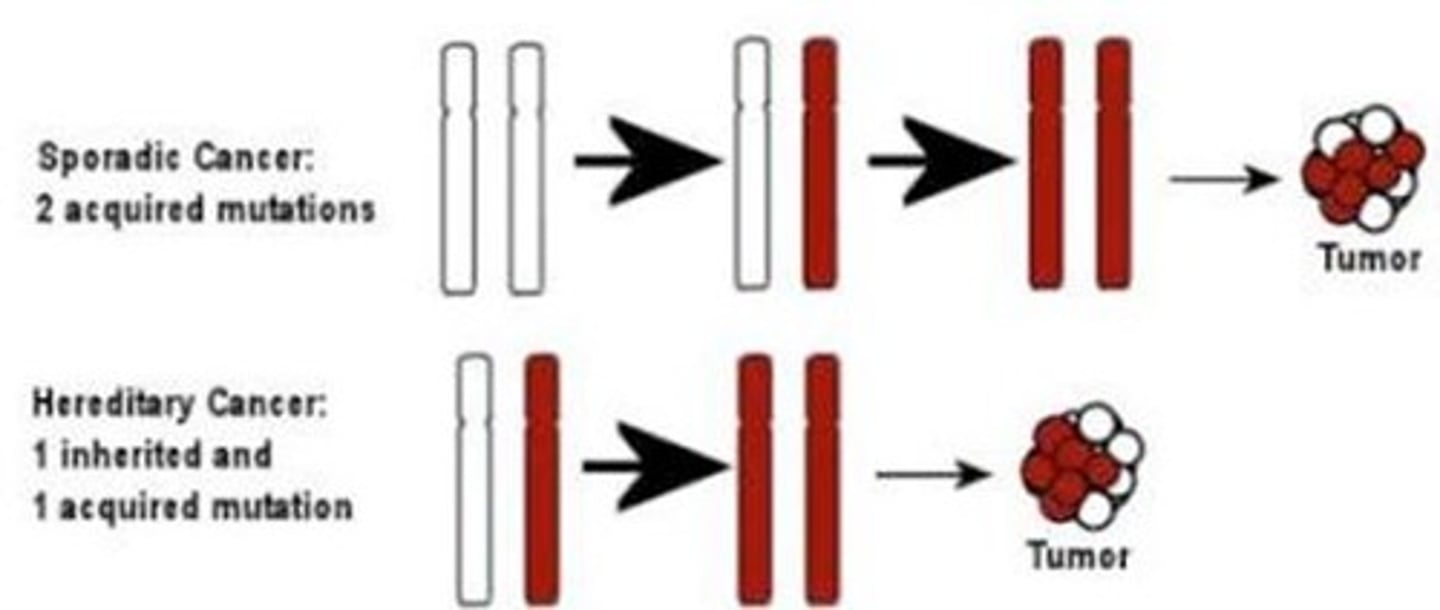
in the two hit hypothesis, initially all cells are ___
heterozygous
cell cycle control is normal
1 hit
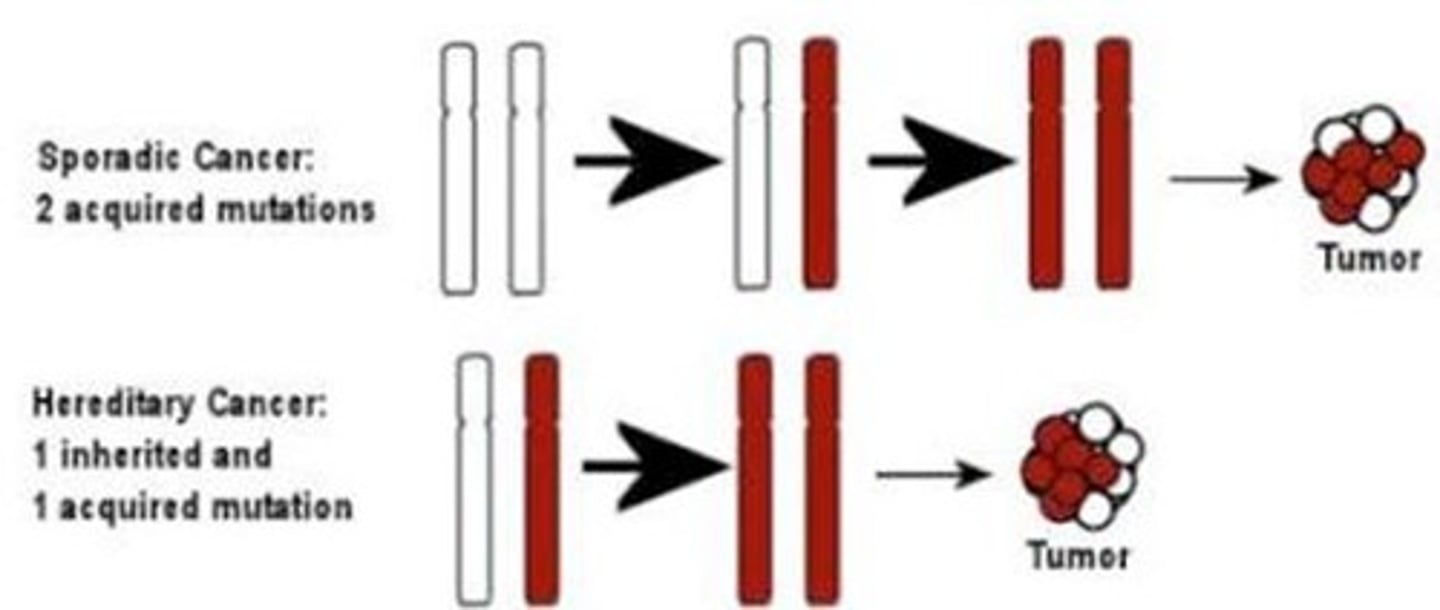
in the two hit hypothesis, a mutation becomes ___
homozygous
cell cycle control is abnormal
inherited predisposition
For most inherited pre-dispositions to cancer the mutation is recessive at the cellular level, but leads to a dominant inheritance pattern.
Why?
cancer is familial
two hit hypothesis
Spectrum of genetic instability in cancer
Nucleotide-level alterations
- DNA damage repair pathways
- MMR or NER deficiency
- herediatry or somatic
Gross Chromosomal Rearrangements
- telomere erosion
- ionizing radiation
- non-allelic HR
- replication stress
- chromothripsis
Whole Chromosome Instability
- loss/gain of whole chrom
- spindle checkpoint dysfunction
- centrosome overduplication
- chromatid cohesion defects
- merotelic attachments
Discuss the evidence that most variability in cancer incidence between tissues is due to numbers of cell division undertaken
PAPER #0
Random mutations occurring during normal DNA replication of stem cell division explains the variation in cancer risk among tissues.
This is shown by the strong correlation between the number of stem cell divisions and the lifetime risk of cancer.
This explains why certain parts of the body are more likely to develop cancer, regardless of hereditary factors or exposure to environmental mutagens.
why is cell division mutagenic
Most dangerous thing a cell can do: DNA replication
not perfect
- mis-incorporation of bases (mistakes, presence of tautomers in DNA)
- incorporation of base analogs (more tautomeric shifts)
- breakage of unwound DNA (one backbone broken progresses into full DSB)
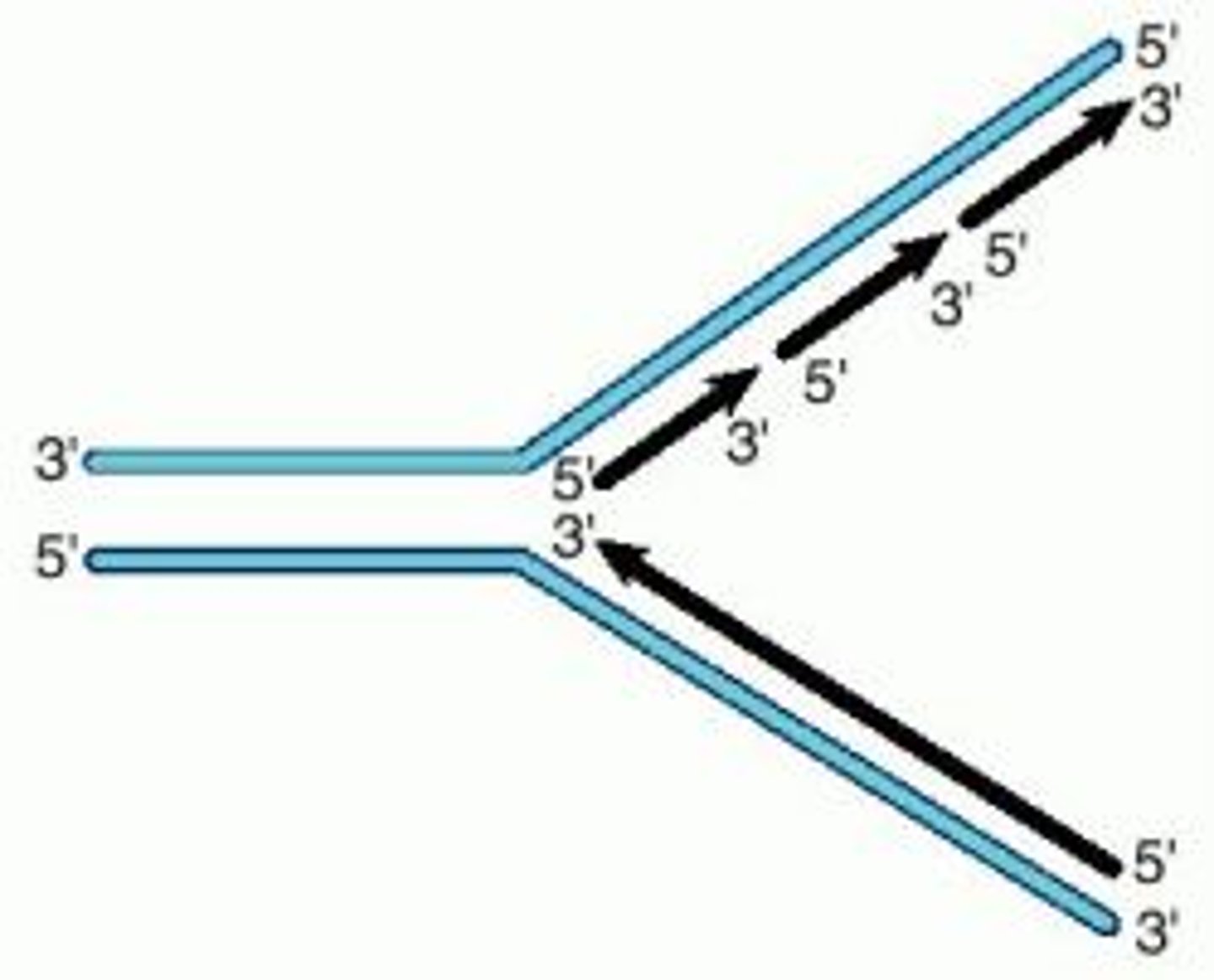
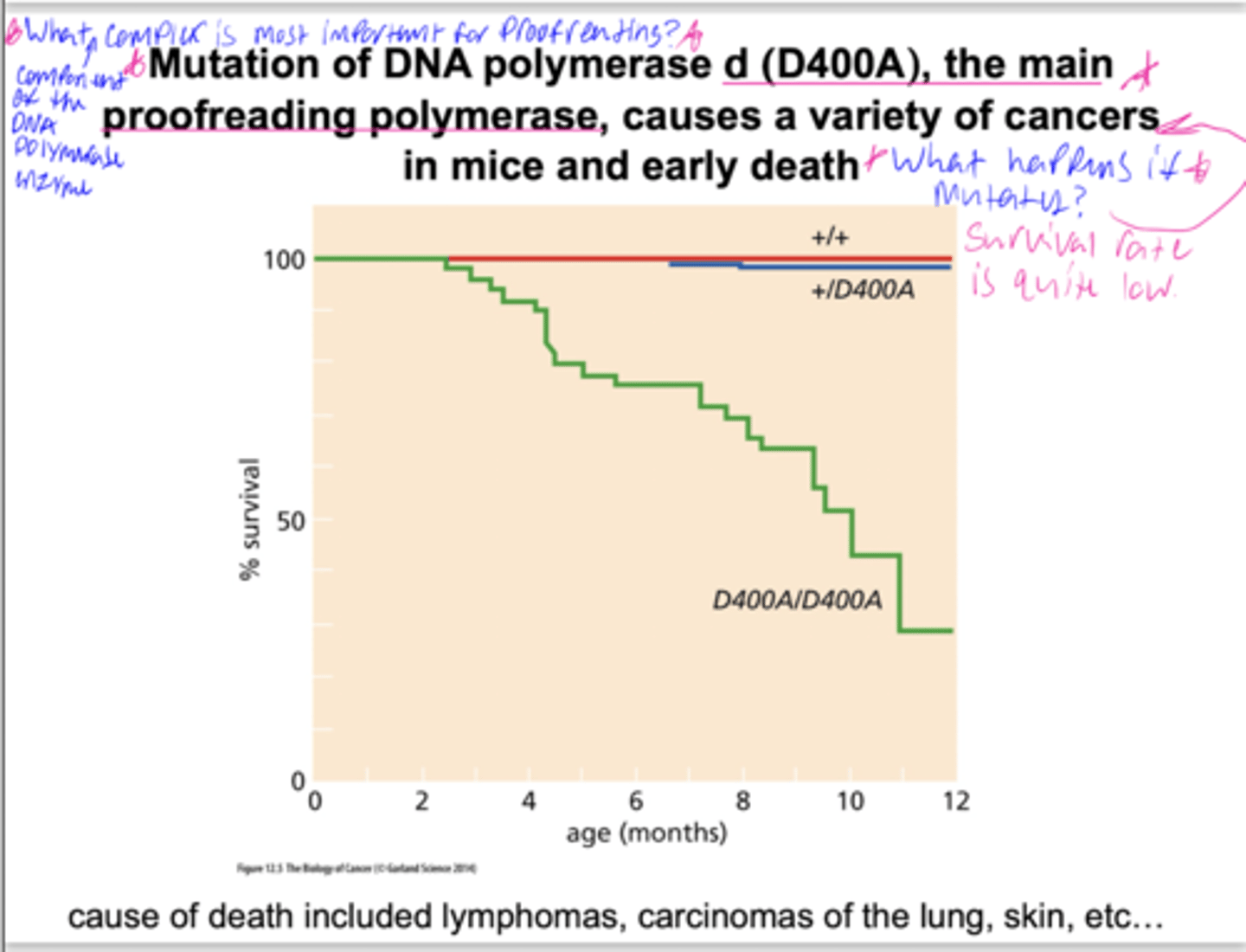
importance of proofreading
B: Pol delt synthesizes lagging strand
Q: Does proofreading activity contribute to genome maintenance?
A: mutate proofreading domain in mice
O: RECESSIVE (need 2 mutant alleles, low survival rate)
C: proofreading in important for survival
Normally, mutations missed by proof-reading can be ____
repaired
Where do cancer causing mutations come from?
- DNA replication
- Inherited
- Induced: mutagen exposure
- spontaneous
categories of mutagen + examples
physical: UV, x-rays, gamma rays, cosmic rays
chemical: ethidium bromide, smoking
infectious: HPV
How do viruses cause cancer?
1. carry oncogenes (RSV and src)
2. insertional mutagenesis
3. illegitimate recombination
How do viruses cause cancer?
1. carry oncogenes
do not have promoter/enhancer/silencer
RSV (Rous sarcoma virus)
has src oncogene - drives human cell to replicate
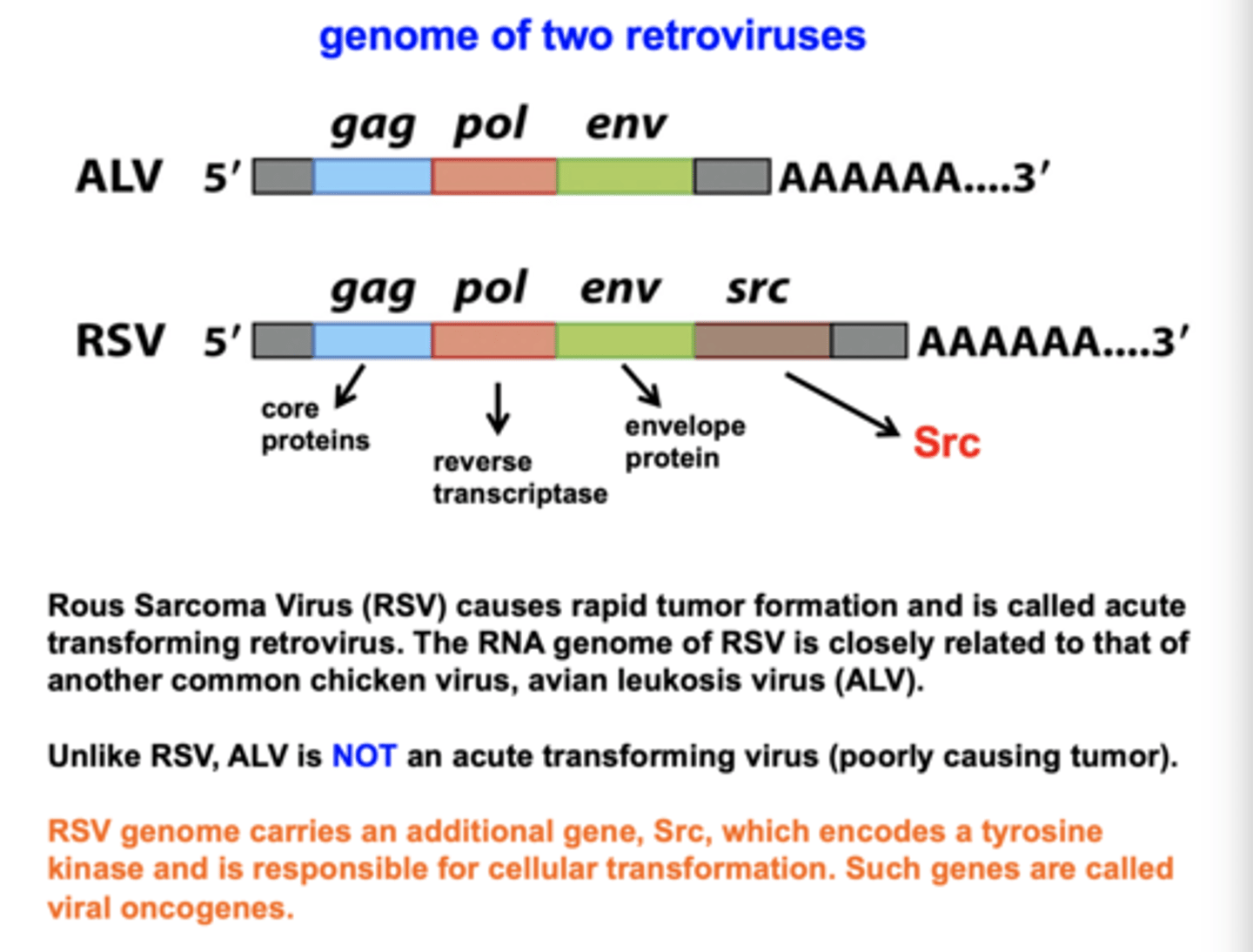
How do viruses cause cancer?
2. insertional mutagenesis
insertion next to, and activation of, cellular proto-oncogenes
random - may be next to genes with no proliferative activity
sometimes convert proto-onco to oncogene
Myc oncoprotein dysregulation
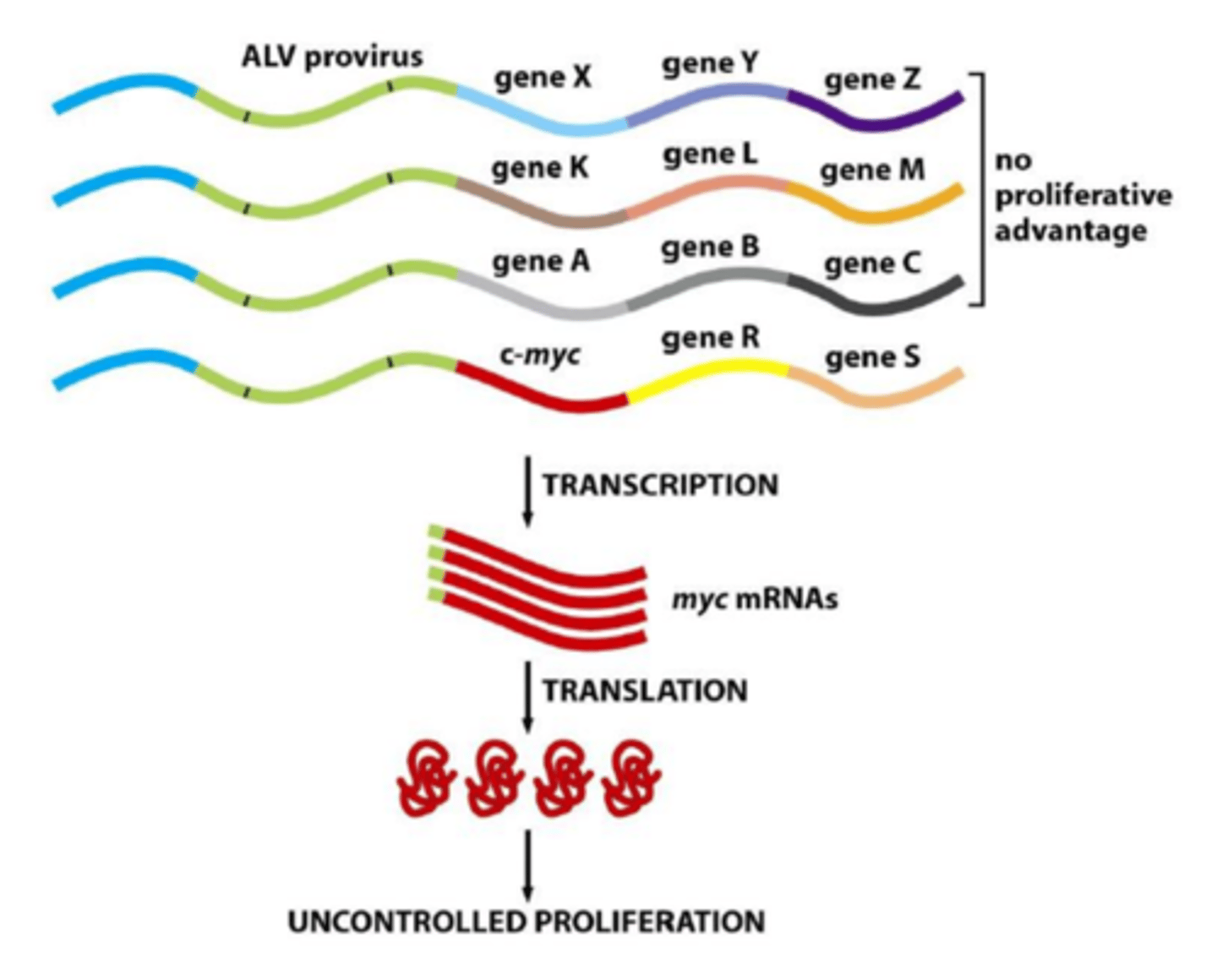
How do viruses cause cancer?
3. illegitimate recombination
full/partial integration of viral genome
eg. HPV = misregulation of viral genes
episome - circular extrachromosomal DNA
linearization & recomb with host cell
The nature of viral propagation affects the way in which a virus causes cancer
2 types of cancer causing viruses:
- Retrovirus: RNA genome. [Copied to DNA and
inserts into host chr]
TRANSCRIPTION
- DNA tumor viruses: DNA genome carried in host
cell, doesn't integrate into genome
TRANSLATION
Physical and Chemical mutagens:
Types of changes
Chemical:
endogenous - produced in body
1. metabolic products, reactive oxygen species
2. breakdown of toxins, EtOH -> formaldehyde
exogenous - external
Procarcinogens undergo _________ and become ______
cellular processing = ultimate carcinogen
Mutational signatures
- Pattern of mutations produced by a particular
mutational process
- Mutational process = DNA damage PLUS DNA repair/replication
- Different damaging agents -> different pattern
- History of past exposure to specific chemicals can sometimes be inferred by the mutational signature observed
EX: smokers have more G:C to T:A
Exposure to a mutagen may leave a
specific _______ in the DNA
signature
1) How has the profile of mutations been altered by smoking?
- more G:C to T:A
2) Come up with an explanation.
chemicals in smoke interact with those specific bases and produce a mutational signature
Where are mutations found
Within the body?
epithelial ells
exposed to mutagens
# of divisions
Where are mutations found
Within the genome?
CpG islands
- methylates C's get deaminated and looks like T
late-replicating genes - less time to be replicated = accumulate mut
regions associated with repressive chromatin marks - transcription coupled repair (better NER, nucleotide excision repair)
- NER mutations lead to other muts spread across genome
transcription factor binding sites - prevent access to repair
Where are mutations found
Within the genome?
CpG islands
methylates C's get deaminated and looks like T
Where are mutations found
Within the genome?
late-replicating genes
less time to be replicated = accumulate mutation
Where are mutations found
Within the genome?
repressive chromatin marks
regions associated with repressive chromatin marks
- transcription coupled repair (better NER, nucleotide excision repair)
- NER mutations lead to other muts spread across genome
Where are mutations found
Within the genome?
transcription factor binding sites
prevent access to repair
Does this graph show that some
nucleotides are more prone to damage
than others?
NO
sequencing of cancer
if mutation is neutral, there is no advtg for cancer
if mutation decreases function of p53, there is an advtg
= accumulation of inactive p53
= selection, more likely to have this
evolutionary process
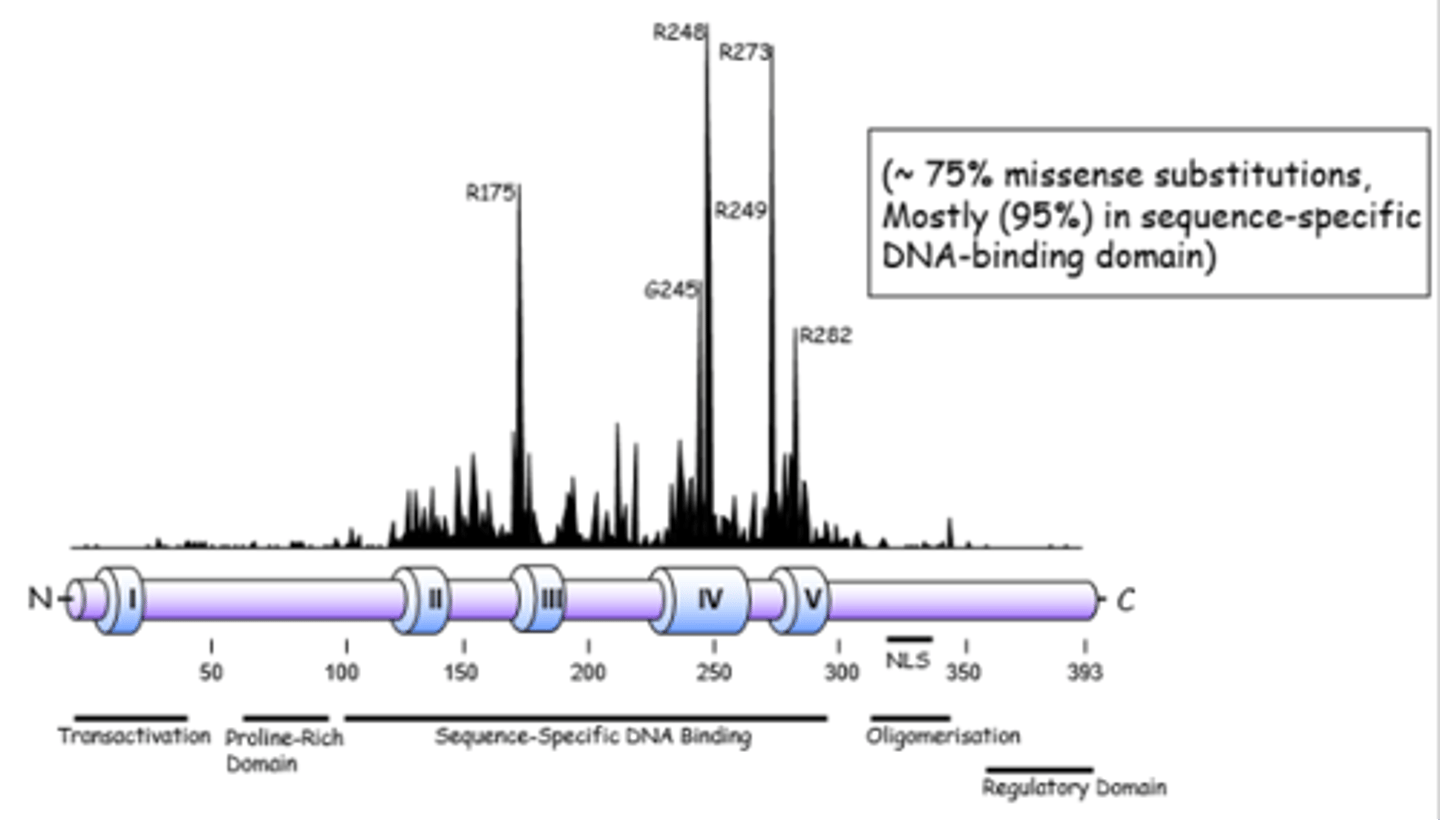
tumor suppressor
brakes
inhibit cell growth
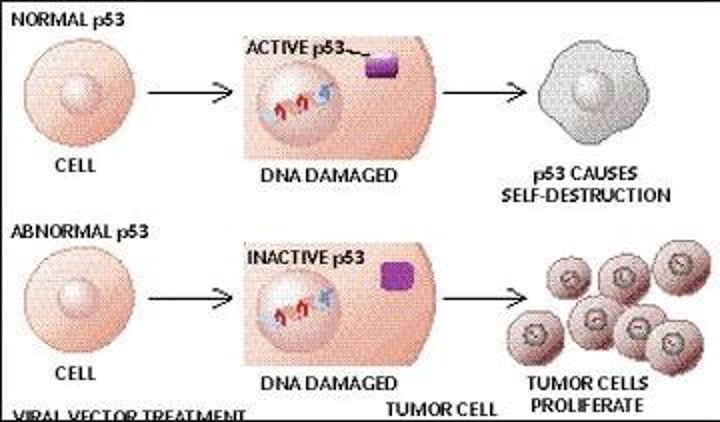
oncogene examples
Ras-Raf-MEK-ERK
protooncogenes
Normal cellular genes that regulate cell proliferation and differentiation that can become oncogenes.
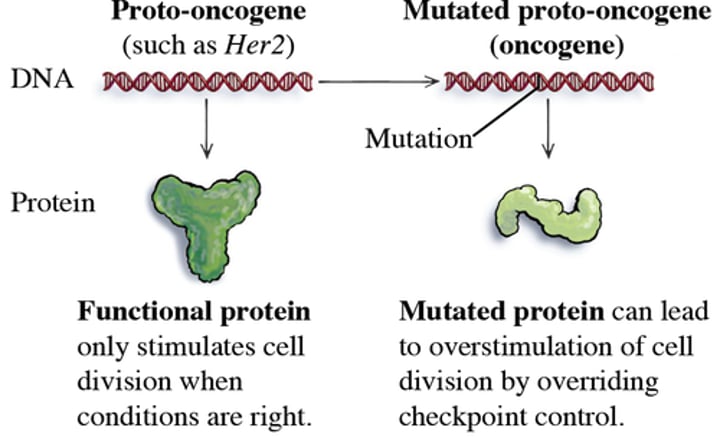
Mutator Hypothesis
Loss of DNA repair genes leads to genome instability
increase mutation rate
cellular transformation
Modern sequencing techniques suggest that defects in DNA repair contribute to cancer when
repair pathway
- inactivated
- temporarily overwhelmed (high doses)
DNA editing
- enzymes deregulated
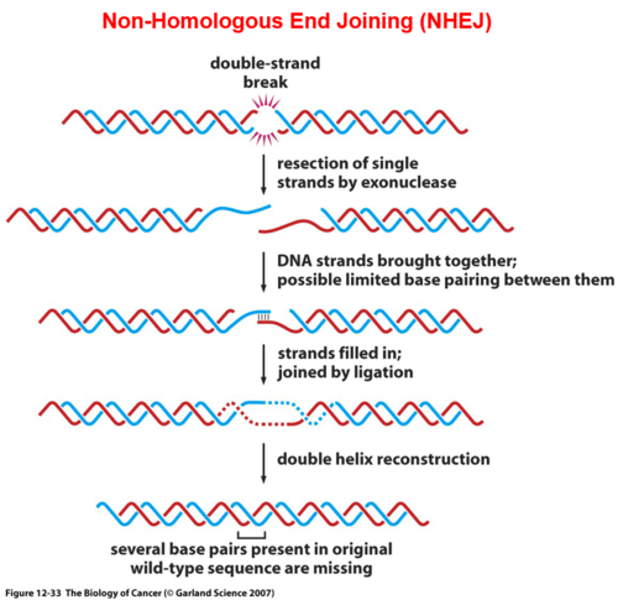
2 ways DNA double strand breaks can be repaired by ____
DSB rejoining (NHEJ, non-homologous end joining)
- used when no sister chromatid available (G1, S phase)
homologous recomb (HDR, homology directed repair)
- late S, G2, mitosis
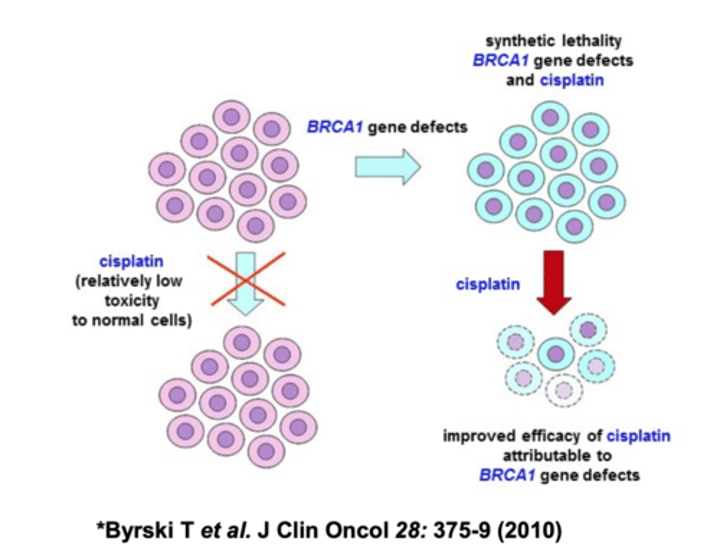
BRCA1
NHEJ
non-homologous end joining
- error prone
- alignment between segments getting joined isnt informed in WT
- used when no sister chromatid/template is available (G1, S phase)
• Ends are recognized and processed
• Ends are bridged and then ligated
HDR
Homology Directed Repair
- Use undamaged homologous DNA in
sister chromatid
- used during late S, G2, mitosis
- Break recognized
- Resection (3'overhang)
- Rad51 binds ssDNA = invasion
BRCA1
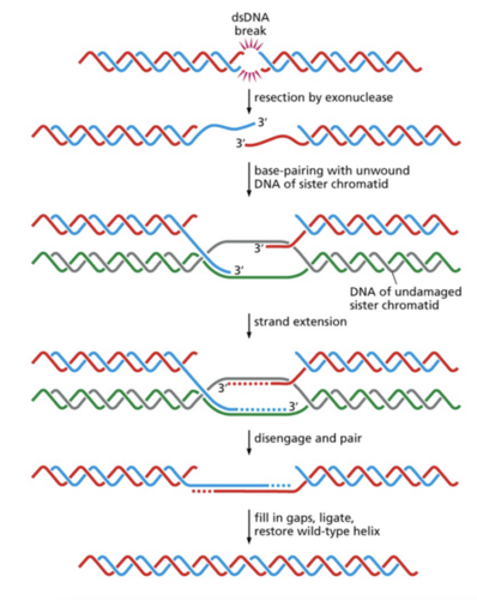
BRCA1 functions in DSB repair
Evidence
1. localization
2. expose cells to DNA damaging agents (cisplatin) = death
3. LOF mutation in mice = illegitimate recombination
4. interaction (immunoprecipitation)
BRCA1 helps cells choose _____
NHEJ
- default if ∆BRCA1
- 1st proteins jump on, then pause, and BRCA kicks them off
or
HDR
- more common
diagram DNA repair pathways
Removal of NORMAL bases
- MisMatch Repair (MMR)
Repair of ABNORMAL bases
- Direct Repair eg. MGMT
- BER
- NER
diagram DNA repair pathways:
Removal of NORMAL bases
Mismatch repair
- Recognize normal structure, wrong location
- Especially important for repairing mistakes made by strand slippage
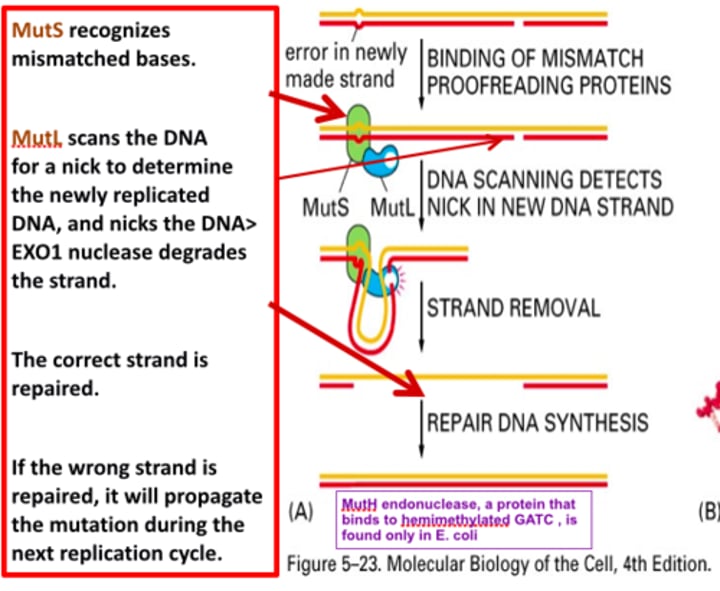
diagram DNA repair pathways
MisMatch Repair (MMR)
- Recognize mismatch - MutSα
- Identify recently synthesized strand - MutLα
- Excise nucleotide
- Resynthesize
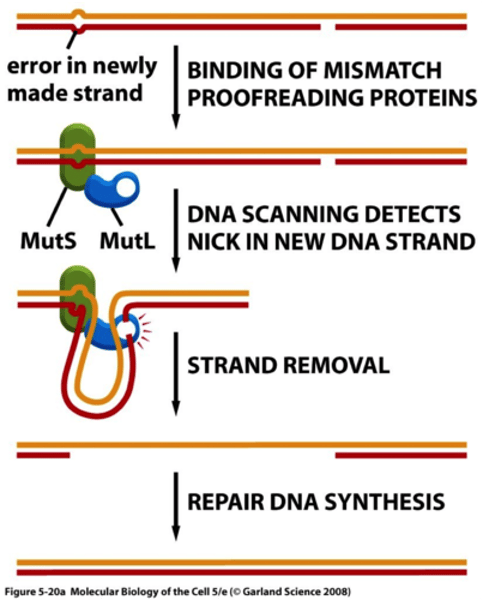
diagram DNA repair pathways
Repair of ABNORMAL bases
Direct repair
Direct Repair
- MGMT - DNA alkyltransferase
- suicide enzyme - only used once
- expression in mice = resistance to MNU
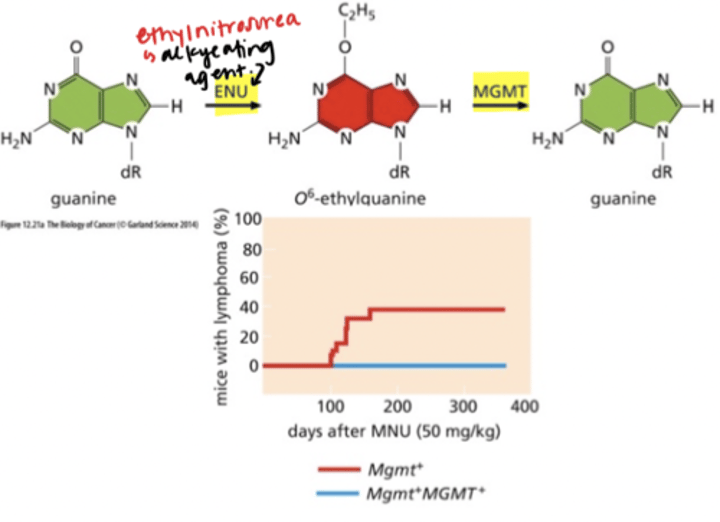
diagram DNA repair pathways
Repair of ABNORMAL bases
BER
Base excision repair (BER)
- Recognizes altered bases with minimal helix distorting effects
- DNA glycosylases specialized to recognize a specific abnormal base
- Cleavage: APE 5', AP lyase 3'
- DNA pol, ligase
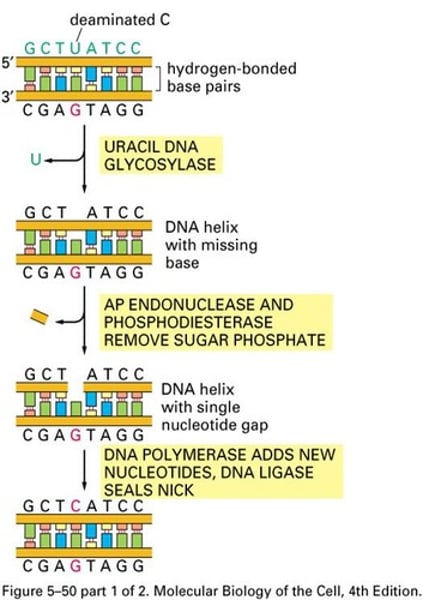
diagram DNA repair pathways
Repair of ABNORMAL bases
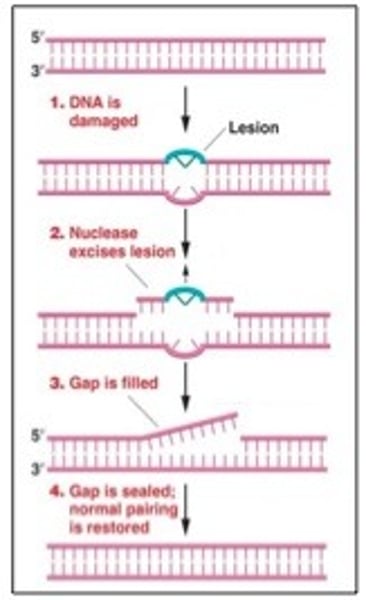
NER
- Entire nucleotide cut out
- Repairs lesions caused by exogenous agents eg. UV
- Large multiprotein complex.
- DNA pol, ligase
THYMINE DIMERS - bulky
Determining karyotype using the metaphase
spread/FISH technique
find sequence of interest in nucleus
1. block cells in metaphase
2. fix using chemical
- metaphase spread - break apart on glass slide
3. (myc. gene) probe from sequence
- normal or amplified?
DNA seq → probe → DNA comp. seq → label → fluorscent dye
*denature so probe can bind
should have: 4 dots, 2 copies of each gene
X X
(1) (1)
polyploid
genomes in which chromosomes are present in more than 2 copies
triploid
addition of haploid # of chromosomes
tetraploid
addition of diploid # of chromosomes
aneuploid
loss or gain of chromosomes
many cancer cells are aneuploid
- 90% of solid tumors, 75% hematopoietic cancers
- some are STABLY aneuploid - gained/lost but happy
- others UNSTABLE
(chromosome instability - CIN)
constantly restructuring
unstable karyotype
effects of aneuploidy
gene expression
Gene dosage imbalance
Protein deregulation
Metabolic alterations
Replication stress
Senescence
effects of aneuploidy
gene expression
Chromosome gains and losses typically result in a proportional change in the expression of genes on an affected chromosome
- i.e. an increase in gene expression from trisomic chromosomes
- a decrease in expression from monosomies
effects of aneuploidy
protein deregulation
- proteins function in complexes
- lead to proteotoxic stress inducing protein aggregation
effects of aneuploidy
replication stress
- more DNA that needs to be replicated (no time)
- hyper-recombination
- chromosome mis-segregation
effects of aneuploidy
senescence
- removed from cell cycle but not dead
- secretes molecules, modulates microenvironment
Does aneuploidy provide a selective
advantage?
trisomy 21
trisomy 21 - ALL CELLS ARE ANEUPLOID
tumor suppressive
- dec angiogenesis
- dec risk solid tumor
oncogenic
- inc myeloproliferation
- inc risk haematopoetic tumor
Does aneuploidy provide a selective
advantage?
its complicated
tumor suppressive
- Aneuploid cells are slowgrowing/unhealthy
oncogenic
- many CIN mouse models are cancer prone
How can Aneuploidy be both pro- and anti-
tumorigenic?
Aneuploid cells likely accumulate mutations to restore their fitness.
How might Aneuploidy aid tumorigenesis?
- Evolutionary flexibility: Increased genomic instability
-> increased genomic diversity
-> rare combos
-> selective advantage
• Senescent cells modify microenvironment
• Amplification of oncogenes/normal genes.
• Loss of tumor suppressors
• neoantigen-independent mechanism to promote tumorigenesis
(change protein structures, immune system doesn't recognize itself)
evidence that aneuploidy causes cancer
- aneuploidy appears early in tumor transformation
- CIN mice - cancer prone, dont have to treat with other mutagens
- mut that cause chrom missegreation in tumor cells are inc in inherited cancers
How does aneuploidy arise?
mis-segregation of chromosomes during mitosis.
Mechanisms that aid appropriate chr
segregation
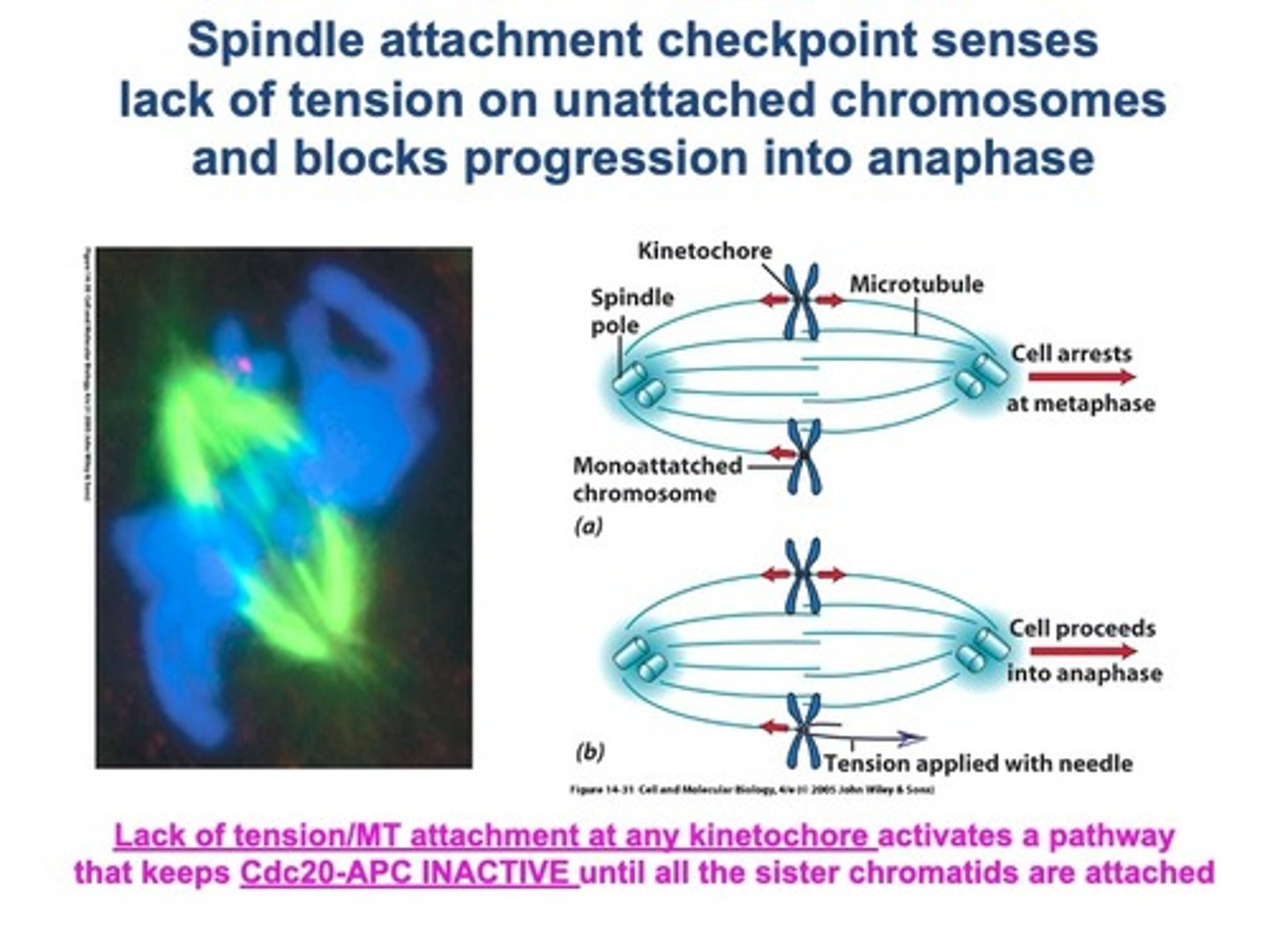
• The SAC remains active until all chr are aligned.
• Sister chromatids stay attached until anaphase.
• Bipolar attachments of chr.
• Bipolar spindle allows equal chr segregation to 2 daughter cells.
• Cell cycle regulation